Exploring the Role of the Microbiome in Rheumatoid Arthritis—A Critical Review
Abstract
:1. Introduction
2. The Microbiome and the Autoimmunity
3. Lung Microbiome in Rheumatoid Arthritis
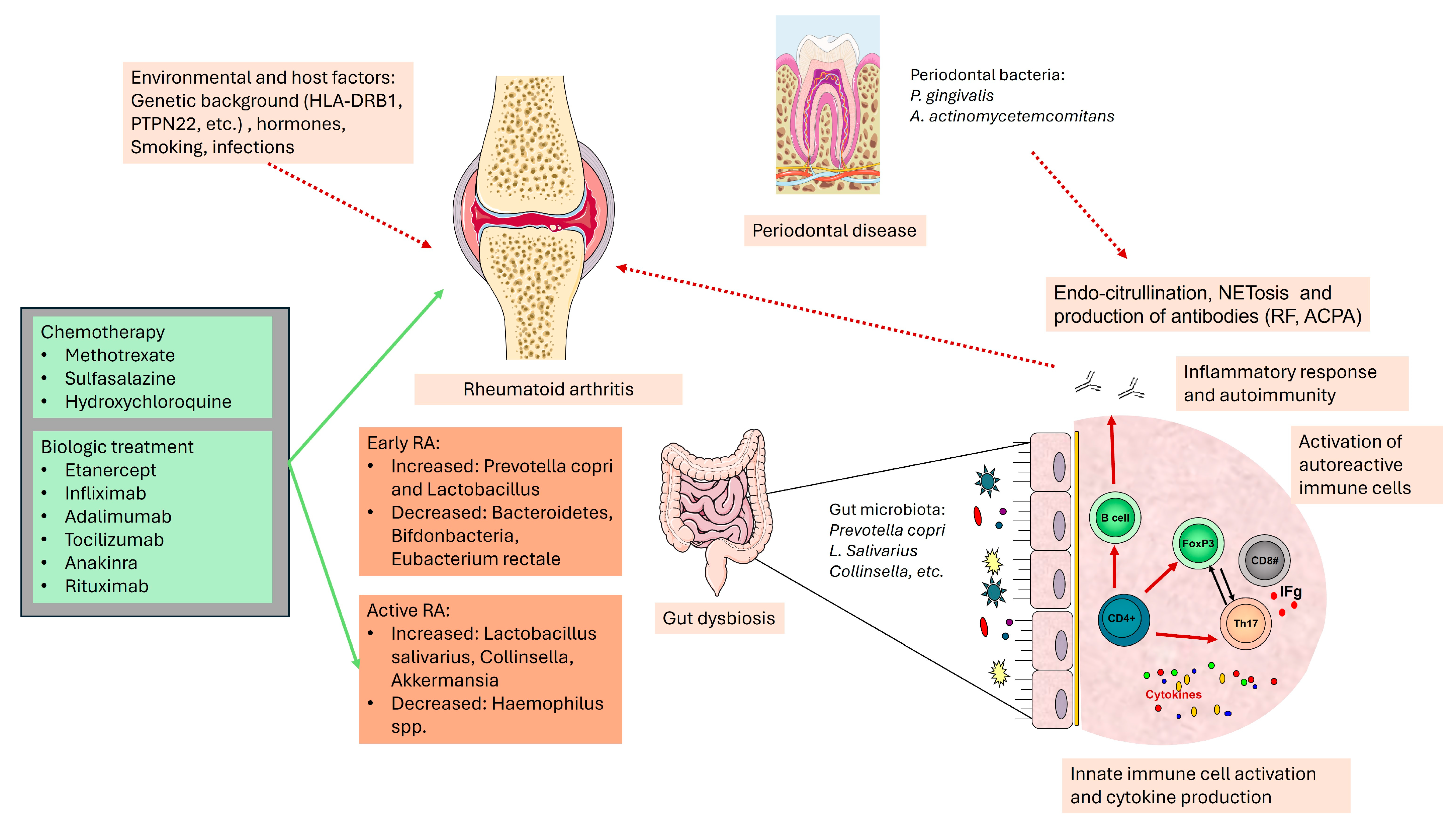
4. Oral Microbiome in Rheumatoid Arthritis
5. Gut Microbiome in Rheumatoid Arthritis
5.1. Gastrointestinal Microbiota and the Immune System Interactions
5.2. Genetic Factors That Influence the Gut Microbiota
5.3. Gut Microbiota Development during Life
5.4. Immunopathogenesis of RA: Molecular Mimicry
5.5. Immunopathogenesis of RA: Microbiota Metabolites
5.6. Immunopathogenesis of RA: Intestinal Permeability and Cell Signaling
5.7. Immunopathogenesis of RA: Autoantibodies, miRNAs and Microbiome
5.8. Immunopathogenesis of RA: Role of HLA Alleles and Gut Microbiome
5.9. Gut Microbiome-Based Interventions in RA
6. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Shumnalieva, R.; Kachakova, D.; Monov, S.; Kaneva, R.; Kolarov, Z.; Rashkov, R. AB0142 Mir-223 Expression Profile as A Biomarker in Rheumatoid Arthritis. Ann. Rheum. Dis. 2014, 73, 850. [Google Scholar] [CrossRef]
- Shumnalieva, R.; Kachakova, D.; Monov, S.; Kaneva, R.; Kolarov, Z.; Rashkov, R. Synovial fluid MIRNAs multimarker analysis in patients with rheumatoid arthritis. Ann. Rheum. Dis. 2017, 76, 208. [Google Scholar]
- Kolarov, Z.; Altunkova, I.; Baleva, M.; Martinova, F.; Monov, S.; Shumnalieva, R. Relation between serum and synovial fluid levels of IL-1-alpha, TNF-alpha, IL-6, IFN-gamma and sIL-6r and clinical, immunological and genetic factors in rheumatoid arthritis patients. Ann. Rheum. Dis. 2015, 74, 914. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Yang, H.-Y.; Luo, S.-F.; Lai, J.-H. From Rheumatoid Factor to Anti-Citrullinated Protein Antibodies and Anti-Carbamylated Protein Antibodies for Diagnosis and Prognosis Prediction in Patients with Rheumatoid Arthritis. Int. J. Mol. Sci. 2021, 22, 686. [Google Scholar] [CrossRef] [PubMed]
- Kay, J.; Upchurch, K.S. ACR/EULAR 2010 rheumatoid arthritis classification criteria. Rheumatology 2012, 51 (Suppl. 6), vi5–vi9. [Google Scholar] [CrossRef] [PubMed]
- Kolarov, Z.; Altankova, I.; Baleva, M.; Monov, S.; Shumnalieva, R. Serum and synovial fluid concentration of RF subclasses in characterization of the inflammatory reaction in rheumatoid arthritis. Ann. Rheum. Dis. 2016, 75, A32. [Google Scholar] [CrossRef]
- Nguyen, H.; James, E.A. Immune recognition of citrullinated epitopes. Immunology 2016, 149, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Darrah, E.; Andrade, F. Rheumatoid arthritis and citrullination. Curr Opin Rheumatol. 2018, 30, 72–78. [Google Scholar] [CrossRef]
- Sorice, M.; Iannuccelli, C.; Manganelli, V.; Capozzi, A.; Alessandri, C.; Lococo, E.; Garofalo, T.; Di Franco, M.; Bombardieri, M.; Nerviani, A.; et al. Autophagy generates citrullinated peptides in human synoviocytes: A possible trigger for anti-citrullinated peptide antibodies. Rheumatology 2016, 55, 1374–1385. [Google Scholar] [CrossRef]
- Vossenaar, E.R.; Radstake, T.R.; van der Heijden, A.; van Mansum, M.A.; Dieteren, C.; de Rooij, D.J.; Barrera, P.; Zendman, A.J.; van Venrooij, W.J. Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Ann. Rheum. Dis. 2004, 63, 373–381. [Google Scholar] [CrossRef]
- Konig, M.F.; Andrade, F. A Critical Reappraisal of Neutrophil Extracellular Traps and NETosis Mimics Based on Differential Requirements for Protein Citrullination. Front. Immunol. 2016, 7, 461. [Google Scholar] [CrossRef]
