Impact of Biotic and Abiotic Factors on Listeria monocytogenes, Salmonella enterica, and Enterohemorrhagic Escherichia coli in Agricultural Soil Extracts
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain Preparation
2.2. Soil Sample Collection and Extract Preparation
2.2.1. Phase 1
2.2.2. Phase 2
2.3. Inoculation of Soil Extracts
2.4. Enumeration of Pathogens from Soil Extracts
2.5. Enumeration of Mesophilic Aerobic Microbes
2.6. Chemical Analysis of Soil Extracts
2.7. DNA Extraction
2.8. PCR and Sequencing
2.9. Statistical Analysis and Predictive Modelling
2.9.1. Pathogen Survival
2.9.2. 16S Sequence Analysis
2.9.3. Dimensionality Reduction for Predictive Modeling
2.9.4. Development and Evaluation of Predictive Models
3. Results
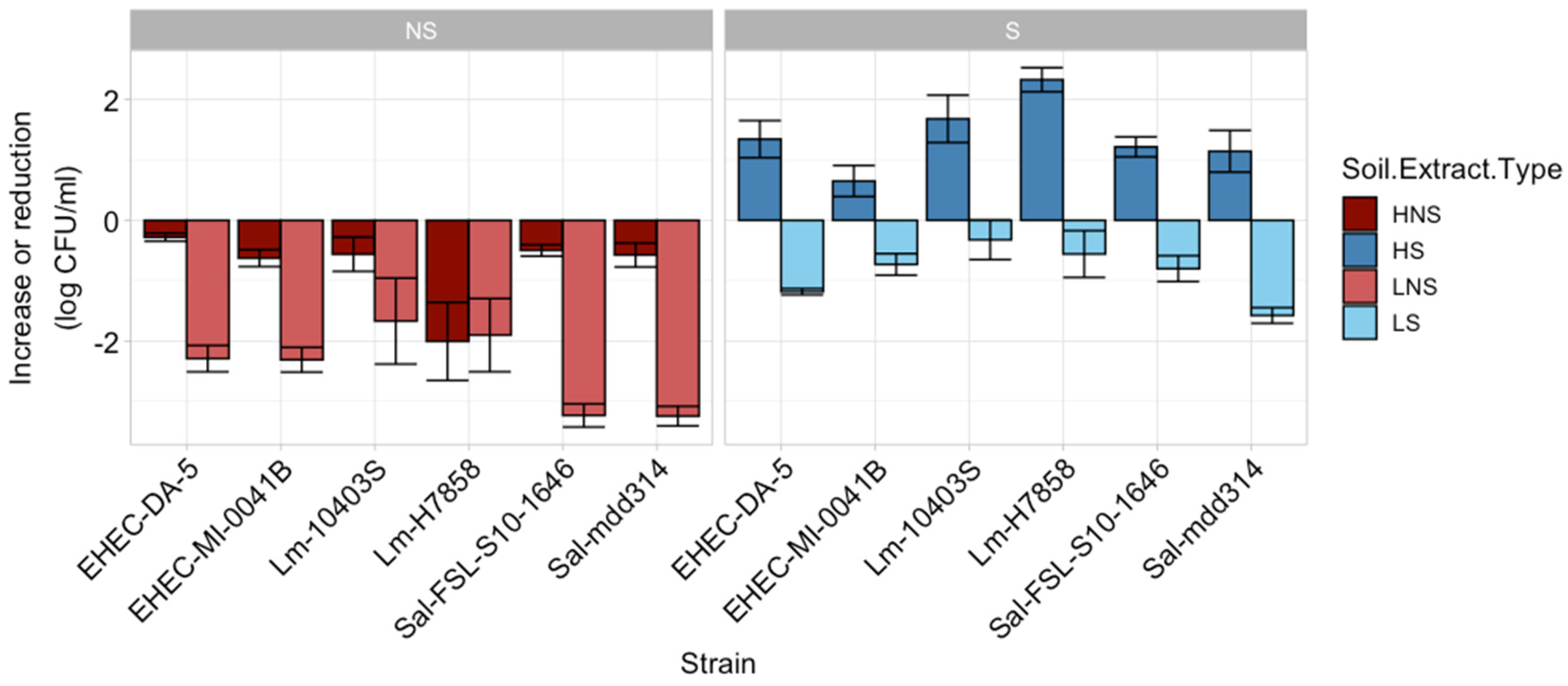
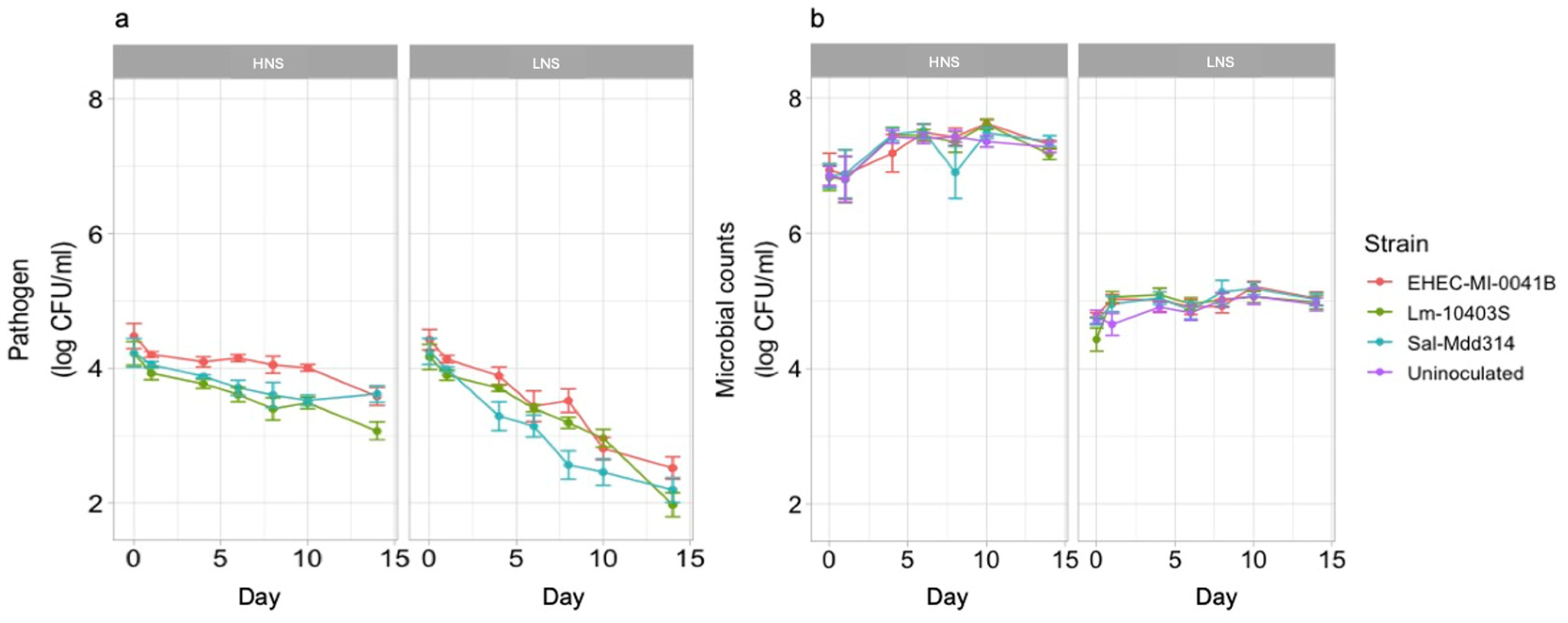
3.1. Pathogen Behavior Is Influenced by Soil Extract Chemistry and Presence of Native Microbiome
