Discovery and Characterization of Epichloë Fungal Endophytes from Elymus spp. in Northwest China
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Materials
2.2. Isolation of Epichloë Endophyte
2.3. Morphological Examination
2.4. DNA Isolation and PCR Amplification of Endophytes
2.5. Phylogenetic Analysis
2.6. Alkaloid Gene and Mating-Type Analysis
3. Results
3.1. Morphological Characteristics
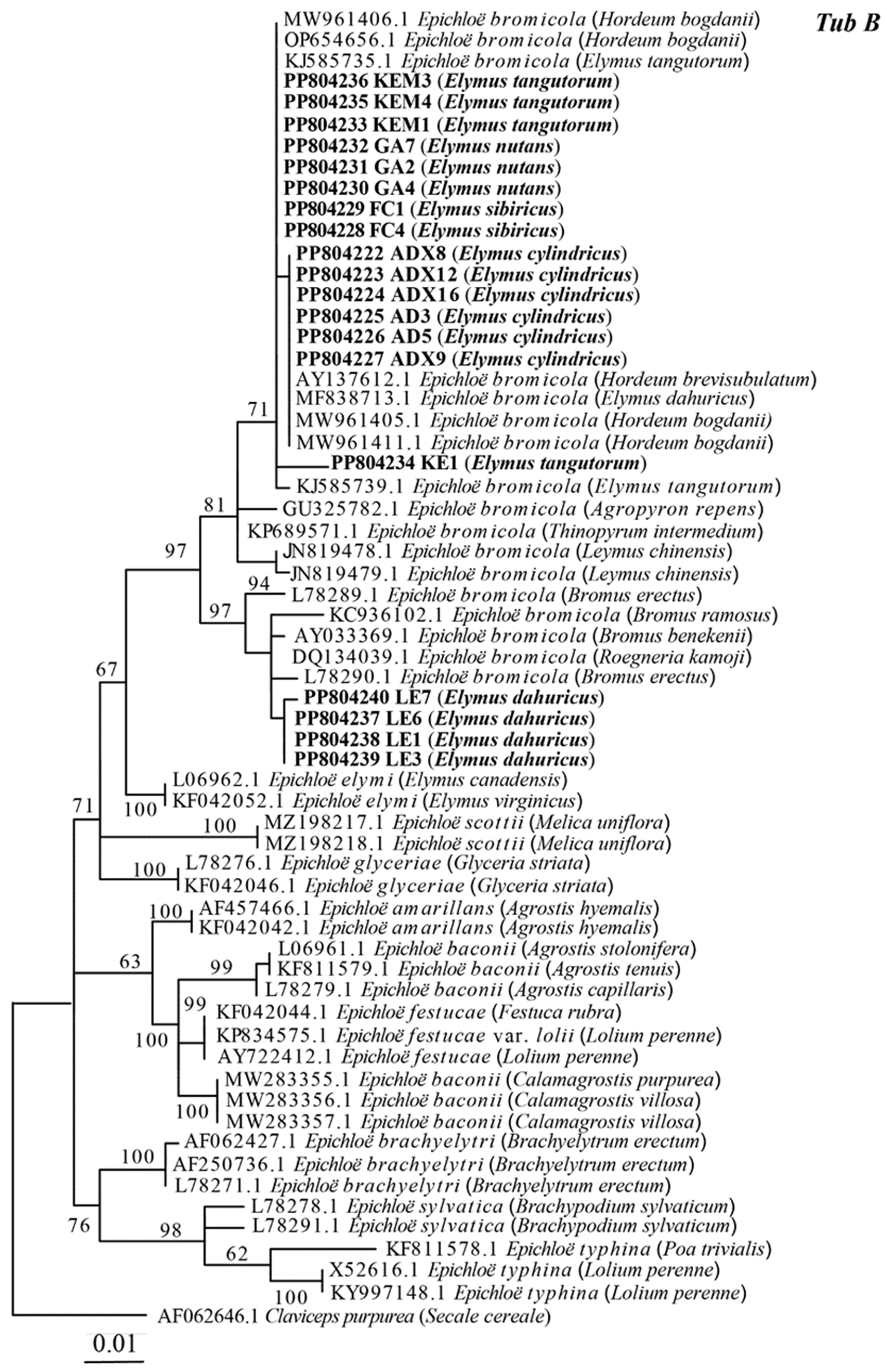
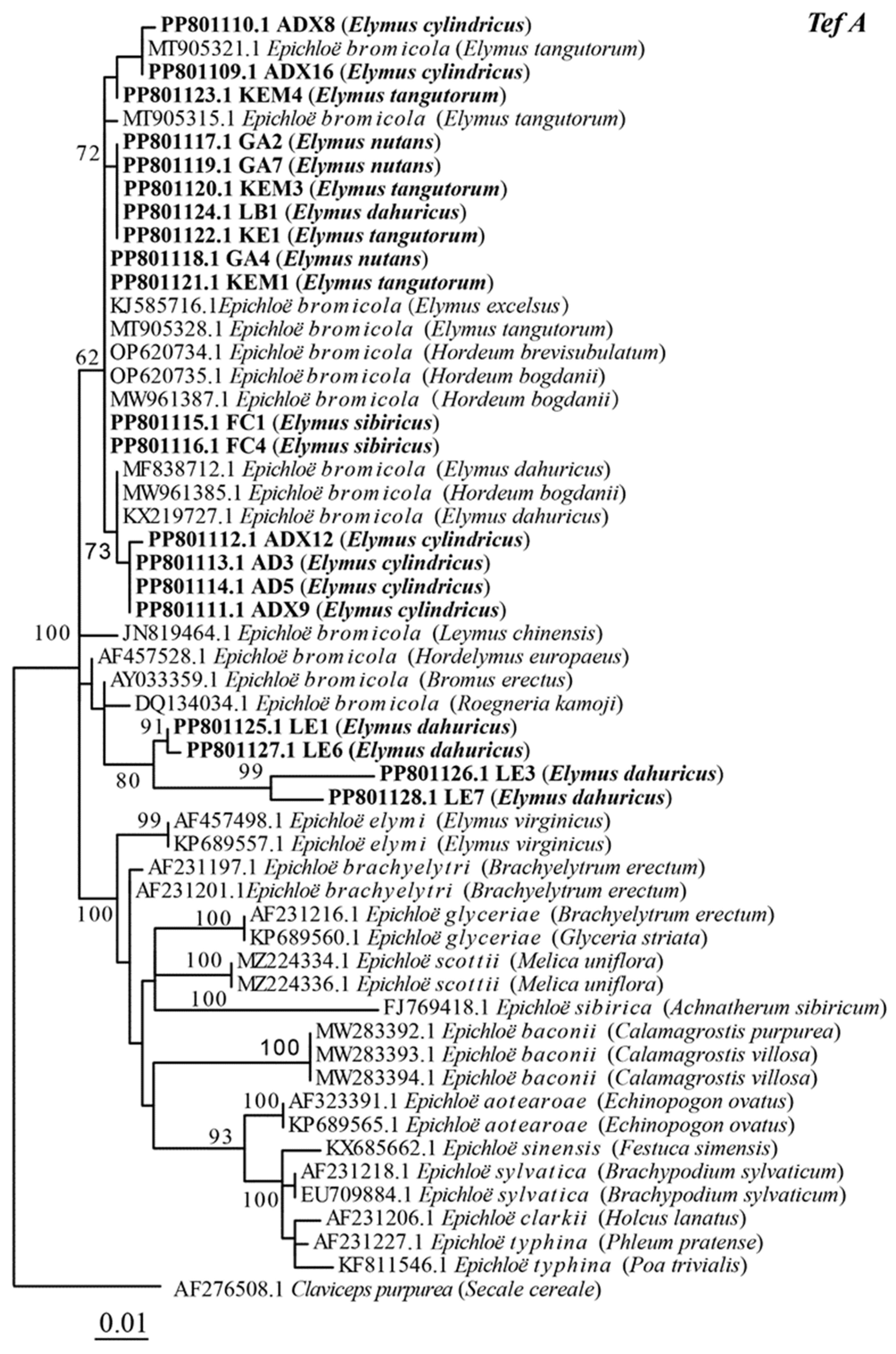
3.2. Phylogenetic Analyses
3.3. Alkaloid Gene Profiling and Mating-Type Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, M.R.; Dahlman, D.L.; Bush, L.P. The role of endophytic fungi in grasses-new approaches to biological-control of pests. Abstr. Pap. Am. Chem. Soc. 1987, 194, 247. [Google Scholar]
- Kuldau, G.; Bacon, C. Clavicipitaceous endophytes: Their ability to enhance plant resistance to multiple stresses. Phytopathology 2005, 956, 138. [Google Scholar] [CrossRef]
- Vogel, K.P.; Hopkins, A.A.; Moore, K.J.; Johnson, K.D.; Carlson, I.T. Genetic variation among canada wildrye accessions from midwest USA remnant prairies for biomass yield and other traits. Crop Sci. 2006, 466, 2348–2353. [Google Scholar] [CrossRef]
- Iannone, L.J.; Novas, M.V.; Young, C.A.; De Battista, J.P.; Schardl, C.L. Endophytes of native grasses from south america: Biodiversity and ecology. Fungal Ecol. 2012, 53, 357–363. [Google Scholar] [CrossRef]
- Johnson, L.J.; de Bonth, A.C.M.; Briggs, L.R.; Caradus, J.R.; Finch, S.C.; Fleetwood, D.J.; Fletcher, L.R.; Hume, D.E.; Johnson, R.D.; Popay, A.J.; et al. The exploitation of Epichloë endophytes for agricultural benefit. Fungal Divers. 2013, 601, 171–188. [Google Scholar] [CrossRef]
- Schardl, C.L.; Craven, K.D.; Speakman, S.; Stromberg, A.; Lindstrom, A.; Yoshida, R. A novel test for host-symbiont codivergence indicates ancient origin of fungal endophytes in grasses. Syst. Biol. 2008, 573, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Casas, C.; Torretta, J.P.; Exeler, N.; Omacini, M. What happens next? Legacy effects induced by grazing and grass-endophyte symbiosis on thistle plants and their floral visitors. Plant Soil 2016, 4051–4052, 211–229. [Google Scholar] [CrossRef]
- Li, F.; Guo, Y.E.; Christensen, M.J.; Gao, P.; Li, Y.Z.; Duan, T.Y. An arbuscular mycorrhizal fungus and Epichloë festucae var. lolii reduce Bipolaris sorokiniana disease incidence and improve perennial ryegrass growth. Mycorrhiza 2018, 282, 159–169. [Google Scholar]
- Chen, T.X.; Johnson, R.; Chen, S.H.; Lv, H.; Zhou, J.L.; Li, C.J. Infection by the fungal endophyte Epichloë bromicola enhances the tolerance of wild barley (Hordeum brevisubulatum) to salt and alkali stresses. Plant Soil 2018, 4281–4282, 353–370. [Google Scholar] [CrossRef]
- Song, M.L.; Li, X.Z.; Saikkonen, K.; Li, C.J.; Nan, Z.B. An asexual Epichloë endophyte enhances waterlogging tolerance of Hordeum brevisubulatum. Fungal Ecol. 2015, 13, 44–52. [Google Scholar] [CrossRef]
