Retrospective Detection of Ophidiomyces ophidiicola from Snake Moults Collected in Bieszczady Mountains, Poland
Abstract
1. Introduction
2. Materials and Methods
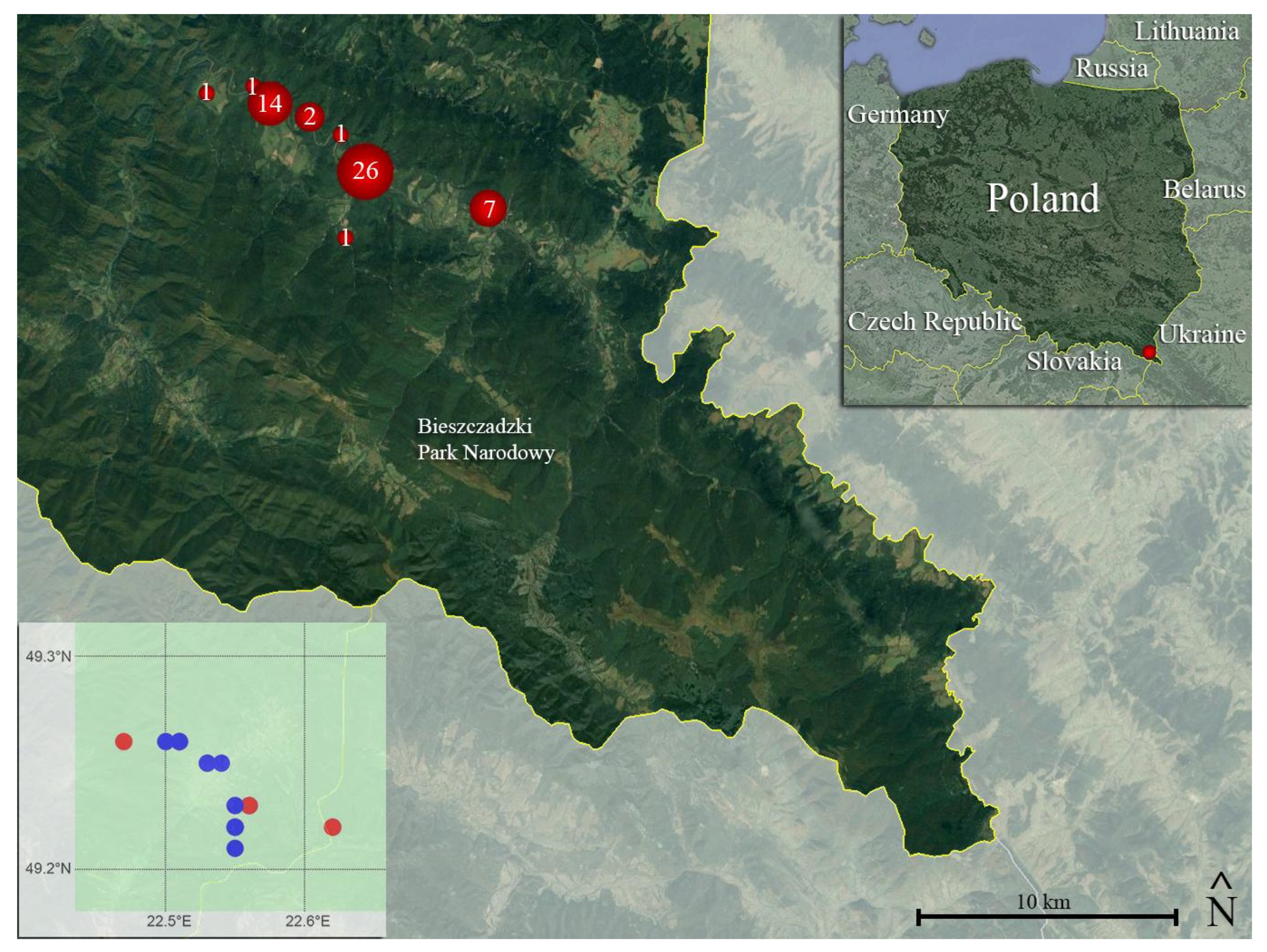
2.1. Study Species, Study Site, and Moult Collection
2.2. Shed Sampling
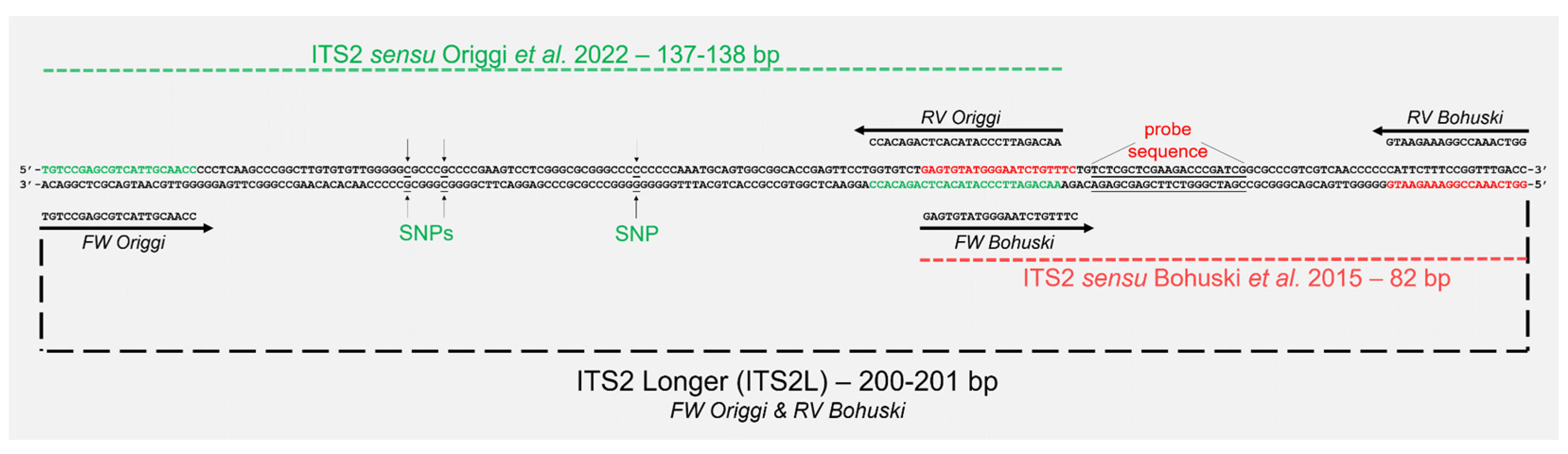
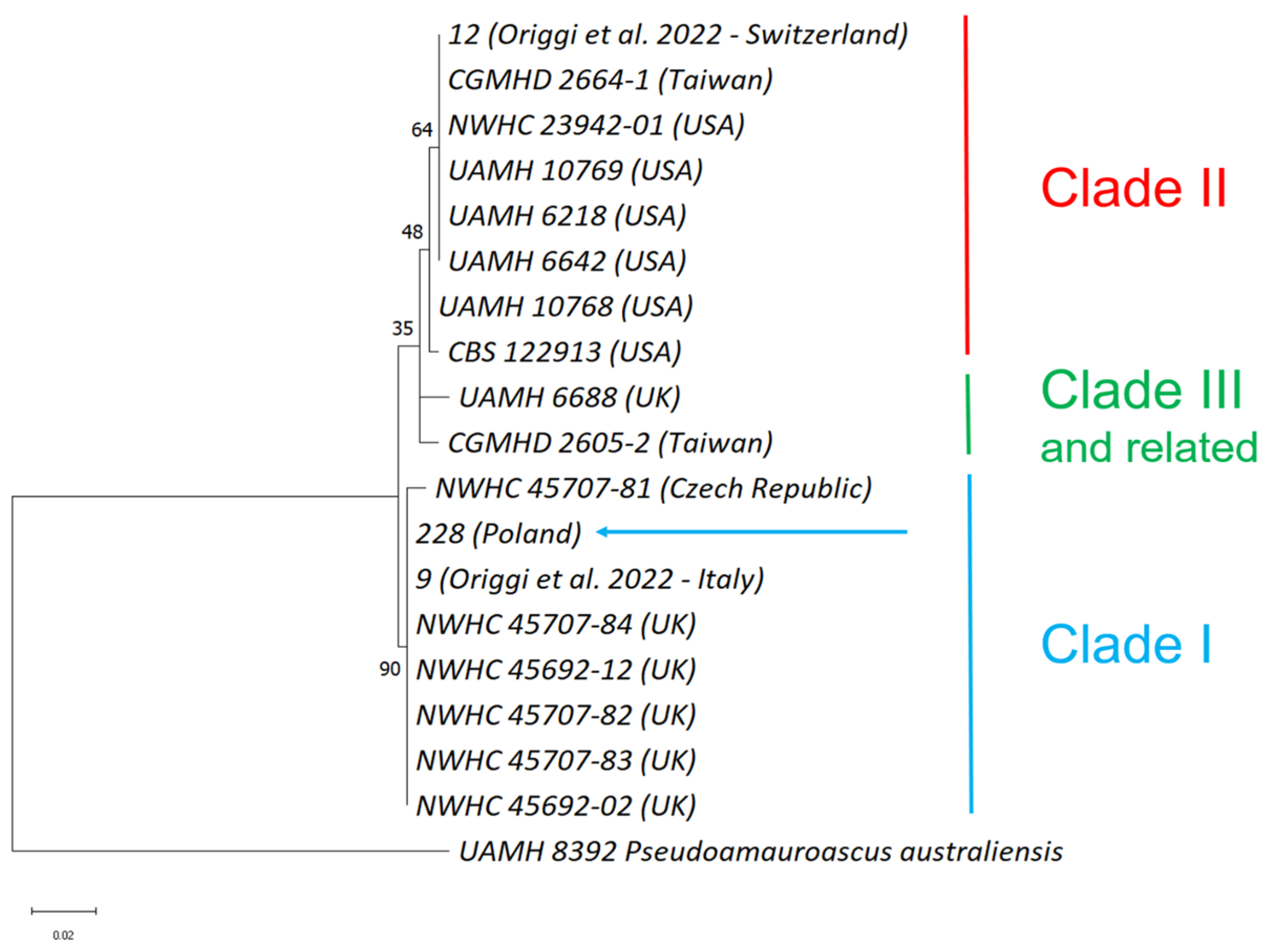
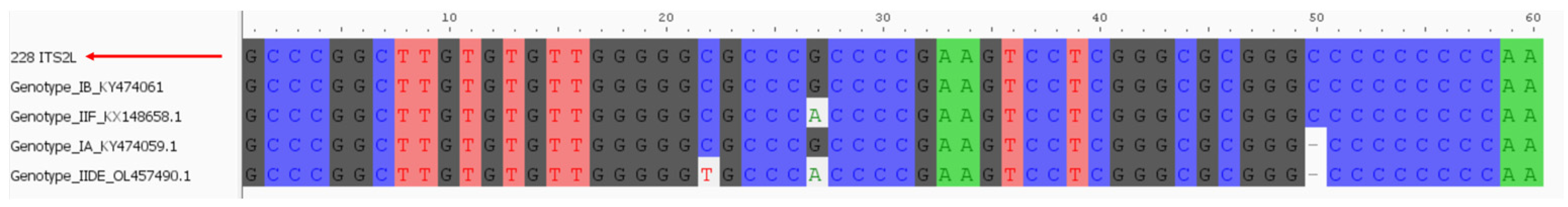
2.3. Molecular Analysis and Clade Characterisation
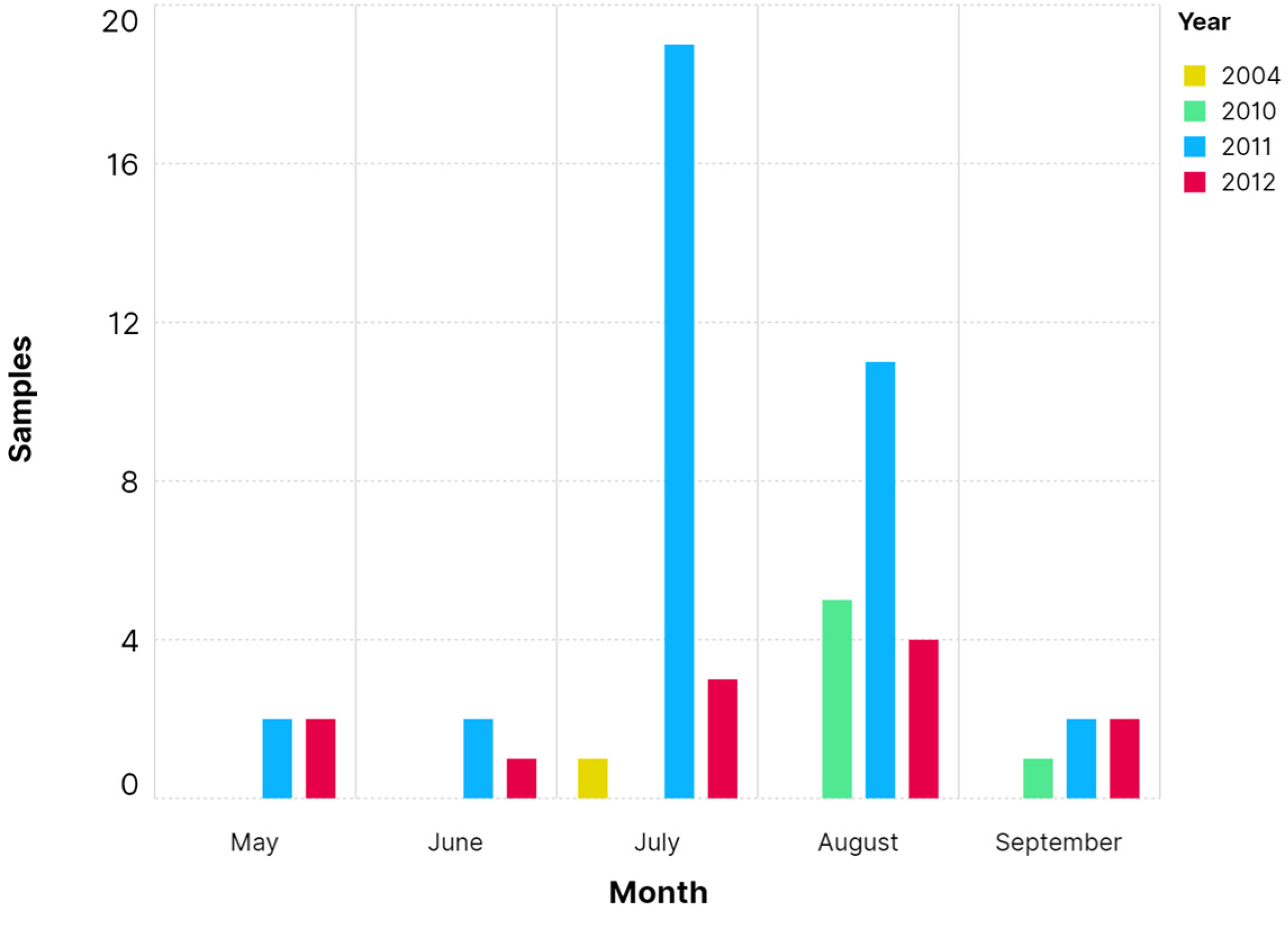
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging Fungal Threats to Animal, Plant and Ecosystem Health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef]
- Reynolds, H.T.; Raudabaugh, D.; Lilje, O.; Allender, M.; Miller, A.N.; Gleason, F.H. Emerging Mycoses and Fungus-Like Diseases of Vertebrate Wildlife. In The Fungal Community: Its Organization and Role in the Ecosystem, 4th ed.; Dighton, J., White, J.F., Eds.; CRC Press: Boca Raton, FL, USA, 2017; pp. 286–403. ISBN 1498706673. [Google Scholar]
- Langwig, K.E.; Frick, W.F.; Hoyt, J.R.; Parise, K.L.; Drees, K.P.; Kunz, T.H.; Foster, J.T.; Kilpatrick, A.M. Drivers of Variation in Species Impacts for a Multi-Host Fungal Disease of Bats. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150456. [Google Scholar] [CrossRef]
- Stegen, G.; Pasmans, F.; Schmidt, B.R.; Rouffaer, L.O.; Van Praet, S.; Schaub, M.; Canessa, S.; Laudelout, A.; Kinet, T.; Adriaensen, C.; et al. Drivers of Salamander Extirpation Mediated by Batrachochytrium salamandrivorans. Nature 2017, 544, 353–356. [Google Scholar] [CrossRef]
- O’Hanlon, S.J.; Rieux, A.; Farrer, R.A.; Rosa, G.M.; Waldman, B.; Bataille, A.; Kosch, T.A.; Murray, K.A.; Brankovics, B.; Fumagalli, M.; et al. Recent Asian Origin of Chytrid Fungi Causing Global Amphibian Declines. Science 2018, 360, 621–627. [Google Scholar] [CrossRef]
- Böhm, M.; Collen, B.; Baillie, J.E.M.; Bowles, P.; Chanson, J.; Cox, N.; Hammerson, G.; Hoffmann, M.; Livingstone, S.R.; Ram, M.; et al. The Conservation Status of the World’s Reptiles. Biol. Conserv. 2013, 157, 372–385. [Google Scholar] [CrossRef]
- Paré, J.A.; Conley, K.J. Mycotic Diseases of Reptiles [Infectious Diseases and Pathology of Reptiles]. In Infectious Diseases and Pathology of Reptiles; CRC Press: Boca Raton, FL, USA, 2020; pp. 795–858. [Google Scholar]
- Picquet, P.; Heckers, K.O.; Kolesnik, E.; Heusinger, A.; Marschang, R.E. Detection Of Ophidiomyces ophiodiicola In Two Captive Bocourt Water Snakes (Subsessor bocourti) And One Captive Pueblan Milk Snake (Lampropeltis triangulum campbelli). J. Zoo Wildl. Med. Off. Publ. Am. Assoc. Zoo Vet. 2018, 49, 219–222. [Google Scholar] [CrossRef]
- Allain, S.J.R.; Duffus, A.L.J. Emerging Infectious Disease Threats to European Herpetofauna. Herpetol. J. 2019, 29, 189–206. [Google Scholar] [CrossRef]
- Peterson, N.R.; Rose, K.; Shaw, S.; Hyndman, T.H.; Sigler, L.; Kurtböke, D.İ.; Llinas, J.; Littleford-Colquhoun, B.L.; Cristescu, R.; Frère, C. Cross-Continental Emergence of Nannizziopsis barbatae Disease May Threaten Wild Australian Lizards. Sci. Rep. 2020, 10, 20976. [Google Scholar] [CrossRef]