3.2. Levels of Mesophilic Aerobic Microbes and Pathogens Are Affected by Soil Extract Nutrient Composition
3.3. Influence of Soil Extract Composition on the Microbiome
3.4. Soil Extract Microbiome Diversity Was Not Affected by Pathogen Inoculation, but Was Influenced by Soil Extract Type
3.5. Identification of Key Variables by PCA
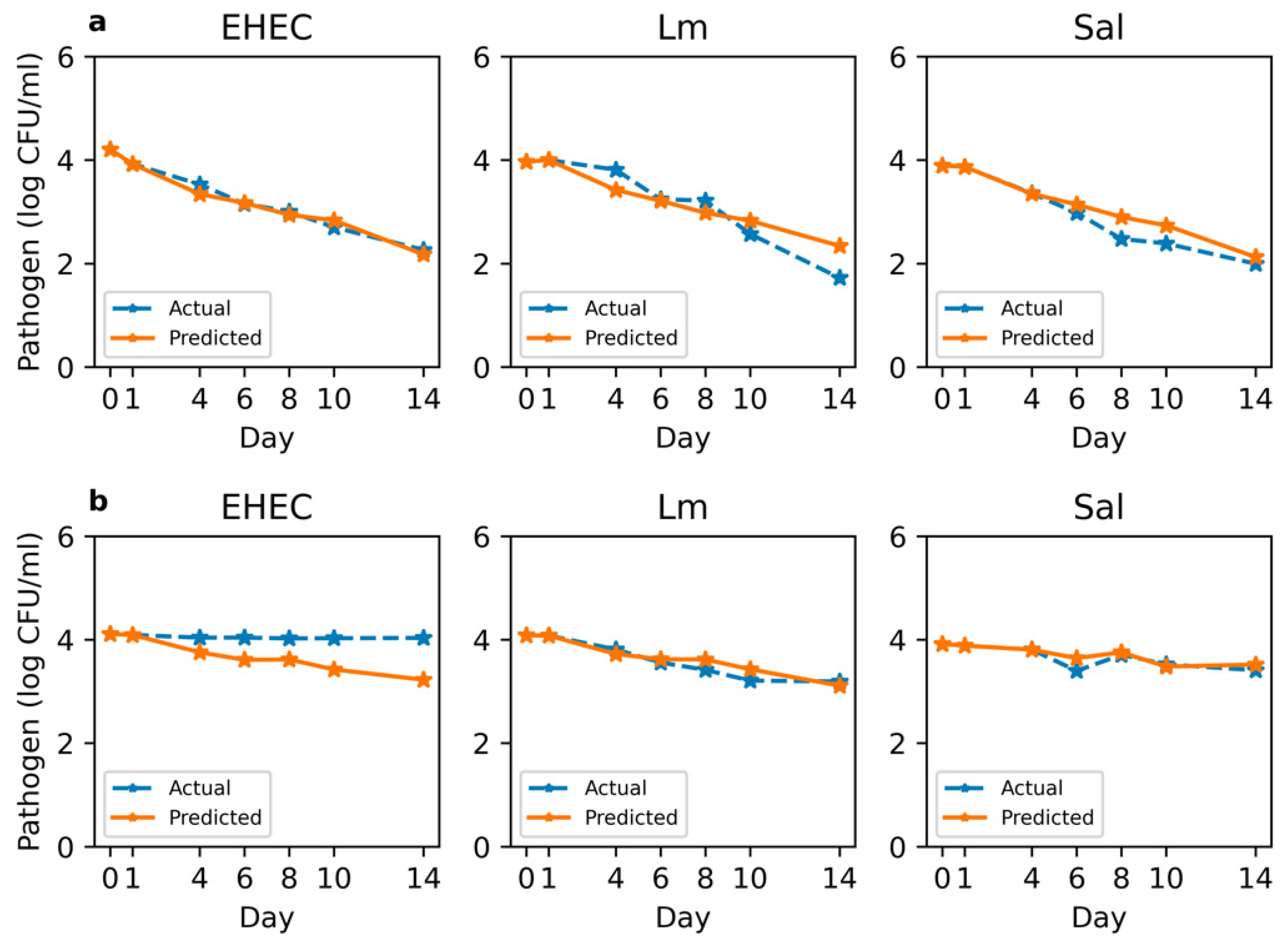
3.6. Prediction of Pathogen Survival Based on Soil Chemistry and Microbial Composition
4. Discussion
4.1. Significant Differences in Soil Extract Chemistry Were Associated with Differences in Pathogen Survival and Native Microbiome Diversity
4.2. AI Models Can Predict Pathogen Survival Given Key Soil Variables
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Turner, K.; Nou Moua, C.; Hajmeer, M.; Barnes, A.; Needham, M. Overview of Leafy Greens-Related Food Safety Incidents with a California Link: 1996 to 2016. J. Food Prot. 2019, 82, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Self, J.L.; Conrad, A.; Stroika, S.; Jackson, A.; Whitlock, L.; Jackson, K.A.; Beal, J.; Wellman, A.; Fatica, M.K.; Bidol, S.; et