- Konig, M.F.; Abusleme, L.; Reinholdt, J.; Palmer, R.J.; Teles, R.P.; Sampson, K.; Rosen, A.; Nigrovic, P.A.; Sokolove, J.; Giles, J.T.; et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci. Transl. Med. 2016, 8, 369ra176. [Google Scholar] [CrossRef]
- Navarro-Millán, I.; Darrah, E.; Westfall, A.O.; Mikuls, T.R.; Reynolds, R.J.; Danila, M.I.; Curtis, J.R.; CLEAR Investigators; Rosen, A.; Bridges, S.L., Jr. Association of anti-peptidyl arginine deiminase antibodies with radiographic severity of rheumatoid arthritis in African Americans. Arthritis. Res. Ther. 2016, 18, 241. [Google Scholar] [CrossRef]
- Darrah, E.; Giles, J.T.; Ols, M.L.; Bull, H.G.; Andrade, F.; Rosen, A. Erosive rheumatoid arthritis is associated with antibodies that activate PAD4 by increasing calcium sensitivity. Sci. Transl. Med. 2013, 5, 186ra65. [Google Scholar] [CrossRef]
- Regueiro, C.; Rodriguez-Rodriguez, L.; Lopez-Mejias, R.; Nuño, L.; Triguero-Martinez, A.; Perez-Pampin, E.; Corrales, A.; Villalba, A.; Lopez-Golan, Y.; Abasolo, L.; et al. A predominant involvement of the triple seropositive patients and others with rheumatoid factor in the association of smoking with rheumatoid arthritis. Sci. Rep. 2020, 10, 3355, Erratum in Sci. Rep. 2020, 10, 18372. [Google Scholar] [CrossRef]
- Curran, A.M.; Girgis, A.A.; Jang, Y.; Crawford, J.D.; Thomas, M.A.; Kawalerski, R.; Coller, J.; Bingham, C.O., 3rd; Na, C.H.; Darrah, E. Citrullination modulates antigen processing and presentation by revealing cryptic epitopes in rheumatoid arthritis. Nat. Commun. 2023, 14, 1061. [Google Scholar] [CrossRef] [PubMed]
- Kurowska, W.; Kuca-Warnawin, E.H.; Radzikowska, A.; Maśliński, W. The role of anti-citrullinated protein antibodies (ACPA) in the pathogenesis of rheumatoid arthritis. Cent. -Eur. J. Immunol. 2017, 42, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Rasouli-Saravani, A.; Jahankhani, K.; Moradi, S.; Gorgani, M.; Shafaghat, Z.; Mirsanei, Z.; Mehmandar, A.; Mirzaei, R. Role of microbiota short-chain fatty acids in the pathogenesis of autoimmune diseases. Biomed. Pharmacother. 2023, 162, 114620. [Google Scholar] [CrossRef] [PubMed]
- Mahroum, N.; Seida, R.; Shoenfeld, Y. Triggers and regulation: The gut microbiome in rheumatoid arthritis. Expert Rev. Clin. Immunol. 2023, 19, 1449–1456. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hong, Q.; Zhang, X.; Liu, Z. Rheumatoid arthritis and the intestinal microbiome: Probiotics as a potential therapy. Front. Immunol. 2024, 15, 1331486. [Google Scholar] [CrossRef]
- Longo, U.G.; Lalli, A.; Bandini, B.; de Sire, R.; Angeletti, S.; Lustig, S.; Ammendolia, A.; Budhiparama, N.C.; de Sire, A. Role of the Gut Microbiota in Osteoarthritis, Rheumatoid Arthritis, and Spondylarthritis: An Update on the Gut-Joint Axis. Int. J. Mol. Sci. 2024, 25, 3242. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Coradduzza, D.; Bo, M.; Congiargiu, A.; Azara, E.; De Miglio, M.R.; Erre, G.L.; Carru, C. Decoding the Microbiome’s Influence on Rheumatoid Arthritis. Microorganisms 2023, 11, 2170. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70 (Suppl. 1), S38–S44. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, T.; Wei, Y.; Zhu, Y.; Xie, Z.; Hai, Q.; Li, Z.; Qin, D. Gut microbiota and rheumatoid arthritis: From pathogenesis to novel therapeutic opportunities. Front. Immunol. 2022, 13, 1007165. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yu, Y.; Yue, Y.; Zhang, Z.; Su, K. Microbial infection and rheumatoid arthritis. J. Clin. Cell. Immunol. 2013, 4, 174. [Google Scholar] [PubMed]