- Paré, J.A.; Sigler, L. An Overview of Reptile Fungal Pathogens in the Genera Nannizziopsis, Paranannizziopsis, and Ophidiomyces. J. Herpetol. Med. Surg. 2016, 26, 46. [Google Scholar] [CrossRef]
- Di Nicola, M.R.; Coppari, L.; Notomista, T.; Marini, D. Ophidiomyces ophidiicola Detection and Infection: A Global Review on a Potential Threat to the World’s Snake Populations. Eur. J. Wildl. Res. 2022, 68, 64. [Google Scholar] [CrossRef]
- Sigler, L.; Hambleton, S.; Paré, J.A. Molecular Characterization of Reptile Pathogens Currently Known as Members of the Chrysosporium anamorph of Nannizziopsis vriesii Complex and Relationship with Some Human-Associated Isolates. J. Clin. Microbiol. 2013, 51, 3338–3357. [Google Scholar] [CrossRef] [PubMed]
- Stchigel, A.M.; Sutton, D.A.; Cano-Lira, J.F.; Cabañes, F.J.; Abarca, L.; Tintelnot, K.; Wickes, B.L.; García, D.; Guarro, J. Phylogeny of Chrysosporia Infecting Reptiles: Proposal of the New Family Nannizziopsiaceae and Five New Species. Persoonia Mol. Phylogeny Evol. Fungi 2013, 31, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Woodburn, D.B.; Miller, A.N.; Allender, M.C.; Maddox, C.W.; Terio, K.A. Emydomyces Testavorans, a New Genus and Species of Onygenalean Fungus Isolated from Shell Lesions of Freshwater Aquatic Turtles. J. Clin. Microbiol. 2019, 57, e00628-18. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, C.M.; Oesterle, P.T.; Stevens, B.; Shirose, L.; Mastromonaco, G.F.; Lillie, B.N.; Davy, C.M.; Jardine, C.M.; Nemeth, N.M. Ophidiomycosis in Red Cornsnakes (Pantherophis guttatus): Potential Roles of Brumation and Temperature on Pathogenesis and Transmission. Vet. Pathol. 2020, 57, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.J.; Burger, J.; Zappalorti, R.T.; Bunnell, J.F.; Winzeler, M.E.; Taylor, D.R.; Lorch, J.M. Soil Reservoir Dynamics of Ophidiomyces ophidiicola, the Causative Agent of Snake Fungal Disease. J. Fungi 2021, 7, 461. [Google Scholar] [CrossRef] [PubMed]
- Allender, M.C.; Raudabaugh, D.B.; Gleason, F.H.; Miller, A.N. The Natural History, Ecology, and Epidemiology of Ophidiomyces ophiodiicola and Its Potential Impact on Free-Ranging Snake Populations. Fungal Ecol. 2015, 17, 187–196. [Google Scholar] [CrossRef]
- Lorch, J.M.; Lankton, J.; Werner, K.; Falendysz, E.A.; McCurley, K.; Blehert, D.S. Experimental Infection of Snakes with Ophidiomyces ophiodiicola Causes Pathological Changes That Typify Snake Fungal Disease. mBio 2015, 6, e01534-15. [Google Scholar] [CrossRef] [PubMed]
- Britton, M.; Allender, M.C.; Hsiao, S.-H.; Baker, S.J. Postnatal Mortality in Neonate Rattlesnakes Associated with Ophidiomyces ophiodiicola. J. Zoo Wildl. Med. 2019, 50, 672. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.W.; Marchand, M.N.; Clifford, B.J.; Stechert, R.; Stephens, S. Decline of an Isolated Timber Rattlesnake (Crotalus horridus) Population: Interactions between Climate Change, Disease, and Loss of Genetic Diversity. Biol. Conserv. 2011, 144, 886–891. [Google Scholar] [CrossRef]