- Wang, J.J.; Zhou, Y.P.; Lin, W.H.; Li, M.M.; Wang, M.N.; Wang, Z.G.; Kuang, Y.; Tian, P. Effect of an Epichloë endophyte on adaptability to water stress in Festuca sinensis. Fungal Ecol. 2017, 30, 39–47. [Google Scholar] [CrossRef]
- Wang, Z.F.; Li, C.J.; White, J. Effects of Epichloë endophyte infection on growth, physiological properties and seed germination of wild barley under saline conditions. J. Agron. Crop Sci. 2020, 2061, 43–51. [Google Scholar] [CrossRef]
- Chen, Z.J.; White, J.F.; Malik, K.; Chen, H.; Jin, Y.Y.; Yao, X.; Wei, X.K.; Li, C.J.; Nan, Z.B. Soil nutrient dynamics relate to Epichloë endophyte mutualism and nitrogen turnover in a low nitrogen environment. Soil Biol. Biochem. 2022, 174, 108832. [Google Scholar] [CrossRef]
- Clay, K.; Schardl, C. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am. Nat. 2002, 160, 99–127. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Xie, L.; Zeng, J.; Xie, J. Biosynthesis and regulation of bioprotective alkaloids in the Gramineae endophytic fungi with implications for herbivores deterrents. Curr. Microbiol. 2015, 716, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Young, C.A.; Faulkner, J.R.; Florea, S.; Pan, J. Chemotypic diversity of Epichloë, fungal symbionts of grasses. Fungal Ecol. 2012, 53, 331–344. [Google Scholar] [CrossRef]
- Siegel, M.R.; Latch, G.C.M.; Bush, L.P.; Fannin, F.F.; Rowan, D.D.; Tapper, B.A.; Bacon, C.W.; Johnson, M.C. Fungal endophyte-infected grasses-alkaloid accumulation and aphid response. J. Chem. Ecol. 1990, 1612, 3301–3315. [Google Scholar] [CrossRef] [PubMed]
- Clay, K.; Hardy, T.N.; Hammond, A.M. Fungal endophytes grasses and their effects on an insect herbivore. Oecologia 1985, 661, 1–5. [Google Scholar] [CrossRef]
- Bacon, C.W.; Porter, J.K.; Robbins, J.D.; Luttrell, E.S. Epichloë typhina from toxic tall fescue grasses. Appl. Environ. Microbiol. 1977, 345, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Richardson, K.A.; de Bonth, A.C.M.; Beechey-Gradwell, Z.; Kadam, S.; Cooney, L.J.; Nelson, K.A.; Cookson, R.; Winichayakul, S.; Reid, M.; Anderson, P.; et al. Epichloë fungal endophyte interactions in perennial ryegrass (Lolium perenne L.) modified to accumulate foliar lipids for increased energy density. BMC Plant Biol. 2023, 23, 636. [Google Scholar] [CrossRef]
- Wille, P.; Boller, T.; Kaltz, O. Mixed inoculation alters infection success of strains of the endophyte Epichloë bromicola on its grass host Bromus erectus. Proc. R. Soc. B Biol. Sci. 2002, 2691489, 397–402. [Google Scholar] [CrossRef]
- Fleetwood, D.J.; Khan, A.K.; Johnson, R.D.; Young, C.A.; Mittal, S.; Wrenn, R.E.; Hesse, U.; Foster, S.J.; Schardl, C.L.; Scott, B. Abundant degenerate miniature inverted-repeat transposable elements in genomes of Epichloë fungal endophytes of grasses. Genome Biol. Evol. 2011, 3, 1253–1264. [Google Scholar] [CrossRef]