- Rooney, C.M.; Mankia, K.; Emery, P. The Role of the Microbiome in Driving RA-Related Autoimmunity. Front. Cell Dev. Biol. 2020, 8, 538130. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brown, K.K. Rheumatoid lung disease. Proc. Am. Thorac. Soc. 2007, 4, 443–448. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Demoruelle, M.K.; Weisman, M.H.; Simonian, P.L.; Lynch, D.A.; Sachs, P.B.; Pedraza, I.F.; Harrington, A.R.; Kolfenbach, J.R.; Striebich, C.C.; Pham, Q.N.; et al. Brief report: Airways abnormalities and rheumatoid arthritis-related autoantibodies in subjects without arthritis: Early injury or initiating site of autoimmunity? Arthritis. Rheum. 2012, 64, 1756–1761. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, D.; Zhang, J.; Lau, J.; Wang, S.; Taneja, V.; Matteson, E.L.; Vassallo, R. Mechanisms of lung disease development in rheumatoid arthritis. Nat. Rev. Rheumatol. 2019, 15, 581–596. [Google Scholar] [CrossRef]
- Reynisdottir, G.; Karimi, R.; Joshua, V.; Olsen, H.; Hensvold, A.H.; Harju, A.; Engström, M.; Grunewald, J.; Nyren, S.; Eklund, A.; et al. Structural changes and antibody enrichment in the lungs are early features of anti-citrullinated protein antibody-positive rheumatoid arthritis. Arthritis Rheumatol. 2014, 66, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Willis, V.C.; Demoruelle, M.K.; Derber, L.A.; Chartier-Logan, C.J.; Parish, M.C.; Pedraza, I.F.; Weisman, M.H.; Norris, J.M.; Holers, V.M.; Deane, K.D. Sputum autoantibodies in patients with established rheumatoid arthritis and subjects at risk of future clinically apparent disease. Arthritis Rheum. 2013, 65, 2545–2554. [Google Scholar] [CrossRef] [PubMed]
- Ytterberg, A.J.; Joshua, V.; Reynisdottir, G.; Tarasova, N.K.; Rutishauser, D.; Ossipova, E.; Haj Hensvold, A.; Eklund, A.; Sköld, C.M.; Grunewald, J.; et al. Shared immunological targets in the lungs and joints of patients with rheumatoid arthritis: Identification and validation. Ann. Rheum. Dis. 2015, 74, 1772–1777. [Google Scholar] [CrossRef] [PubMed]
- Scher, J.U.; Joshua, V.; Artacho, A.; Abdollahi-Roodsaz, S.; Öckinger, J.; Kullberg, S.; Sköld, M.; Eklund, A.; Grunewald, J.; Clemente, J.C.; et al. The lung microbiota in early rheumatoid arthritis and autoimmunity. Microbiome 2016, 4, 60. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lou, Y.; Wei, Q.; Fan, B.; Zhang, L.; Wang, X.; Chen, Z.; Tan, X.; Zheng, Y. The composition of the lung microbiome differs between patients with dermatomyositis and rheumatoid arthritis associated with interstitial lung disease. FEBS Open Bio. 2022, 12, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Meade, J.; Mankia, K.; Emery, P.; Devine, D.A. Periodontal disease and periodontal bacteria as triggers for rheumatoid arthritis. Best Pract. Res. Clin. Rheumatol. 2017, 31, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Kroese, J.M.; Brandt, B.W.; Buijs, M.J.; Crielaard, W.; Lobbezoo, F.; Loos, B.G.; van Boheemen, L.; van Schaardenburg, D.; Zaura, E.; Volgenant, C.M.C. Differences in the Oral Microbiome in Patients With Early Rheumatoid Arthritis and Individuals at Risk of Rheumatoid Arthritis Compared to Healthy Individuals. Arthritis Rheumatol. 2021, 73, 1986–1993. [Google Scholar] [CrossRef] [PubMed]
- Mysak, J.; Podzimek, S.; Sommerova, P.; Lyuya-Mi, Y.; Bartova, J.; Janatova, T.; Prochazkova, J.; Duskova, J. Porphyromonas gingivalis: Major periodontopathic pathogen overview. J. Immunol. Res. 2014, 2014, 476068. [Google Scholar] [CrossRef] [PubMed]