- Allender, M.C.; Dreslik, M.; Wylie, S.; Phillips, C.; Wylie, D.B.; Maddox, C.; Delaney, M.A.; Kinsel, M.J. Chrysosporium Sp. Infection in Eastern Massasauga Rattlesnakes. Emerg. Infect. Dis. 2011, 17, 2383–2384. [Google Scholar] [CrossRef]
- Dolinski, A.C.; Allender, M.C.; Hsiao, V.; Maddox, C.W. Systemic Ophidiomyces ophiodiicola Infection in a Free-Ranging Plains Garter Snake (Thamnophis radix). J. Herpetol. Med. Surg. 2014, 24, 7. [Google Scholar] [CrossRef]
- Steeil, J.C.; Hope, K.L.; Evans, M.; Peters, A.; Cartoceti, A. Multifocal Ophidiomyces ophiodiicola Infection in an Eastern Diamondback Rattlesnake (Crotalus adamanteus) without the Presence of Skin Lesions. J. Herpetol. Med. Surg. 2018, 28, 76. [Google Scholar] [CrossRef]
- Agugliaro, J.; Lind, C.M.; Lorch, J.M.; Farrell, T.M. An Emerging Fungal Pathogen Is Associated with Increased Resting Metabolic Rate and Total Evaporative Water Loss Rate in a Winter-active Snake. Funct. Ecol. 2020, 34, 486–496. [Google Scholar] [CrossRef]
- Lind, C.M.; Agugliaro, J.; Lorch, J.M.; Farrell, T.M. Ophidiomycosis Is Related to Seasonal Patterns of Reproduction, Ecdysis, and Thermoregulatory Behavior in a Free-living Snake Species. J. Zool. 2023, 319, 54–62. [Google Scholar] [CrossRef]
- Tetzlaff, S.J.; Ravesi, M.J.; Allender, M.C.; Carter, E.T.; DeGregorio, B.A.; Josimovich, J.M.; Kingsbury, B.A. Snake Fungal Disease Affects Behavior of Free-Ranging Massasauga Rattlesnakes (Sistrurus catenatus). Herpetol. Conserv. Biol. 2017, 12, 624–634. [Google Scholar]
- Lind, C.M.; Lorch, J.M.; Moore, I.T.; Vernasco, B.J.; Farrell, T.M. Seasonal Sex Steroids Indicate Reproductive Costs Associated with Snake Fungal Disease. J. Zool. 2019, 307, 104–110. [Google Scholar] [CrossRef]
- Meier, G.; Notomista, T.; Marini, D.; Ferri, V. First Case of Snake Fungal Disease Affecting a Free-Ranging Natrix natrix (Linnaeus, 1758) in Ticino Canton, Switzerland. Herpetol. Notes 2018, 11, 885–891. [Google Scholar]
- Franklinos, L.H.V.; Lorch, J.M.; Bohuski, E.; Rodriguez-Ramos Fernandez, J.; Wright, O.N.; Fitzpatrick, L.; Petrovan, S.; Durrant, C.; Linton, C.; Baláž, V.; et al. Emerging Fungal Pathogen Ophidiomyces ophiodiicola in Wild European Snakes. Sci. Rep. 2017, 7, 3844. [Google Scholar] [CrossRef]
- Sun, P.-L.; Yang, C.-K.; Li, W.-T.; Lai, W.-Y.; Fan, Y.-C.; Huang, H.-C.; Yu, P.-H. Infection with Nannizziopsis guarroi and Ophidiomyces ophiodiicola in Reptiles in Taiwan. Transbound. Emerg. Dis. 2022, 69, 764–775. [Google Scholar] [CrossRef]
- Ladner, J.T.; Palmer, J.M.; Ettinger, C.L.; Stajich, J.E.; Farrell, T.M.; Glorioso, B.M.; Lawson, B.; Price, S.J.; Stengle, A.G.; Grear, D.A.; et al. The Population Genetics of the Causative Agent of Snake Fungal Disease Indicate Recent Introductions to the USA. PLoS Biol. 2022, 20, e3001676. [Google Scholar] [CrossRef]