- Schardl, C.L.; Young, C.A.; Hesse, U.; Amyotte, S.G.; Andreeva, K.; Calie, P.J.; Fleetwood, D.J.; Haws, D.C.; Moore, N.; Oeser, B.; et al. Plant-symbiotic fungi as chemical engineers: Multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013, 92, e1003323. [Google Scholar] [CrossRef]
- Spiering, M.J.; Moon, C.D.; Wilkinson, H.H.; Schardl, C.L. Gene clusters for insecticidal loline alkaloids in the grass-endophytic fungus Neotyphodium uncinatum. Genetics 2005, 1693, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Machado, C.; Panaccione, D.G.; Tsai, H.F.; Schardl, C.L. The determinant step in ergot alkaloid biosynthesis by an endophyte of perennial ryegrass. Fungal Genet. Biol. 2004, 412, 189–198. [Google Scholar] [CrossRef]
- Pan, J. Ether Bridge Formation and Chemical Diversification in Loline Alkaloid Biosynthesis. Ph.D. Thesis, University of Kentucky, Lexington, KY, USA, 2014. [Google Scholar]
- Takach, J.E.; Young, C.A. Alkaloid genotype diversity of tall fescue endophytes. Crop Sci. 2014, 542, 667–678. [Google Scholar] [CrossRef]
- Song, H.; Nan, Z. Origin, divergence, and phylogeny of asexual Epichloë endophyte in Elymus species from Western China. PLoS ONE 2015, 105, e0127096. [Google Scholar] [CrossRef]
- Charlton, N.D.; Craven, K.D.; Mittal, S.; Hopkins, A.A.; Young, C.A. Epichloë canadensis, a new interspecific Epichloë hybrid symbiotic with Canada wildrye (Elymus canadensis). Mycologia 2012, 1045, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Leuchtmann, A. Three new species of Epichloë symbiotic with North American grasses. Mycologia 1999, 911, 95–107. [Google Scholar] [CrossRef]
- White, J.F. Endophyte-host associations in grasses.17. Ecological and physiological features characterizing Epichloë typhina and some anamorphic varieties in England. Mycologia 1992, 843, 431–441. [Google Scholar] [CrossRef]
- White, J.F.; Bultman, T.L. Endophyte-host associations in forage grasses. 8. Heterothallism in Epichloë typhina. Am. J. Bot. 1987, 7411, 1716–1721. [Google Scholar] [CrossRef]
- White, J.F.; Chambless, D.A. Endophyte-host associations in forage grasses. 15. Clustering of stromata-bearing individuals of agrostis infected by Epichloë typhina. Am. J. Bot. 1991, 784, 527–533. [Google Scholar] [CrossRef]
- White, J.F.; Morrow, A.C.; Morganjones, G.; Chambless, D.A. Endophyte-host associations in forage grasses. 14. Primary stromata formation and seed transmission in Epichloë typhina—Developmental and regulatory aspects. Mycologia 1991, 831, 72–81. [Google Scholar] [CrossRef]
- Li, W.; Ji, Y.L.; Yu, H.S.; Wang, Z.W. A new species of Epichloë symbiotic with Chinese grasses. Mycologia 2006, 984, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Li, C. Biological and Ecological Characteristics of Achnatherum inebrians/Neotyphodium Endophyte Symbiont. Ph.D. Thesis, Lanzhou University, Lanzhou, China, 2005. [Google Scholar]
- Moon, C.D.; Guillaumin, J.J.; Ravel, C.; Li, C.; Craven, K.D.; Schardl, C.L. New Neotyphodium endophyte species from the grass tribes stipeae and meliceae. Mycologia 2007, 996, 895–905. [Google Scholar] [CrossRef]