- Malmström, V.; Catrina, A.I.; Klareskog, L. The immunopathogenesis of seropositive rheumatoid arthritis: From triggering to targeting. Nat. Rev. Immunol. 2017, 17, 60–75. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, K.; Kinloch, A.; Fisher, B.A.; Wegner, N.; Wait, R.; Charles, P.; Mikuls, T.R.; Venables, P.J. Antibodies to citrullinated alpha-enolase peptide 1 are specific for rheumatoid arthritis and cross-react with bacterial enolase. Arthritis Rheum. 2008, 58, 3009–3019. [Google Scholar] [CrossRef]
- Kinloch, A.; Tatzer, V.; Wait, R.; Peston, D.; Lundberg, K.; Donatien, P.; Moyes, D.; Taylor, P.C.; Venables, P.J. Identification of citrullinated alpha-enolase as a candidate autoantigen in rheumatoid arthritis. Arthritis Res. Ther. 2005, 7, R1421–R1429. [Google Scholar] [CrossRef]
- de Pablo, P.; Dietrich, T.; Chapple, I.L.C.; Milward, M.; Chowdhury, M.; Charles, P.J.; Buckley, C.D.; Venables, P.J. The autoantibody repertoire in periodontitis: A role in the induction of autoimmunity to citrullinated proteins in rheumatoid arthritis? Ann. Rheum. Dis. 2014, 73, 580–586. [Google Scholar] [CrossRef]
- Kozhakhmetov, S.; Babenko, D.; Issilbayeva, A.; Nurgaziyev, M.; Kozhakhmetova, S.; Meiramova, A.; Akhmetova, Z.; Kunz, J.; Ainabekova, B.; Marotta, F.; et al. Oral Microbial Signature of Rheumatoid Arthritis in Female Patients. J. Clin. Med. 2023, 26, 3694. [Google Scholar] [CrossRef] [PubMed]
- Khor, B.; Snow, M.; Herrman, E.; Ray, N.; Mansukhani, K.; Patel, K.A.; Said-Al-Naief, N.; Maier, T.; Machida, C.A. Interconnections Between the Oral and Gut Microbiomes: Reversal of Microbial Dysbiosis and the Balance Between Systemic Health and Disease. Microorganisms 2021, 26, 496. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, J.M.; Bandiaky, O.N.; Le Goff, B.; Amador, G.; Chaux, A.G.; Soueidan, A.; Denis, F. Another Look at the Contribution of Oral Microbiota to the Pathogenesis of Rheumatoid Arthritis: A Narrative Review. Microorganisms 2021, 10, 59. [Google Scholar] [CrossRef]
- Maeda, Y.; Takeda, K. Role of gut microbiota in rheumatoid arthritis. J. Clin. Med. 2017, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Tsetseri, M.-N.; Silman, A.J.; Keene, D.J.; Dakin, S.G. The role of the microbiome in rheumatoid arthritis: A review. Rheumatol. Adv. Pract. 2023, 7, rkad034. [Google Scholar] [CrossRef]
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef]
- Podolsky, D.; Lynch-Devaney, K.; Stow, J.; Oates, P.; Murgue, B.; DeBeaumont, M.; Sands, B.; Mahida, Y. Identification of human intestinal trefoil factor. Goblet cell-specific expression of a peptide targeted for apical secretion. J. Biol. Chem. 1993, 268, 6694–6702. [Google Scholar] [CrossRef]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef]
- Hooper, L.V. Do symbiotic bacteria subvert host immunity? Nat. Rev. Microbiol. 2009, 7, 367–374. [Google Scholar] [CrossRef]
- Cullen, T.W.; Schofield, W.B.; Barry, N.A.; Putnam, E.E.; Rundell, E.A.; Trent, M.S.; Degnan, P.H.; Booth, C.J.; Yu, H.; Goodman, A.L. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 2015, 347, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Randal Bollinger, R.; Everett, M.L.; Palestrant, D.; Love, S.D.; Lin, S.S.; Parker, W. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 2003, 109, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Rawls, J.F.; Mahowald, M.A.; Ley, R.E.; Gordon, J.I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 2006, 127, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.E. The role of microRNA in carcinogenesis and biomarker selection: A methodological perspective. Exp. Rev. Mol. Diag. 2007, 7, 569–603. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Weiner, H.L. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microbes Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef]