- Origgi, F.C.; Pisano, S.R.R.; Glaizot, O.; Hertwig, S.T.; Schmitz, A.; Ursenbacher, S. Ophiodimyces ophiodiicola, Etiologic Agent of Snake Fungal Disease, in Europe since Late 1950s. Emerg. Infect. Dis. 2022, 28, 2064–2068. [Google Scholar] [CrossRef] [PubMed]
- Schüler, L.; Lenz, S.; Marschang, E.R. Ophidiomyces ophidiicola bei wildlebenden Würfelnattern (Natrix tessellata) in Deutschland im Jahre 2021. Tierárztliche Prax. Ausg. K Kleintiere/Heimtiere 2022, 50, 151. [Google Scholar]
- Schüler, L.; Lenz, S.; Mittenzwei, F.; Gletscher, I.; Müller, E.; Heckers, K.; Marschang, R.E. Ophidiomycosis in Wild Dice Snakes (Natrix tessellata) in Germany. J. Herpetol. Med. Surg. 2024, 34, 11–15. [Google Scholar] [CrossRef]
- Marini, D.; Di Nicola, M.R.; Crocchianti, V.; Notomista, T.; Iversen, D.; Coppari, L.; Di Criscio, M.; Brouard, V.; Dorne, J.-L.C.M.; Rüegg, J.; et al. Pilot Survey Reveals Ophidiomycosis in Dice Snakes Natrix tessellata from Lake Garda, Italy. Vet. Res. Commun. 2023, 47, 1707–1719. [Google Scholar] [CrossRef] [PubMed]
- Přibyl, M.; Kabelka, R.; Hanzlík, P.M.; Mikulíček, P.; Folk, N.; Piaček, V.; Pikula, J.; Baláž, V. Ophidiomyces ophidiicola in Free-Ranging and Captive Snakes in the Czech and Slovak Republics. J. Vertebr. Biol. 2023, 72, 23050. [Google Scholar] [CrossRef]
- Joudrier, N.; Blanvillain, G.; Meier, G.; Hoyt, J.; Chèvre, M.; Dubey, S.; Origgi, F.C.; Ursenbacher, S. Unravelling the Disease Ecology of Snake Fungal Disease: High Genetic Variability and Ecological Features of Ophidiomyces ophidiicola in Switzerland. Amphib.-Reptil. 2024, 45, 85–98. [Google Scholar] [CrossRef]
- Joudrier, N.; Blanvillain, G.; Ursenbacher, S. First Detection of Apparent Ophidiomycosis in the Vipera Genus in Europe: Findings on Two Asp Vipers, Vipera aspis (Linnaeus, 1758), in Switzerland. Herpetol. Notes 2024, 17, 311–314. [Google Scholar]
- Allain, S.J.R.; Leech, D.I.; Hopkins, K.; Seilern-Moy, K.; Rodriguez-Ramos Fernandez, J.; Griffiths, R.A.; Lawson, B. Characterisation, Prevalence and Severity of Skin Lesions Caused by Ophidiomycosis in a Population of Wild Snakes. Sci. Rep. 2024, 14, 5162. [Google Scholar] [CrossRef] [PubMed]
- Blanvillain, G.; Lorch, J.M.; Joudrier, N.; Bury, S.; Cuenot, T.; Franzen, M.; Martínez-Freiría, F.; Guiller, G.; Halpern, B.; Kolanek, A.; et al. Contribution of Host Species and Pathogen Clade to Snake Fungal Disease Hotspots in Europe. Commun. Biol. 2024, 7, 440. [Google Scholar] [CrossRef]
- Stark, T.; Beukema, W.; Gilbert, M.J.; Goverse, E.; Spitzen-van der Sluijs, A.; Struijk, R.; Verbrugghe, E.; Pasmans, F.; Martel, A. Detection of Ophidiomyces ophidiicola in Wild Barred Grass Snakes (Natrix helvetica) in the Netherlands. Vlaams Diergeneeskd. Tijdschr. 2024, 93, 79–84. [Google Scholar] [CrossRef]
- Martinez-Silvestre, A.; Blanvillain, G.; Gonzalez, J.; Ribo, J. First Record of Ophidiomycosis in a Wild Aesculapian Snake, Zamenis longissimus (Laurenti, 1768), in Spain. Herpetol. Notes 2024, 17, 423–426. [Google Scholar]