- Moon, C.D.; Miles, C.O.; Jarlfors, U.; Schardl, C.L. The evolutionary origins of three new Neotyphodium endophyte species from grasses indigenous to the southern hemisphere. Mycologia 2002, 944, 694–711. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. Phylosuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 201, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Dambe6: New tools for microbial genomics, phylogenetics, and molecular evolution. J. Hered. 2017, 1084, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.; Chernomor, O.; Schrempf, D.; Woodhams, M.; von Haeseler, A.; Lanfear, R. Iq-tree 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 375, 1530–1534. [Google Scholar] [CrossRef]
- Tanaka, A.; Tapper, B.A.; Popay, A.; Parker, E.J.; Scott, B. A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol. Microbiol. 2005, 574, 1036–1050. [Google Scholar] [CrossRef]
- Young, C.A.; Bryant, M.K.; Christensen, M.J.; Tapper, B.A.; Bryan, G.T.; Scott, B. Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Mol. Genet. Genom. 2005, 2741, 13–29. [Google Scholar] [CrossRef]
- Young, C.A.; Felitti, S.; Shields, K.; Spangenberg, G.; Johnson, R.D.; Bryan, G.T.; Saikia, S.; Scott, B. A complex gene cluster for indole-diterpene biosynthesis in the grass endophyte Neotyphodium lolii. Fungal Genet. Biol. 2006, 4310, 679–693. [Google Scholar] [CrossRef]
- Schardl, C.L.; Panaccione, D.G.; Tudzynski, P. Ergot Alkaloids—Biology and molecular biology. In Alkaloids: Chemistry and Biology; Cordell, G.A., Ed.; Academic Press: Cambridge, MA, USA, 2006; Volume 63, pp. 45–86. [Google Scholar]
- Berry, D.; Takach, J.E.; Schardl, C.L.; Charlton, N.D.; Scott, B.; Young, C.A. Disparate independent genetic events disrupt the secondary metabolism gene perA in certain symbiotic Epichloë species. Appl. Environ. Microbiol. 2015, 818, 2797–2807. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, X.Z.; Li, C.J.; Swoboda, G.A.; Young, C.A.; Sugawara, K.; Leuchtmann, A.; Schardl, C.L. Two distinct Epichloë species symbiotic with Achnatherum inebrians, drunken horse grass. Mycologia 2015, 1074, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.X. Physiological Mechanism of Epichloë Endophyte Infection to Enhance Salt Tolerance of Wild Barley. Ph.D. Thesis, Lanzhou University, Lanzhou, China, 2019. [Google Scholar]
- Moon, C.D.; Scott, B.; Schardl, C.L.; Christensen, M.J. The evolutionary origins of Epichloë endophytes from annual ryegrasses. Mycologia 2000, 926, 1103–1118. [Google Scholar] [CrossRef]
- Schardl, C.L.; Florea, S.; Pan, J.; Nagabhyru, P.; Bec, S.; Calie, P.J. The Epichloë: Alkaloid diversity and roles in symbiosis with grasses. Curr. Opin. Plant Biol. 2013, 164, 480–488. [Google Scholar] [CrossRef]