- Yoshioka, H.; Iseki, K.-I.; Fujita, K. Development and differences of intestinal flora in the neonatal period in breastfed and bottle-fed infants. Pediatrics 1983, 72, 317–321. [Google Scholar] [CrossRef]
- Poretsky, R.; Rodriguez-R, L.M.; Luo, C.; Tsementzi, D.; Konstantinidis, K.T.; Rodriguez-Valera, F. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE 2014, 9, e93827. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841 . [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2015, 14, 20–32 . [Google Scholar] [CrossRef]
- Konig, M.F. The microbiome in autoimmune rheumatic disease. Best Pract. Res. Clin. Rheumatol. 2020, 34, 101473. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.W.; Pang, X.T.; Zhang, H.; Gao, H.X.; Leng, Y.F.; Chen, F.Q.; Sun, Z.L. Gut microbial dysbiosis in rheumatoid arthritis: A systematic review protocol of case-control studies. BMJ Open 2022, 12, e052021. [Google Scholar] [CrossRef] [PubMed]
- Horta-Baas, G.; Romero-Figueroa MD, S.; Montiel-Jarquín, A.J.; Pizano-Zárate, M.L.; García-Mena, J.; Ramírez-Durán, N. Intestinal dysbiosis and rheumatoid arthritis: A link between gut microbiota and the pathogenesis of rheumatoid arthritis. J. Immunol. Res. 2017, 2017, 4835189. [Google Scholar] [CrossRef]
- Pianta, A.; Arvikar, S.L.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C.E.; Steere, A.C. Two rheumatoid arthritis-specific autoantigens correlate microbial immunity with autoimmune responses in joints. J. Clin. Investig. 2017, 127, 2946–2956. [Google Scholar] [CrossRef] [PubMed]
- Alcaide-Ruggiero, L.; Molina-Hernandez, V.; Domínguez, J.M. Main and minor types of collagens in the articular cartilage: The role of collagens in repair tissue evaluation in chondral defects. Int. J. Mol. Sci. 2021, 22, 13329. [Google Scholar] [CrossRef]
- Lin, L.; Zhang, K.; Xiong, Q.; Zhang, J.; Cai, B.; Huang, Z.; Yang, B.; Wei, B.; Chen, J.; Niu, Q. Gut microbiota in pre-clinical rheumatoid arthritis: From pathogenesis to preventing. J. Autoimmun. 2023, 141, 103001. [Google Scholar] [CrossRef]
- Maeda, Y.; Kurakawa, T.; Umemoto, E.; Motooka, D.; Ito, Y.; Gotoh, K.; Hirota, K.; Matsushita, M.; Furuta, Y.; Narazaki, M.; et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016, 68, 2646–2661. [Google Scholar] [CrossRef]
- Martinsson, K.; Dürholz, K.; Schett, G.; Zaiss, M.M.; Kastbom, A. Higher serum levels of short-chain fatty acids are associated with non-progression to arthritis in individuals at increased risk of RA. Ann. Rheum. Dis. 2021, 81, 445–447. [Google Scholar] [CrossRef]
- Luu, M.; Pautz, S.; Kohl, V.; Singh, R.; Romero, R.; Lucas, S.; Hofmann, J.; Raifer, H.; Vachharajani, N.; Carrascosa, L.C.; et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic epigenetic crosstalk in lymphocytes. Nat. Commun. 2019, 10, 760. [Google Scholar] [CrossRef] [PubMed]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Chu, Y.; Li, J.; Meng, Q.; Liu, Y.; Jin, J.; Wang, Y.; Wang, J.; Huang, B.; Shi, L.; et al. Intestinal butyrate-metabolizing species contribute to autoantibody production and bone erosion in rheumatoid arthritis. Sci. Adv. 2022, 8, eabm1511. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Du, J.; Pu, X.; Zheng, L.; Chen, S.; Wang, N.; Li, J.; Chen, S.; Pan, S.; Shen, B. The gut microbiome and metabolites are altered and interrelated in patients with rheumatoid arthritis. Front. Cell. Infect. Microbiol. 2021, 11, 763507. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yu, Y.; Liao, J.; Hu, W.; Bian, X.; Wu, J.; Zhu, Y.Z. S-Propargyl-Cysteine remodels the gut microbiota to alleviate rheumatoid arthritis by regulating bile acid metabolism. Front. Cell. Infect. Microbiol. 2021, 11, 670593. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cheng, M.; Zou, L.; Yin, L.; Zhong, C.; Zha, Y.; Zhu, X.; Zhang, L.; Ning, K.; Han, J. Hidden link in gut-joint axis: Gut microbes promote rheumatoid arthritis at early stage by enhancing ascorbate degradation. Gut 2021, 71, 1041–1043. [Google Scholar] [CrossRef] [PubMed]
- Matei, D.E.; Menon, M.; Alber, D.G.; Smith, A.M.; Nedjat-Shokouhi, B.; Fasano, A.; Magill, L.; Duhlin, A.; Bitoun, S.; Gleizes, A.; et al. Intestinal barrier dysfunction plays an integral role in arthritis pathology and can be targeted to ameliorate disease. Med 2021, 2, 864–883.e9. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wright, K.; Davis, J.M.; Jeraldo, P.; Marietta, E.V.; Murray, J.; Nelson, H.; Matteson, E.L.; Taneja, V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Kinashi, Y.; Hase, K. Partners in Leaky Gut Syndrome: Intestinal Dysbiosis and Autoimmunity. Front. Immunol. 2021, 12, 673708. [Google Scholar] [CrossRef]
- Arvonen, M.; Berntson, L.; Pokka, T.; Karttunen, T.J.; Vähäsalo, P.; Stoll, M.L. Gut microbiota-host interactions and juvenile idiopathic arthritis. Pediatr. Rheumatol. 2016, 14, 44. [Google Scholar] [CrossRef]
- Oh, H.; Ghosh, S. NF-κB: Roles and regulation in different CD4(+) T-cell subsets. Immunol. Rev. 2013, 252, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, S.-X.; Chang, M.-J.; Qiao, J.; Wang, C.-H.; Li, X.-F.; Yu, Q.; He, P.-F. Characteristics of the Gut Microbiome and Its Relationship With Peripheral CD4+ T Cell Subpopulations and Cytokines in Rheumatoid Arthritis. Front. Microbiol. 2022, 13, 799602. [Google Scholar] [CrossRef] [PubMed]
- Pianta, A.; Arvikar, S.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C.E.; Steere, A.C. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 2017, 69, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Velikova, T.; Shumnalieva, R.; Kachakova, D.; Tumangelova-Yuzeir, K.; Ivanova-Todorova, E.; Kyurkchiev, D.; Monov, S. IL-17A—A promising biomarker for disease activity in rheumatoid arthritis patients. J. Biomed. Sci. Technol. 2018, 1, 6–20. [Google Scholar]
- Mangalea, M.R.; Paez-Espino, D.; Kieft, K.; Chatterjee, A.; Chriswell, M.E.; Seifert, J.A.; Feser, M.L.; Demoruelle, M.K.; Sakatos, A.; Anantharaman, K.; et al. Individuals at risk for rheumatoid arthritis harbor differential intestinal bacteriophage communities with distinct metabolic potential. Cell Host Microbe 2021, 29, 726–739.e5. [Google Scholar] [CrossRef] [PubMed]
- Shumnalieva, R.; Kachakova, D.; Kaneva, R.; Kolarov, Z.; Monov, S. AB0017 Comparison between the altered peripheral blood miRNA expression in patients with rheumatoid arthritis and systemic lupus erythematosus. Ann. Rheum. Dis. 2019, 78, 1475. [Google Scholar]
- Cunningham, C.C.; Wade, S.; Floudas, A.; Orr, C.; McGarry, T.; Wade, S.; Cregan, S.; Fearon, U.; Veale, D.J. Serum miRNA signature in rheumatoid arthritis and “At-Risk individuals”. Front. Immunol. 2021, 12, 633201. [Google Scholar] [CrossRef]