- Marini, D.; Di Nicola, M.R.; Crocchianti, V.; Di Criscio, M.; Rüegg, J.; Marenzoni, M.L. Ophidiomyces ophidiicola from Lake Garda (Italy) Belongs to Clade, I. In Proceedings of the 5th International Conference of the European College of Veterinary Microbiology (5th ICECVM), Bled, Slovenia, 21–23 September 2023; p. 48. [Google Scholar]
- Musilová, R.; Zavadil, V.; Kotlík, P. Isolated Populations of Zamenis longissimus (Reptilia: Squamata) above the Northern Limit of the Continuous Range in Europe: Origin and Conservation Status. Acta Soc. Zool. Bohem. 2007, 71, 197–208. [Google Scholar]
- Gomille, A. Die Äskulapnatter-Elaphe Longissima: Verbreitung und Lebensweise in Mitteleuropa; Chimaira: Frankfurt, Germany, 2002. [Google Scholar]
- Mikátová, B.; Vlašín, M. Distribution and Biology of Aesculapian Snake (Zamenis longissimus) in the Territory of the Podyji and Thayatal National Parks and in Their Neighbourhood. Thayensia Znojmo 2012, 9, 51–81. [Google Scholar]
- Waitzmann, M. Zur Situation Der Äskulapnatter Elaphe longissima (Laurenti, 1768) in Der Bundesrepublik Deutschland. Mertensiella 1993, 3, 115–133. [Google Scholar]
- Janoušek, K.; Musilová, R. Aesculapian Snake in the Czech Republic. ZOO Rep. Brno 2009, 4, 1–4. [Google Scholar]
- Kurek, K.; Król, W.; Najberek, K.; Ćmiel, A.M.; Solarz, W.; Bury, S.; Baś, G.; Najbar, B.; Okarma, H. Habitat Use of the Aesculapian Snake at Different Spatial Scales. J. Wildl. Manag. 2018, 82, 1746–1755. [Google Scholar] [CrossRef]
- Edgar, P.; Bird, D.R. Action Plan for the Conservation of the Aesculapian Snake (Zamenis longissimus) in Europe. In Proceedings of the Convention on the Conservation of European Wildlife and Natural Habitats, Strasbourg, France, 27–30 November 2006; Council of Europe: Strasbourg, France, 2006. [Google Scholar]
- Najbar, B. Aesculapian Snake Elaphe (Zamenis) longissima (Laurenti 1768) in Western Bieszczady; Oficyna Wydawnicza Uniwersytetu Zielonogórskiego: Zielona Góra, Poland, 2004; p. 140. [Google Scholar]
- Kurek, K.; Najberek, K.; Zając, B.; Bury, S.; Ćmiel, A.M.; Baś, G.; Najbar, B. Changes in Distribution of Aesculapian Snake and Implications for Its Active Conservation in Poland. Pol. J. Ecol. 2017, 65, 422–431. [Google Scholar] [CrossRef]
- Zając, B. Wąż Eskulapa Zamenis longissimus w Beskidzie Niskim. Przegląd Przyr. 2023, 34, 72–75. [Google Scholar]
- Kurek, K.; Bury, S.; Baś, G.; Najberek, K.; Kaczmarski, M.; Śnieżko, S. Telemetry Studies of the Aesculapian Snake in the Bieszczady Mountains—Preliminary Results and Evaluation of the Methods Used. Chrońmy Przyr. Ojczystą 2014, 70, 3–15. [Google Scholar]
- Kurek, K.; Ćmiel, A.; Bury, S.; Zając, B.; Najberek, K.; Babiasz, R.; Musilová, R.; Baś, G.; Najbar, B. What Has Happened to the Females? Population Trends in the Aesculapian Snake at Its Northern Range Limit. Glob. Ecol. Conserv. 2019, 17, e00550. [Google Scholar] [CrossRef]
- Kotula, F. Po Rzeszowskim Podgorzu Błądząc: Reportaź Historyczny; Wydawn Literackie: Krakow, Poland, 1974. [Google Scholar]