- Kaur, J.; Ekanayake, P.N.; Tian, P.; de Jong, E.v.Z.; Dobrowolski, M.P.; Rochfort, S.J.; Mann, R.C.; Smith, K.F.; Forster, J.W.; Guthridge, K.M.; et al. Discovery and characterisation of novel asexual Epichloë endophytes from perennial ryegrass (Lolium perenne L.). Crop Pasture Sci. 2015, 6610, 1058–1070. [Google Scholar] [CrossRef]
- Rasmussen, S.; Parsons, A.J.; Fraser, K.; Xue, H.; Newman, J.A. Metabolic profiles of Lolium perenne are differentially affected by nitrogen supply, carbohydrate content, and fungal endophyte infection. Plant Physiol. 2008, 1463, 1440–1453. [Google Scholar] [CrossRef]
- Vázquez-De-Alldana, B.R.; Zabalgogeazcoa, I.; García-Ciudad, A.; García-Criado, B. Fungal alkaloids in populations of endophyte-infected Festuca rubra subsp. pruinosa. Grass Forage Sci. 2007, 623, 364–371. [Google Scholar] [CrossRef]
- Shi, C.; An, S.; Yao, Z.; Young, C.A.; Panaccione, D.G.; Lee, S.T.; Schardl, C.L.; Li, C. Toxin-producing Epichloë bromicola strains symbiotic with the forage grass Elymus dahuricus in China. Mycologia 2017, 1096, 847–859. [Google Scholar] [CrossRef]
- Berry, D.; Mace, W.; Grage, K.; Wesche, F.; Gore, S.; Schardl, C.L.; Young, C.A.; Dijkwel, P.P.; Leuchtmann, A.; Bode, H.B.; et al. Efficient nonenzymatic cyclization and domain shuffling drive pyrrolopyrazine diversity from truncated variants of a fungal nrps. Proc. Natl. Acad. Sci. USA 2019, 11651, 25614–25623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.P.; Nan, Z.B. Growth and anti-oxidative systems changes in Elymus dahuricus is affected by Neotyphodium endophyte under contrasting water availability. J. Agron. Crop Sci. 2007, 1936, 377–386. [Google Scholar] [CrossRef]
- Rudgers, J.A.; Swafford, A.L. Benefits of a fungal endophyte in Elymus virginicus decline under drought stress. Basic Appl. Ecol. 2009, 101, 43–51. [Google Scholar] [CrossRef]
- Zhang, X.X.; Li, C.J.; Nan, Z.B. Effects of cadmium stress on seed germination and seedling growth of Elymus dahuricus infected with the Neotyphodium endophyte. Sci. China-Life Sci. 2012, 559, 793–799. [Google Scholar] [CrossRef] [PubMed]
- Caradus, J.R. Epichloë fungal endophytes—A vital component for perennial ryegrass survival in New Zealand. N. Z. J. Agric. Res. 2023, 502, 279–298. [Google Scholar] [CrossRef]
- Li, C.J.; Wang, Z.F.; Chen, T.X.; Nan, Z.B. Creation of novel barley germplasm using an Epichloë endophyte. Chin. Sci. Bull. 2021, 6620, 2608–2617. [Google Scholar] [CrossRef]
- Wang, Z.F.; Liu, J.; White, J.F.; Li, C.J. Epichloë bromicola from wild barley improves salt-tolerance of cultivated barley by altering physiological responses to salt stress. Front. Microbiol. 2022, 13, 1044735. [Google Scholar] [CrossRef]
- Xie, X.G.; Lu, W.L.; Feng, K.M.; Zheng, C.J.; Yang, Y.; Jia, M.; Wu, Y.S.; Shi, Y.Z.; Han, T.; Qin, L.P. Mechanisms of Epichloë bromicola to promote plant growth and its potential application for Coix lacryma-jobi L. cultivation. Curr. Microbiol. 2023, 809, 306. [Google Scholar] [CrossRef]
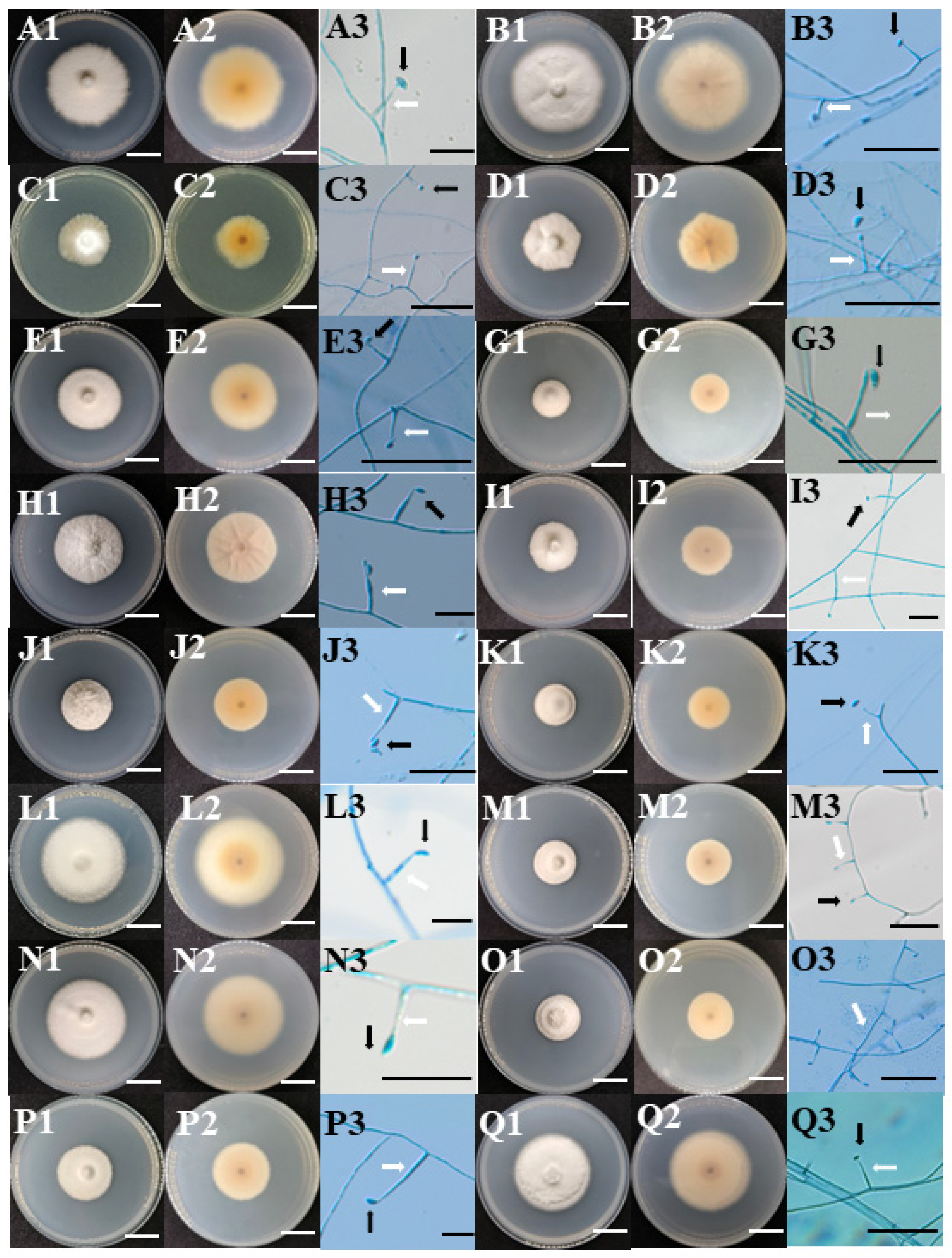

| Location | Altitude (m) | Longitude | Latitude | Host | No. of Samples | No. of Infected Samples | Infection Frequency (%) | Total No. of Strains |