al. Multistate outbreak of listeriosis associated with packaged leafy green salads, United States and Canada, 2015–2016. Emerg. Infect. Dis. 2019, 25, 1461. [Google Scholar] [CrossRef] [PubMed]
- Krishnasamy, V.P.; Marshall, K.; Dewey-Mattia, D.; Wise, M. Outbreak Characteristics and Epidemic Curves for Multistate Outbreaks of Salmonella Infections Associated with Produce: United States, 2009–2015. Foodborne Pathog. Dis 2020, 17, 15. [Google Scholar] [CrossRef] [PubMed]
- CDC Foodborne Germs and Illnesses. Available online: https://www.cdc.gov/foodsafety/foodborne-germs.html (accessed on 5 January 2024).
- Brandl, M.T. Fitness of Human Enteric Pathogens on Plants and Implications for Food Safety. Annu. Rev. Phytopathol. 2006, 44, 367–392. [Google Scholar] [CrossRef] [PubMed]
- Bergholz, P.W.; Strawn, L.K.; Ryan, G.T.; Warchocki, S.; Wiedmann, M. Spatiotemporal Analysis of Microbiological Contamination in New York State Produce Fields Following Extensive Flooding from Hurricane Irene, August 2011. J. Food Prot. 2016, 79, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.H.; Wolny, J.; Kase, J.A.; Unno, T.; Pachepsky, Y. Interactions of E. Coli with Algae and Aquatic Vegetation in Natural Waters. Water Res. 2021, 209, 117952. [Google Scholar] [CrossRef]
- FSMA Final Rule on Pre-Harvest Agricultural Water FSMA Final Rule on Pre-Harvest Agricultural Water|FDA. Available online: https://www.fda.gov/food/food-safety-modernization-act-fsma/fsma-final-rule-pre-harvest-agricultural-water (accessed on 16 May 2024).
- California Leafy Greens Marketing Agreement California Leafy Greens Marketing Agreement|Home Page. Available online: https://lgma.ca.gov/ (accessed on 28 April 2024).
- Food Safety Program, 2024 California Leafy Greens Marketing Agreement|LGMA Connect. Available online: https://lgma.ca.gov/lgma-connect (accessed on 28 April 2024).