- Kovar, R.; Brabec, M.; Vita, R.; Vodicka, R.; Bogdan, V. Habitat Use of the Aesculapian Snake, Zamenis longissimus at the Northern Extreme of Its Range in Northwest Bohemia. Herpetol. Bull. 2016, 136, 35–36. [Google Scholar]
- Kovar, R.; Brabec, M.; Vita, R.; Vodicka, R.; Bogdan, V. Nesting and Over-Wintering Sites of Aesculapian Snake, Zamenis longissimus, in an Anthropogenic Landscape in the Northern Extreme of Its Range. Herpetol. Bull. 2016, 136, 35–36. [Google Scholar]
- Gherghel, I.; Strugariu, A.; Sahlean, T.C.; Zamfirescu, O. Anthropogenic Impact or Anthropogenic Accommodation? Distribution Range Expansion of the Common Wall Lizard (Podarcis muralis) by Means of Artificial Habitats in the North-Eastern Limits of Its Distribution Range. Acta Herpetol. 2009, 4, 183–189. [Google Scholar] [CrossRef]
- Löwenborg, K.; Shine, R.; Kärvemo, S.; Hagman, M. Grass Snakes Exploit Anthropogenic Heat Sources to Overcome Distributional Limits Imposed by Oviparity. Funct. Ecol. 2010, 24, 1095–1102. [Google Scholar] [CrossRef]
- Najbar, B. Aesculapian Snake. Monografie Przyrodnicze; Wydawnictwo Klubu Przyrodników: Świebodzin, Poland, 2004; p. 104. [Google Scholar]
- Kaźmierczak, T. Distribution of the Aesculapian Snake Elaphe longissima (Laur.) in Poland. Przeg Zool 1965, 9, 380–385. [Google Scholar]
- Najbar, B. The Aesculapian Snake Elaphe l. longissima [Laurenti, 1768] Population in Bieszczady [Poland] between 1990–1998. Bull. Pol. Acad. Sci. Biol. Sci. 2000, 48, 41–51. [Google Scholar]
- Najbar, B. The State of the Aesculapian Snake Elaphe l. longissima [Laurenti, 1768] Population in Poland. Bull. Pol. Acad. Sci. Biol. Sci. 2000, 48, 53–62. [Google Scholar]
- Kurek, K.; Holuk, J.; Bury, S.; Piotrowski, M. Best Practices Manual for Reptile Protection; Centrum Koordynacji Projektów Środowiskowych: Warszawa, Poland, 2014. [Google Scholar]
- Lelièvre, H.; Blouin-Demers, G.; Bonnet, X.; Lourdais, O. Thermal Benefits of Artificial Shelters in Snakes: A Radiotelemetric Study of Two Sympatric Colubrids. J. Therm. Biol. 2010, 35, 324–331. [Google Scholar] [CrossRef]
- Bohuski, E.; Lorch, J.M.; Griffin, K.M.; Blehert, D.S. TaqMan Real-Time Polymerase Chain Reaction for Detection of Ophidiomyces ophiodiicola, the Fungus Associated with Snake Fungal Disease. BMC Vet. Res. 2015, 11, 95. [Google Scholar] [CrossRef]
- Lorch, J.M.; Price, S.J.; Lankton, J.S.; Drayer, A.N. Confirmed Cases of Ophidiomycosis in Museum Specimens from as Early as 1945, United States. Emerg. Infect. Dis. 2021, 27, 1986–1989. [Google Scholar] [CrossRef]
- Crossley, B.M.; Bai, J.; Glaser, A.; Maes, R.; Porter, E.; Killian, M.L.; Clement, T.; Toohey-Kurth, K. Guidelines for Sanger Sequencing and Molecular Assay Monitoring. J. Vet. Diagn. Investig. 2020, 32, 767–775. [Google Scholar] [CrossRef]
- Shine, R.; Bonnet, X. 6. Reproductive Biology, Population Viability, and Options for Field Management. In Snakes: Ecology and Conservation; Mullin, S.J., Seigel, R.A., Eds.; Cornell University Press: Ithaca, NY, USA, 2009; pp. 172–200. [Google Scholar] [CrossRef]