|---|---|---|---|---|---|---|---|---|
| Rangtang | 3571 | 33°24′02″ | 32°20′27″ | Elymus tangutorum | 28 | 8 | 28.57 | 4 |
| Elymus dahuricus | 7 | 4 | 57.14 | 4 | ||||
| Hongyuan | 3494 | 102°31′31″ | 33°16′41″ | Elymus cylindricus | 44 | 36 | 81.82 | 6 |
| Tumotezuo | 1150 | 111°34′00″ | 40°34′00″ | Elymus sibiricus | 12 | 2 | 16.67 | 2 |
| Maqu | 3600 | 102°39′20″ | 33°09′24″ | Elymus nutans | 12 | 1 | 8.33 | 3 |
| Huachi | 1337 | 107°45′32″ | 36°42′53″ | Elymus dahuricus | 6 | 4 | 66.67 | 1 |
| Endophyte | Host | Conidia Size (µm) | Length of Conidiogenous Cell (µm) | Growth Rate (mm/d) | Significance | ||
|---|---|---|---|---|---|---|---|
| AD3 | Elymus cylindricus | 4.16 ± 0.07 (c) | 1.68 ± 0.04 (a) | 10.75 ± 0.76 (c) | 1.25 ± 0.03 b (ab) | 1.01 ± 0.04 e (a) | * |
| AD5 | Elymus cylindricus | 3.79 ± 0.09 (b) | 1.66 ± 0.04 (a) | 14.75 ± 0.62 (a) | 1.25 ± 0.12 bc (b) | 1.44 ± 0.10 e (a) | ns |
| AD16 | Elymus cylindricus | 3.57 ± 0.04 (a) | 1.75 ± 0.03 (a) | 12.74 ± 0.63 (b) | 1.24 ± 0.02 bc (b) | 1.36 ± 0.01 e (a) | ns |
| ADX8 | Elymus cylindricus | 3.78 ± 0.06 (b) | 1.76 ± 0.03 (a) | 11.09 ± 0.55 (bc) | 1.17 ± 0.09 bc (b) | 1.29 ± 0.09 e (a) | ns |
| ADX9 | Elymus cylindricus | nd | nd | nd | 0.75 ± 0.04 d (c) | 1.06 ± 0.07 e (a) | * |
| ADX12 | Elymus cylindricus | nd | nd | nd | 1.49 ± 0.04 b (a) | 1.29 ± 0.01 e (a) | ns |
| FC1 | Elymus sibiricus | 3.78 ± 0.09 (a) | 1.71 ± 0.04 (a) | 13.59 ± 0.91 (b) | 0.59 ± 0.04 de (a) | 1.13 ± 0.03 e (b) | * |
| FC4 | Elymus sibiricus | 3.77 ± 0.08 (a) | 1.54 ± 0.04 (a) | 21.79 ± 1.19 (a) | 0.70 ± 0.03 d (a) | 1.27 ± 0.04 e (a) | * |
| GA2 | Elymus nutans | 3.75 ± 0.10 | 1.71 ± 0.05 | 10.76 ± 0.47 | 0.71 ± 0.02 d (c) | 0.83 ± 0.03 e (c) | ns |
| GA4 | Elymus nutans | nd | nd | nd | 1.14 ± 0.01 bc (a) | 1.33 ± 0.03 e (a) | * |
| GA7 | Elymus nutans | nd | nd | nd | 0.79 ± 0.01 cd (b) | 1.22 ± 0.03 e (b) | * |
| KE1 | Elymus tangutorum | 5.09 ± 0.13 (a) | 1.44 ± 0.03 (b) | 13.33 ± 0.62 (a) | 0.92 ± 0.05 cd (ab) | 1.29 ± 0.09 e (b) | * |
| KEM1 | Elymus tangutorum | 4.16 ± 0.09 (c) | 1.38 ± 0.02 (b) | 11.90 ± 0.66 (ab) | 0.93 ± 0.04 cd (ab) | 1.72 ± 0.06 cd (a) | * |
| KEM3 | Elymus tangutorum | 4.35 ± 0.09 (c) | 1.40 ± 0.03 (b) | 11.86 ± 0.56 (ab) | 1.03 ± 0.02 bc (a) | 1.18 ± 0.04 e (b) | ns |
| KEM4 | Elymus tangutorum | 4.67 ± 0.10 (b) | 1.54 ± 0.03 (a) | 10.54 ± 0.71 (b) | 0.80 ± 0.03 cd (b) | 1.38 ± 0.07 e (b) | ns |
| LB1 | Elymus dahuricus (Huachi) | 3.92 ± 0.06 (ab) | 1.75 ± 0.03 (a) | 13.30 ± 0.56 (c) | 1.40 ± 0.02 b (b) | 1.69 ± 0.01 d (b) | * |
| LE1 | Elymus dahuricus (Rangtang) | 3.33 ± 0.09 (c) | 1.60 ± 0.05 (b) | 11.85 ± 0.70 (cd) | 1.98 ± 0.05 ab (a) | 2.32 ± 0.06 b (a) | * |
| LE3 | Elymus dahuricus (Rangtang) | 3.40 ± 0.07 (c) | 1.71 ± 0.06 (a) | 10.27 ± 0.50 (d) | 1.88 ± 0.13 ab (ab) | 1.89 ± 0.24 c (ab) | ns |