- Bach, S.J.; Stanford, K.; Mcallister, T.A. Survival of Escherichia Coli O157:H7 in Feces from Corn- and Barley-Fed Steers. FEMS Microbiol. Lett. 2005, 252, 25–33. [Google Scholar] [CrossRef]
- Shah, M.K.; Bradshaw, R.; Nyarko, E.; Millner, P.D.; Neher, D.; Weicht, T.; Bergholz, T.M.; Sharma, M. Survival and Growth of Wild-Type and RpoS-Deficient Salmonella Newport Strains in Soil Extracts Prepared with Heat-Treated Poultry Pellets. J. Food Prot. 2019, 82, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Locatelli, A.; Spor, A.; Jolivet, C.; Piveteau, P.; Hartmann, A. Biotic and Abiotic Soil Properties Influence Survival of Listeria Monocytogenes in Soil. PLoS ONE 2013, 8, 75969. [Google Scholar] [CrossRef]
- Du¡y, G.; Whiting, R.C.; Sheridan, J.J. The Effect of a Competitive Microflora, PH and Temperature on the Growth Kinetics of Escherichia Coli O157: H7. Food Microbiol. 1999, 16, 299–307. [Google Scholar]
- Shah, M.K.; Bradshaw, R.; Nyarko, E.; Handy, E.T.; East, C.; Millner, P.D.; Bergholz, T.M.; Sharma, M. Salmonella Enterica in Soils Amended with Heat-Treated Poultry Pellets Survived Longer than Bacteria in Unamended Soils and More Readily Transferred to and Persisted on Spinach. Appl. Environ. Microbiol. 2019, 85, 1–14. [Google Scholar] [CrossRef]
- Sharma, M.; Millner, P.D.; Hashem, F.; Vinyard, B.T.; East, C.L.; Handy, E.T.; White, K.; Stonebraker, R.; Cotton, C.P. Survival of Escherichia coli in manure-amended soils is affected by spatiotemporal, agricultural, and weather factors in the Mid-Atlantic United States. Appl. Environ. Microbiol. 2019, 85, e02392-18. [Google Scholar] [CrossRef] [PubMed]
- Neher, D.A.; Cutler, A.J.; Weicht, T.R.; Sharma, M.; Millner, P.D. Composts of poultry litter or dairy manure differentially affect survival of enteric bacteria in fields with spinach. J. Appl. Microbiol. 2019, 126, 1910–1922. [Google Scholar] [CrossRef]
- Hammami, R.; Crippen, T.L.; Strawn, L.K.; Bardsley, C.A.; Weller, D.L.; Ingram, D.T.; Chen, Y.; Oryang, D.; Rideout, S.L. Strain, Soil-Type, Irrigation Regimen, and Poultry Litter Influence Salmonella Survival and Die-off in Agricultural Soils. Front. Microbiol. 2021, 1, 590303. [Google Scholar] [CrossRef]
- Callahan, M.T.; Micallef, S.A.; Sharma, M.; Millner, P.D.; Buchanan, R.L. Metrics Proposed to Prevent the Harvest of Leafy Green Crops Exposed to Flood Water Contaminated with Escherichia Coli. Appl. Environ. Microbiol. 2016, 82, 3746–3753. [Google Scholar] [CrossRef]
- Kim, J.; Shepherd, M.W.; Jiang, X. Evaluating the Effect of Environmental Factors on Pathogen Regrowth in Compost Extract. Microb. Ecol. 2009, 58, 498–508. [Google Scholar] [CrossRef]
- Lin, Y.; Simsek, S.; Bergholz, T.M. Fate of Salmonella and Shiga-Toxin Producing Escherichia Coli on Wheat Grain during Tempering. Food Microbiol. 2023, 111, 104194. [Google Scholar] [CrossRef]
- Kroon, H. Determination of Nitrogen in Water: Comparison of a Continuous-Flow Method with on-Line UV Digestion with the Original Kjeldahl Method. Anal. Chim. Acta 1993, 276, 287–293. [Google Scholar] [CrossRef]
- Zhan, N.; Huang, Y.; Rao, Z.; Zhao, X.-L. Fast Detection of Carbonate and Bicarbonate in Groundwater and Lake Water by Coupled Ion Selective Electrode. Chin. J. Anal. Chem. 2016, 44, 355–360. [Google Scholar] [CrossRef]
- Schriefer, A.E.; Cliften, P.F.; Hibberd, M.C.; Sawyer, C.; Brown-Kennerly, V.; Burcea, L.; Klotz, E.; Crosby, S.D.; Gordon, J.I.; Head, R.D. A Multi-Amplicon 16S RRNA Sequencing and Analysis Method for Improved Taxonomic Profiling of Bacterial Communities. J. Microbiol. Methods 2018, 154, 6–13. [Google Scholar] [CrossRef]
- Bolyen, E.; Ram Rideout, J.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat Biotechnol. 2019, 37, 852–857, Erratum in Nat Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef] [PubMed]
- Package “vegan” Title Community Ecology Package Package “vegan” Title Community Ecology Package. 2024. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 8 May 2024).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury Google, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Adv. Neural Inf. Process. Syst. 2019. Available online: https://papers.nips.cc/paper_files/paper/2019/hash/bdbca288fee7f92f2bfa9f7012727740-Abstract.html (accessed on 8 May 2024).