- McInnes, I.B.; Schett, G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.; Luckey, D.; Yeoman, C.J.; Marietta, E.V.; Berg Miller, M.E.; Murray, J.A.; Taneja, V. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS ONE 2012, 7, e36095. [Google Scholar] [CrossRef]
- Asquith, M.; Sternes, P.R.; Costello, M.; Karstens, L.; Diamond, S.; Martin, T.M.; Li, Z.; Msc, M.S.M.; Spector, T.D.; le Cao, K.; et al. HLA alleles associated with risk of ankylosing spondylitis and rheumatoid arthritis influence the gut microbiome. Arthritis Rheumatol. 2019, 71, 1642–1650. [Google Scholar] [CrossRef]
- Nemoto, N.; Takeda, Y.; Nara, H.; Araki, A.; Gazi, M.Y.; Takakubo, Y.; Asao, H. Analysis of intestinal immunity and flora in a collagen-induced mouse arthritis model: Differences during arthritis progression. Int. Immunol. 2020, 32, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Alpizar-Rodriguez, D.; Lesker, T.R.; Gronow, A.; Gilbert, B.; Raemy, E.; Lamacchia, C.; Gabay, C.; Finckh, A.; Strowig, T. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2019, 78, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Marietta, E.V.; Murray, J.A.; Luckey, D.H.; Jeraldo, P.R.; Lamba, A.; Patel, R.; Luthra, H.S.; Mangalam, A.; Taneja, V. Suppression of inflammatory arthritis by human gut-derived prevotella histicola in humanized mice. Arthritis Rheumatol. 2016, 68, 2878–2888. [Google Scholar] [CrossRef]
- Balakrishnan, B.; Luckey, D.; Bodhke, R.; Chen, J.; Marietta, E.; Jeraldo, P.; Murray, J.; Taneja, V. Prevotella histicola protects from arthritis by expansion of allobaculum and augmenting butyrate production in humanized mice. Front. Immunol. 2021, 12, 609644. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zou, Q.; Zeng, B.; Fang, Y.; Wei, H. Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Curr. Microbiol. 2013, 67, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yao, J.; Deng, Q.; Li, X.; He, Y.; Ren, X.; Zheng, Y.; Song, R.; Zhong, X.; Ma, J.; et al. Relationship between gut microbiota and rheumatoid arthritis: A bibliometric analysis. Front. Immunol. 2023, 14, 1131933. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Yang, B.; Ross, R.P.; Stanton, C.; Zhao, J.; Zhang, H.; Chen, W. The prophylactic effects of different lactobacilli on collagen-induced arthritis in rats. Food Funct. 2020, 11, 3681–3694. [Google Scholar] [CrossRef] [PubMed]
- Forbes, J.D.; Chen, C.Y.; Knox, N.C.; Marrie, R.A.; El-Gabalawy, H.; de Kievit, T.; Van Domselaar, G. A comparative study of the gut microbiota in immune-mediated inflammatory diseases—Does a common dysbiosis exist? Microbiome 2018, 6, 221. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; Charneca, S.; Dourado, E.; Guerreiro, C.S.; Fonseca, J.E. Probiotic supplementation for rheumatoid arthritis: A promising adjuvant therapy in the gut microbiome era. Front. Pharmacol. 2021, 12, 711788. [Google Scholar] [CrossRef]
- Cleusix, V.; Lacroix, C.; Vollenweider, S.; Duboux, M.; Le Blay, G. Inhibitory activity spectrum of reuterin produced by Lactobacillus reuteri against intestinal bacteria. BMC Microbiol. 2007, 7, 101. [Google Scholar] [CrossRef]
- De Vrese, M.; Marteau, P.R. Probiotics and prebiotics: Effects on diarrhea. J. Nutr. 2017, 137, 803S–811S. [Google Scholar] [CrossRef]
- Alwarith, J.; Kahleova, H.; Rembert, E.; Yonas, W.; Dort, S.; Calcagno, M.; Burgess, N.; Crosby, L.; Barnard, N.D. Nutrition interventions in rheumatoid arthritis: The potential use of plant-based diets. A Review. Front. Nutr. 2019, 6, 141. [Google Scholar] [CrossRef]