| LE6 | Elymus dahuricus (Rangtang) | 3.74 ± 0.09 (b) | 1.54 ± 0.06 (b) | 16.23 ± 1.28 (b) | 2.18 ± 0.04 a (a) | 2.80 ± 0.13 a (a) | ns |
| LE7 | Elymus dahuricus (Rangtang) | 4.06 ± 0.10 (a) | 1.70 ± 0.03 (ab) | 20.21 ± 1.46 (a) | 1.36 ± 0.51 b (b) | 2.15 ± 0.14 b (ab) | ns |
| Endophyte | Mating-Type Genes | Segments of ppzA Gene | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| mtAC | mtBA | A1 | T1 | C | A2 | M | T2 | R | ∆R | |
| ADX8 | + | − | + | + | + | + | + | + | + | − |
| ADX12 | + | − | + | + | + | + | + | + | + | − |
| ADX9 | + | − | + | + | + | + | + | + | + | − |
| AD16 | + | − | + | + | + | + | + | + | + | − |
| FC1 | + | − | + | + | + | + | + | + | − | + |
| FC4 | + | − | + | + | + | + | + | + | − | + |
| LE6 | + | − | + | + | + | + | + | + | + | − |
| LE1 | + | − | + | + | + | + | + | + | + | − |
| LE3 | + | − | + | + | + | + | + | + | + | − |
| LE7 | + | − | + | + | + | + | + | + | + | − |
| GA7 | + | − | + | + | + | + | + | + | − | + |
| GA2 | + | − | + | + | + | + | + | + | − | + |
| AD3 | + | − | + | + | + | + | + | + | + | − |
| AD5 | + | − | + | + | + | + | + | + | + | − |
| GA4 | + | − | + | + | + | + | + | + | − | + |
| KE1 | + | − | + | + | + | + | + | + | − | + |
| KEM1 | + | − | + | + | + | + | + | + | − | + |
| KEM3 | + | − | + | + | + | + | + | + | − | + |
| KEM4 | + | − | + | + | + | + | + | + | − | + |
| LB1 | + | − | + | + | + | + | + | + | − | + |
| Endophyte | Ergot Alkaloid (EAS) Genes | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dmaW | easF | easC | easE | easD | easA | easG | cloA | lpsB | lpsA | easH | lpsC | easO | easP | |
| ADX8 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| ADX12 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| ADX9 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| AD16 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| FC1 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| FC4 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| LE6 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| LE1 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| LE3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| LE7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| GA7 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| GA2 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| AD3 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| AD5 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| GA4 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| KE1 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| KEM1 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| KEM3 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| KEM4 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| LB1 | + | + | + | + | + | + | + | + | + | + | + | − | − | − |
| Endophyte | Indole–Diterpene (IDT/LTM) Genes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| idtG | idtB | idtM | idtC | idtS | idtP | idtQ | idtF | idtK | idtE | idtJ | |
| ADX8 | + | + | + | + | + | + | + | + | + | − | − |