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. TensorFlow: A System for Large-Scale Machine Learning TensorFlow: A System for Large-Scale Machine Learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI ’16), Savannah, GA, USA, 2–4 November 2016; USENIX: Berkeley, CA, USA. [Google Scholar]
- Kim, J.; Jiang, X. The Growth Potential of Escherichia Coli O157:H7, Salmonella Spp. and Listeria Monocytogenes in Dairy Manure-Based Compost in a Greenhouse Setting under Different Seasons. J. Appl. Microbiol. 2010, 109, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Ibekwe, A.M.; Crowley, D.E.; Yang, C.H. Persistence of Escherichia Coli O157:H7 in Major Leafy Green Producing Soils. Environ. Sci. Technol. 2012, 46, 12154–12161. [Google Scholar] [CrossRef] [PubMed]
- Dirk van Elsas, J.; Hill, P.; Chroň, A.; Grekova, M.; Topalova, Y.; Elhottová, D.; Krištůfek, V. Survival of Genetically Marked Escherichia Coli O157:H7 in Soil as Affected by Soil Microbial Community Shifts. ISME J. 2007, 1, 204–214. [Google Scholar] [CrossRef]
- Baker, C.A.; Lee, S.; Deid, J.; Jeong, K.C.; Schneiderid, K.R. Survival of Escherichia Coli O157 in Autoclaved and Natural Sandy Soil Mesocosms. PLoS ONE 2020, 15, e0234562. [Google Scholar] [CrossRef] [PubMed]
- Mallon, C.A.; Dirk Van Elsas, J.; Salles, J.F. Microbial Invasions: The Process, Patterns, and Mechanisms. Trends Microbiol. 2015, 23, 719–729. [Google Scholar] [CrossRef]
- Dirk Van Elsas, J.; Semenov, A.V.; Costa, R.; Trevors, J.T. MINI-REVIEW Survival of Escherichia Coli in the Environment: Fundamental and Public Health Aspects. ISME J. 2011, 5, 173–183. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, X. Isolation and Characterization of Competitive Exclusion Microorganisms from Animal Wastes–Based Composts against Listeria Monocytogenes. J. Appl. Microbiol. 2022, 132, 4531–4543. [Google Scholar] [CrossRef]
- Devarajan, N.; Mcgarvey, J.A.; Scow, K.; Jones, M.S.; Lee, S.; Samaddar, S.; Schmidt, R.; Tran, T.D.; Karp, D.S. Cascading Effects of Composts and Cover Crops on Soil Chemistry, Bacterial Communities and the Survival of Foodborne Pathogens. J. Appl. Microbiol. 2021, 131, 1564–1577. [Google Scholar] [CrossRef]
- Messi, P.; Guerrieri, E.; Bondi, M. Antibiotic Resistance and Antibacterial Activity in Heterotrophic Bacteria of Mineral Water Origin. Sci. Total. Environ. 2005, 346, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, M.B.; Abadias, M.; Anguera, M.; Vi, I. Efficacy of Pseudomonas Graminis CPA-7 against Salmonella Spp. and Listeria Monocytogenes on Fresh-Cut Pear and Setting up of the Conditions for Its Commercial Application. Food Microbiol. 2017, 70, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Bach, E.; Pereira Passaglia, M.; Jiao, J.; Gross, H. Burkholderia in the Genomic Era: From Taxonomy to the Discovery of New Antimicrobial Secondary Metabolites. Crit. Rev. Microbiol. 2021, 48, 121–160. [Google Scholar] [CrossRef]