- Khoruts, A.; Sadowsky, M.J. Understanding the mechanisms of fecal microbiota transplantation. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Borody, T.J.; Warren, E.F.; Leis, S.; Surace, R.; Ashman, O. Treatment of ulcerative colitis using fecal bacteriotherapy. J. Clin. Gastroenterol. 2003, 37, 42–47. [Google Scholar] [CrossRef]
- Zeng, J.; Peng, L.; Zheng, W.; Huang, F.; Zhang, N.; Wu, D.; Yang, Y. Fecal microbiota transplantation for rheumatoid arthritis: A case report. Clin. Case Rep. 2020, 9, 906–909. [Google Scholar] [CrossRef]
- Yan, H.; Su, R.; Xue, H.; Gao, C.; Li, X.; Wang, C. Pharmacomicrobiology of methotrexate in rheumatoid arthritis: Gut microbiome as predictor of therapeutic response. Front. Immunol. 2021, 12, 789334. [Google Scholar] [CrossRef]
- Nayak, R.R.; Alexander, M.; Deshpande, I.; Stapleton-Gray, K.; Rimal, B.; Patterson, A.D.; Ubeda, C.; Scher, J.U.; Turnbaugh, P.J. Methotrexate impacts conserved pathways in diverse human gut bacteria leading to decreased host immune activation. Cell Host Microbe 2021, 29, 362–377.e11. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.R.; de Arruda, J.A.A.; Corrêa, J.D.; Carvalho, V.F.; Medeiros, J.D.; Schneider, A.H.; Machado, C.C.; Duffles, L.F.; Fernandes, G.d.R.; Calderaro, D.C.; et al. Methotrexate and Non-Surgical Periodontal Treatment Change the Oral-Gut Microbiota in Rheumatoid Arthritis: A Prospective Cohort Study. Microorganisms 2023, 12, 68. [Google Scholar] [CrossRef]
- Kanerud, L.; Scheynius, A.; Nord, C.E.; Hafström, I. Effect of sulphasalazine on gastrointestinal microflora and on mucosal heat shock protein expression in patients with rheumatoid arthritis. Br. J. Rheumatol. 1994, 33, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Picchianti-Diamanti, A.; Panebianco, C.; Salemi, S.; Sorgi, M.L.; Di Rosa, R.; Tropea, A.; Sgrulletti, M.; Salerno, G.; Terracciano, F.; D’amelio, R.; et al. Analysis of gut microbiota in rheumatoid arthritis patients: Disease-related dysbiosis and modifications induced by etanercept. Int. J. Mol. Sci. 2018, 19, 2938. [Google Scholar] [CrossRef]
- Bodkhe, R.; Balakrishnan, B.; Taneja, V. The role of microbiome in rheumatoid arthritis treatment. Ther. Adv. Musculoskelet. Dis. 2019, 11, 1759720X19844632. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.H.; Lee, E.H.; Cha, K.H.; Pan, C.-H.; Kim, D.; Kim, W.-U. Factors associated with the composition of the gut microbiome in patients with established rheumatoid arthritis and its value for predicting treatment responses. Arthritis Res. Ther. 2023, 25, 32. [Google Scholar] [CrossRef] [PubMed]
| Mucosal Site | Microbiome Alterations |
|---|---|
| Lung mucosa |
|
| Oral mucosa |
|
| Gut mucosa |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ermencheva, P.; Kotov, G.; Shumnalieva, R.; Velikova, T.; Monov, S. Exploring the Role of the Microbiome in Rheumatoid Arthritis—A Critical Review. Microorganisms 2024, 12, 1387. https://doi.org/10.3390/microorganisms12071387
Ermencheva P, Kotov G, Shumnalieva R, Velikova T, Monov S. Exploring the Role of the Microbiome in Rheumatoid Arthritis—A Critical Review. Microorganisms. 2024; 12(7):1387. https://doi.org/10.3390/microorganisms12071387
Chicago/Turabian StyleErmencheva, Plamena, Georgi Kotov, Russka Shumnalieva, Tsvetelina Velikova, and Simeon Monov. 2024. "Exploring the Role of the Microbiome in Rheumatoid Arthritis—A Critical Review" Microorganisms 12, no. 7: 1387. https://doi.org/10.3390/microorganisms12071387