| ADX12 | + | + | + | + | + | + | + | + | + | − | − |
| ADX9 | + | + | + | + | + | + | + | + | + | − | − |
| AD16 | + | + | + | + | + | + | + | + | + | − | − |
| FC1 | + | + | + | + | + | − | + | + | + | − | − |
| FC4 | + | + | + | + | + | − | + | + | + | − | − |
| LE6 | − | − | − | − | − | − | − | − | − | − | − |
| LE1 | − | − | − | − | − | − | − | − | − | − | − |
| LE3 | − | − | − | − | − | − | − | − | − | − | − |
| LE7 | − | − | − | − | − | − | − | − | − | − | − |
| GA7 | + | + | + | + | + | − | + | + | + | − | − |
| GA2 | + | + | + | + | + | − | + | + | + | − | − |
| AD3 | + | + | + | + | + | + | + | + | + | − | − |
| AD5 | + | + | + | + | + | + | + | + | + | − | − |
| GA4 | + | + | + | + | + | − | + | + | + | − | − |
| KE1 | + | + | + | + | + | + | + | + | + | − | − |
| KEM1 | + | + | + | + | + | + | + | + | + | − | − |
| KEM3 | + | + | + | + | + | + | + | + | + | − | − |
| KEM4 | + | + | + | + | + | + | + | + | + | − | − |
| LB1 | + | + | + | + | + | + | + | + | + | − | − |
| Endophyte | Mating-Type Genes | Alkaloid Synthesis Gene Cluster | Type | Predicted Alkaloid-Producing Type | |||
|---|---|---|---|---|---|---|---|
| ppzA | EAS | IDT | LOL | ||||
| LE6 | mtAC | A | B | A | A | 1 | Per |
| LE1 | mtAC | A | B | A | A | 1 | |
| LE3 | mtAC | A | B | A | A | 1 | |
| LE7 | mtAC | A | B | A | A | 1 | |
| AD3 | mtAC | A | A | B | A | 2 | Per, CC+D-LC+ERV, PAS+PAX+TDK |
| AD5 | mtAC | A | A | B | A | 2 | |
| ADX8 | mtAC | A | A | B | A | 2 | |
| ADX12 | mtAC | A | A | B | A | 2 | |
| ADX9 | mtAC | A | A | B | A | 2 | |
| AD16 | mtAC | A | A | B | A | 2 | |
| GA7 | mtAC | B | A | B | A | 3 | CC+D-LC+ERV, PAS |
| GA2 | mtAC | B | A | B | A | 3 | |
| GA4 | mtAC | B | A | B | A | 3 | |
| FC1 | mtAC | B | A | C | A | 3 | |
| FC4 | mtAC | B | A | C | A | 3 | |
| KE1 | mtAC | B | A | B | A | 4 | CC+D-LC+ERV, PAS+PAX+TDK |
| KEM1 | mtAC | B | A | B | A | 4 | |
| KEM3 | mtAC | B | A | B | A | 4 | |
| KEM4 | mtAC | B | A | B | A | 4 | |
| LB1 | mtAC | B | A | B | A | 4 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, M.; Wang, T.; Li, C.; Chen, T. Discovery and Characterization of Epichloë Fungal Endophytes from Elymus spp. in Northwest China. Microorganisms 2024, 12, 1497. https://doi.org/10.3390/microorganisms12071497
Du M, Wang T, Li C, Chen T. Discovery and Characterization of Epichloë Fungal Endophytes from Elymus spp. in Northwest China. Microorganisms. 2024; 12(7):1497. https://doi.org/10.3390/microorganisms12071497
Chicago/Turabian StyleDu, Mingxiang, Tian Wang, Chunjie Li, and Taixiang Chen. 2024. "Discovery and Characterization of Epichloë Fungal Endophytes from Elymus spp. in Northwest China" Microorganisms 12, no. 7: 1497. https://doi.org/10.3390/microorganisms12071497
APA StyleDu, M., Wang, T., Li, C., & Chen, T. (2024). Discovery and Characterization of Epichloë Fungal Endophytes from Elymus spp. in Northwest China. Microorganisms, 12(7), 1497. https://doi.org/10.3390/microorganisms12071497