- Walker, D.M.; Leys, J.E.; Grisnik, M.; Grajal-Puche, A.; Murray, C.M.; Allender, M.C. Variability in Snake Skin Microbial Assemblages across Spatial Scales and Disease States. ISME J. 2019, 13, 2209–2222. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.; Gochfeld, M.; Zappalorti, R.; Bunnell, J.; Jeitner, C.; Schneider, D.; Ng, K.; DeVito, E.; Lorch, J.M. Prevalence of Ophidiomyces ophidiicola and Epizootiology of Snake Fungal Disease in Free-Ranging Northern Pine Snakes (Pituophis melanoleucus melanoleucus) in New Jersey. Environ. Monit. Assess. 2023, 195, 662. [Google Scholar] [CrossRef] [PubMed]
- Błażuk, J. Herpetofauna Doliny Sanu Pod Otrytem i Terenów przyległycH (BieSzczady zacHodnie). Gady. Rocz. Bieszczadzkie 2007, 15, 181–229. [Google Scholar]
- Palomar, G.; Fernández-Chacón, A.; Bosch, J. Amphibian Survival Compromised by Long-Term Effects of Chytrid Fungus. Biodivers. Conserv. 2023, 32, 793–809. [Google Scholar] [CrossRef]
- Blanvillain, G.; Martínez-Freiría, F.; Hoyt, J.R.; Lorch, J.M.; Martinez-Silvestre, A. Paranannizziopsis Spp. Infection in Wild Vipers, Europe. Emerg. Infect. Dis. 2024, 30, 1000–1003. [Google Scholar] [CrossRef]
| Number | Sample ID | Species | Location | Coordinates | Date | Gross Signs |
|---|---|---|---|---|---|---|
| 1 | 20 | Zl | NA | NA | 200407 | y |
| 2 | 21 | Zl | Koliba | 49.26, 22.51 | 201008 | n |
| 3 | 22 | Nn | NA | NA | NA | n |
| 4 | 23 | Zl | Zatwarnica | 49.23, 22.56 | 201008 | y |
| 5 | 25 | Zl | Kamieniołom | 49.25, 22.53 | 201008 | y |
| 6 | 26 | Zl | Zatwarnica | 49.23, 22.56 | 201008 | y |
| 7 | 28 | Zl | NA | NA | 201008 | n |
| 8 | 50 | Nn | Tartak | 49.21, 22.55 | 201009 | n |
| 9 | 52 | Zl | Koliba | 49.26, 22.51 | 201109 | n |
| 10 | 53 | Nn | NA | NA | NA | n |
| 11 | 59 | Nn | NA | NA | NA | n |
| 12 | 60 | Zl | Kopiec Żaka | 49.23, 22.55 | 201109 | y |
| 13 | 65 | Zl | Druga pasieka | 49.26, 22.50 | 201107 | n |
| 14 | 66 | Zl | Druga pasieka | 49.26, 22.50 | 201107 | y |
| 15 | 69 | Zl | Zatwarnica | 49.22, 22.55 | 201107 | y |
| 16 | 70 | Zl | Zakole Sanu | 49.25, 22.54 | 201107 | y |
| 17 | 71 | Zl | Koliba | 49.26, 22.51 | 201107 | n |
| 18 | 94 | Zl | Zatwarnica | 49.22, 22.55 | 201106 | n |
| 19 | 95 | Zl | Zatwarnica | 49.22, 22.55 | 201106 | n |
| 20 | 106 | Zl | Sękowiec | 49.23, 22.56 | 201107 | n |
| 21 | 107 | Zl | Zatwarnica | 49.22, 22.55 | 201107 | n |
| 22 | 108 | Zl | Zatwarnica | 49.22, 22.55 | 201107 | n |
| 23 | 109 | Zl | Zatwarnica | 49.22, 22.55 | 201107 | n |
| 24 | 111 | Zl | Zatwarnica | 49.22, 22.55 | 201107 | n |
| 25 | 122 | Zl | Koliba | 49.26, 22.51 | 201107 | n |
| 26 | 123 | Zl | Koliba | 49.26, 22.51 | 201107 | n |
| 27 | 124 | Zl | Koliba | 49.26, 22.51 | 201107 | y |
| 28 | 125 | Zl | Koliba | 49.26, 22.51 | 201107 | n |
| 29 | 126 | Zl | Koliba | 49.26, 22.51 | 201107 | n |
| 30 | 127 | Zl | Zatwarnica | 49.22, 22.55 | 201107 | n |
| 31 | 136 | Zl | Sękowiec | NA | 201107 | y |