- Alwosheel, A.; Van Cranenburgh, S.; Chorus, C.G. Is Your Dataset Big Enough? Sample Size Requirements When Using Artificial Neural Networks for Discrete Choice Analysis. J. Choice Model. 2018, 28, 167–182. [Google Scholar] [CrossRef]
- Sagheer, A.; Kotb, M. Time Series Forecasting of Petroleum Production Using Deep LSTM Recurrent Networks. Neurocomputing 2019, 323, 203–213. [Google Scholar] [CrossRef]
- Hiura, S.; Koseki, S.; Koyama, K. Prediction of Population Behavior of Listeria Monocytogenes in Food Using Machine Learning and a Microbial Growth and Survival Database. Sci. Rep. 2021, 11, 10613. [Google Scholar] [CrossRef]
- Hui, W.; Shuyi, Y.; Wei, Z.; Junbo, P.; Haiyun, T.; Chunhao, L.; Jiye, Y. Modeling the Dynamic Changes in Plasmopara Viticola Sporangia Concentration Based on LSTM and Understanding the Impact of Relative Factor Variability. Int. J. Biometeorol. 2023, 67, 993–1002. [Google Scholar] [CrossRef]
- Salinas Valley Flooding and the Testing Trap Salinas Valley Flooding and the Testing Trap|Food Safety. Available online: https://www.food-safety.com/articles/8380-salinas-valley-flooding-and-the-testing-trap (accessed on 8 May 2024).


| Strain | Species | Serogroup (Serovar) | Source |
|---|---|---|---|
| 10403S | Listeria monocytogenes | 1/2a | Skin lesion |
| H7858 | Listeria monocytogenes | 4b | Hot dog |
| MI-0041B | Escherichia coli | O157 | Human |
| DA-5 | Escherichia coli | O121 | Human |
| FSL-S10-1646 | Salmonella enterica | Enteritidis | Environmental, produce |
| Mdd314 | Salmonella enterica | Newport | Tomato |
| Soil Extract Type | Total N (ppm) | Total P (ppm) | Total C (ppm) |
|---|---|---|---|
| High-nutrient (beef barn) | 134.5 ± 25.6 | 22.4 ± 4.4 | 248.7 ± 58.8 |
| Low-nutrient (corn field) | 30.1 ± 8.5 | 2.8 ± 0.1 | 27.5 ± 2.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, D.; Kraft, A.L.; Owade, J.O.; Milicevic, M.; Yi, J.; Bergholz, T.M. Impact of Biotic and Abiotic Factors on Listeria monocytogenes, Salmonella enterica, and Enterohemorrhagic Escherichia coli in Agricultural Soil Extracts. Microorganisms 2024, 12, 1498. https://doi.org/10.3390/microorganisms12071498
Sharma D, Kraft AL, Owade JO, Milicevic M, Yi J, Bergholz TM. Impact of Biotic and Abiotic Factors on Listeria monocytogenes, Salmonella enterica, and Enterohemorrhagic Escherichia coli in Agricultural Soil Extracts. Microorganisms. 2024; 12(7):1498. https://doi.org/10.3390/microorganisms12071498
Chicago/Turabian StyleSharma, Dimple, Autumn L. Kraft, Joshua O. Owade, Mateja Milicevic, Jiyoon Yi, and Teresa M. Bergholz. 2024. "Impact of Biotic and Abiotic Factors on Listeria monocytogenes, Salmonella enterica, and Enterohemorrhagic Escherichia coli in Agricultural Soil Extracts" Microorganisms 12, no. 7: 1498. https://doi.org/10.3390/microorganisms12071498
APA StyleSharma, D., Kraft, A. L., Owade, J. O., Milicevic, M., Yi, J., & Bergholz, T. M. (2024). Impact of Biotic and Abiotic Factors on Listeria monocytogenes, Salmonella enterica, and Enterohemorrhagic Escherichia coli in Agricultural Soil Extracts. Microorganisms, 12(7), 1498. https://doi.org/10.3390/microorganisms12071498