| 32 | 141 | Zl | Zatwarnica | 49.22, 22.55 | 201107 | n |
| 33 | 142 | Zl | Zatwarnica | 49.22, 22.55 | 201107 | y |
| 34 | 144 | Zl | Dwernik | 49.22, 22.62 | 201108 | y |
| 35 | 145 | Zl | Dwernik | 49.22, 22.62 | 201108 | y |
| 36 | 147 | Zl | Zatwarnica | 49.22, 22.55 | 201108 | n |
| 37 | 149 | Zl | Sękowiec | 49.23, 22.56 | 201108 | n |
| 38 | 155 | Zl | Dwernik | 49.22, 22.62 | 201108 | n |
| 39 | 175 | Zl | Dwernik | 49.22, 22.62 | 201108 | n |
| 40 | 179 | Zl | Koliba | 49.26, 22.51 | 201108 | n |
| 41 | 181 | Zl | Koliba | 49.26, 22.51 | 201108 | n |
| 42 | 182 | Zl | Koliba | 49.26, 22.51 | 201108 | n |
| 43 | 186 | Zl | Kamieniołom | 49.25, 22.53 | 201108 | n |
| 44 | 187 | Zl | Koliba | 49.26, 22.51 | 201108 | n |
| 45 | 190 | Zl | Zatwarnica | 49.22, 22.55 | 201105 | n |
| 46 | 191 | Zl | Zatwarnica | 49.22, 22.55 | 201105 | n |
| 47 | 215 | Zl | Zatwarnica | 49.22, 22.55 | 201208 | y |
| 48 | 216 | Zl | Koliba | 49.26, 22.51 | 201205 | n |
| 49 | 218 | Zl | Dwernik | 49.22, 22.62 | 201207 | y |
| 50 | 223 | Zl | Zatwarnica | 49.22, 22.55 | 201206 | n |
| 51 | 224 | Zl | Koliba | 49.26, 22.51 | 201207 | n |
| 52 | 225 | Zl | Zatwarnica | 49.22, 22.55 | 201207 | n |
| 53 | 226 | Zl | Zatwarnica | 49.22, 22.55 | 201208 | y |
| 54 | 227 | Zl | Zatwarnica | 49.22, 22.55 | 201208 | y |
| 55 | 228 | Nn | Tworylne | 49.26, 22.47 | 201208 | n |
| 56 | 229 | Zl | Dwernik | 49.22, 22.62 | 201209 | n |
| 57 | 230 | Zl | Dwernik | 49.22, 22.62 | 201209 | n |
| 58 | 231 | Zl | Zatwarnica | 49.22, 22.55 | NA | n |
| 59 | Martwy 1 | Zl | Zatwarnica | 49.23, 22.55 | 201205 | n |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marini, D.; Szczygieł, P.; Kurek, K.; Di Nicola, M.R.; Dorne, J.-L.C.M.; Marenzoni, M.L.; Rüegg, J.; Bury, S.; Kiraga, Ł. Retrospective Detection of Ophidiomyces ophidiicola from Snake Moults Collected in Bieszczady Mountains, Poland. Microorganisms 2024, 12, 1467. https://doi.org/10.3390/microorganisms12071467
Marini D, Szczygieł P, Kurek K, Di Nicola MR, Dorne J-LCM, Marenzoni ML, Rüegg J, Bury S, Kiraga Ł. Retrospective Detection of Ophidiomyces ophidiicola from Snake Moults Collected in Bieszczady Mountains, Poland. Microorganisms. 2024; 12(7):1467. https://doi.org/10.3390/microorganisms12071467
Chicago/Turabian StyleMarini, Daniele, Piotr Szczygieł, Katarzyna Kurek, Matteo Riccardo Di Nicola, Jean-Lou C. M. Dorne, Maria Luisa Marenzoni, Joëlle Rüegg, Stanisław Bury, and Łukasz Kiraga. 2024. "Retrospective Detection of Ophidiomyces ophidiicola from Snake Moults Collected in Bieszczady Mountains, Poland" Microorganisms 12, no. 7: 1467. https://doi.org/10.3390/microorganisms12071467
APA StyleMarini, D., Szczygieł, P., Kurek, K., Di Nicola, M. R., Dorne, J.-L. C. M., Marenzoni, M. L., Rüegg, J., Bury, S., & Kiraga, Ł. (2024). Retrospective Detection of Ophidiomyces ophidiicola from Snake Moults Collected in Bieszczady Mountains, Poland. Microorganisms, 12(7), 1467. https://doi.org/10.3390/microorganisms12071467






