Relationships between Wheat Development, Soil Properties, and Rhizosphere Mycobiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Wheat Plants and Soil Samples
2.2. Investigation of Wheat Disease Incidence and Growth Characteristics
2.3. Measurement of Rhizosphere Soil pH
2.4. Measurement of Total Nitrogen, Organic Carbon, and Mineral Element Content in Rhizosphere Soil
2.5. ITS Amplicon Sequencing of Rhizosphere Soil
2.6. Sequencing Data Analysis
3. Results
3.1. Variation in Wheat Growth in Saline–Alkali Soil
3.2. Disease Index and Wheat Growth Characteristics
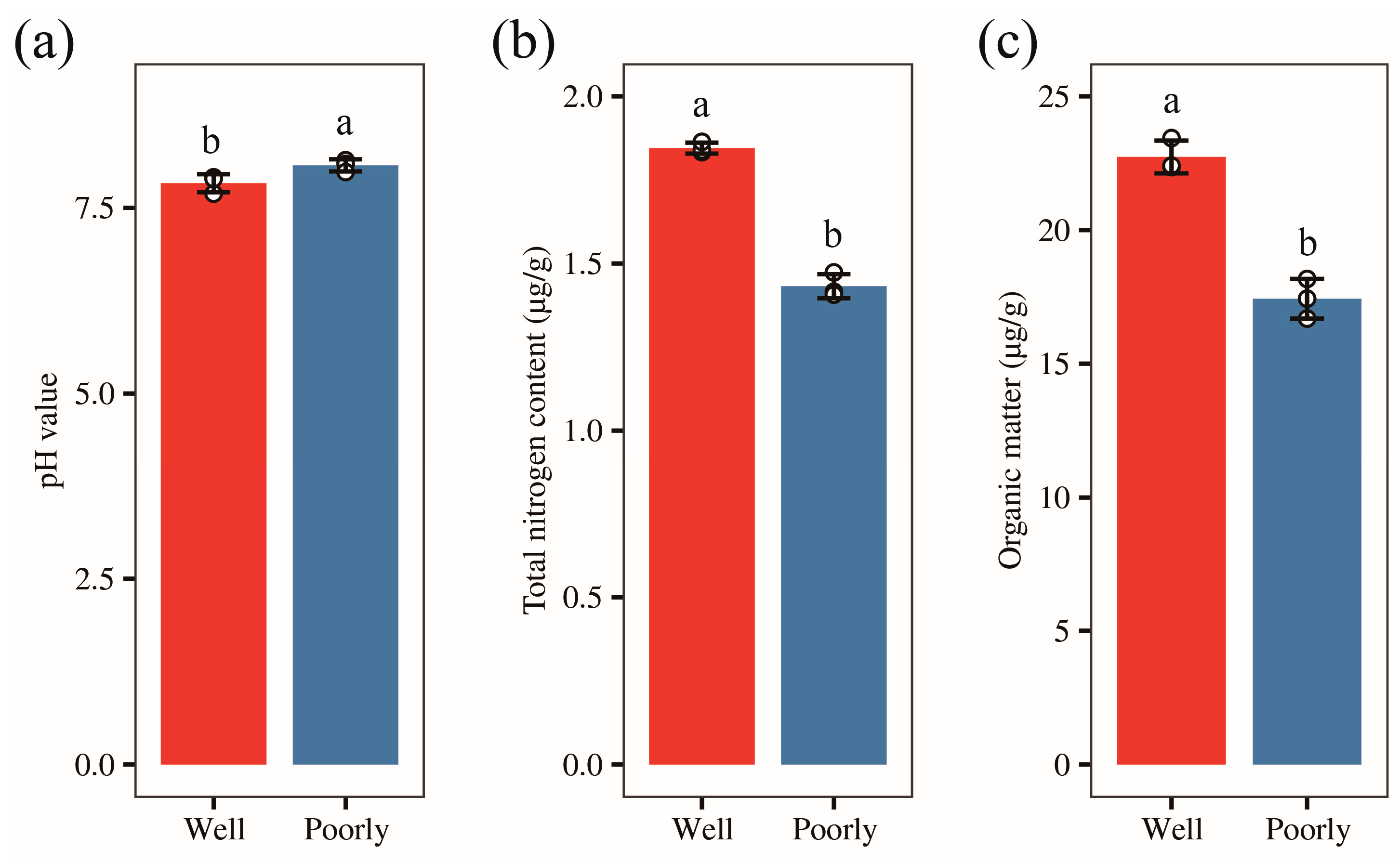
3.3. Determination of Soil pH
3.4. Analysis of Total Nitrogen and Organic Carbon Content in Wheat Rhizosphere Soil
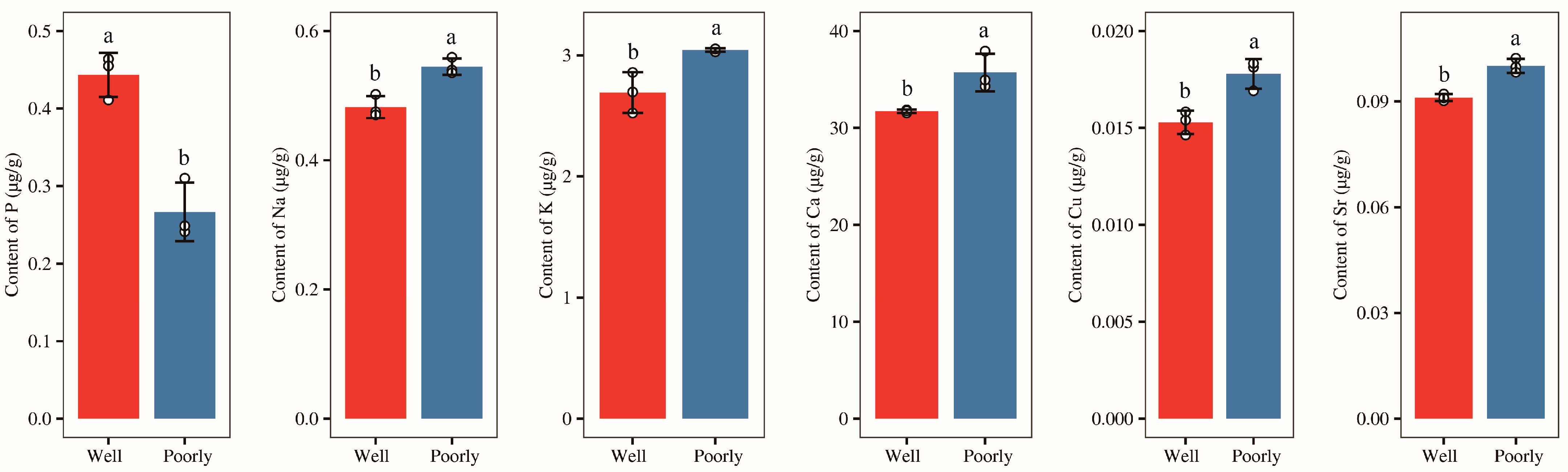
3.5. Mineral Elements Content in Wheat Rhizosphere Soil
3.6. Analysis of Wheat Rhizosphere Microbial Communities
3.6.1. Alpha Diversity Analysis of Microbial Communities
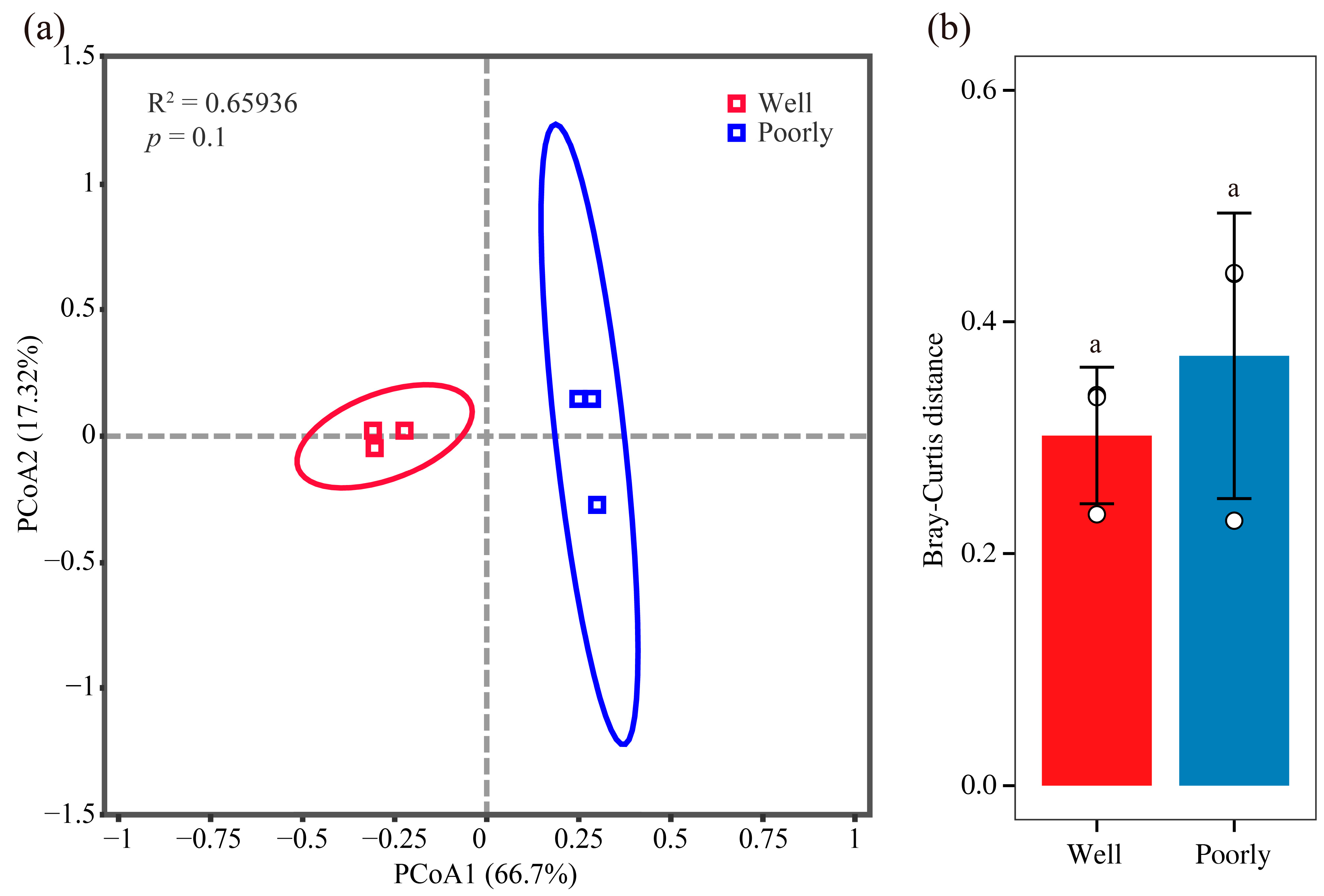
3.6.2. Beta Diversity Analysis of Microbial Communities
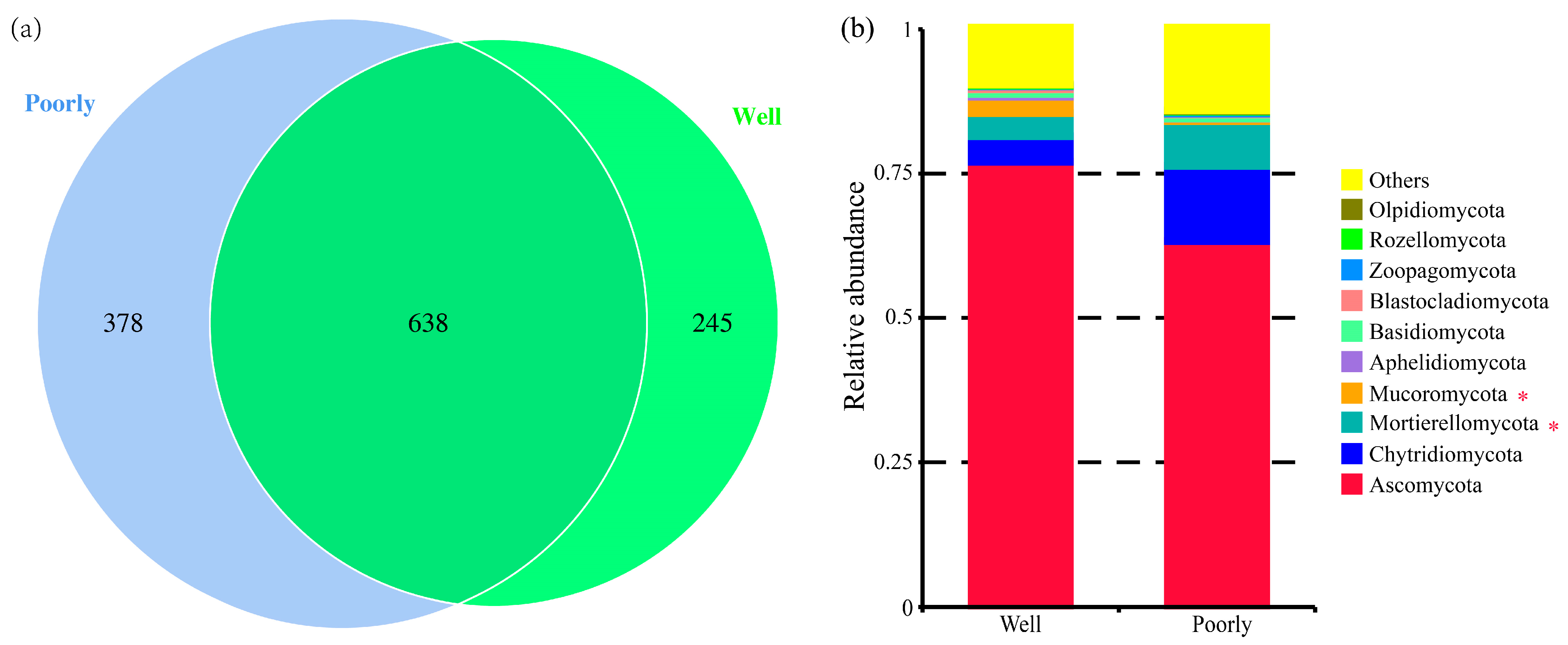
3.6.3. Microbial Community Composition
3.6.4. Differential Analysis of Fungal OTUs in Wheat Rhizosphere Soil
3.6.5. FUNGuild Analysis of Fungal Communities
3.7. Correlation between Microbial Communities and Environmental Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mottaleb, K.A.; Kruseman, G.; Frija, A.; Sonder, K.; Lopez-Ridaura, S. Projecting wheat demand in China and India for 2030 and 2050: Implications for food security. Front. Nutr. 2022, 9, 1077443. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhao, J.; Zhao, S.; Li, M.; Pang, S.; Kang, Z.; Zhen, W.; Chen, S.; Chen, F.; Wang, X. Genetics of resistance to common root rot (spot blotch), Fusarium crown rot, and sharp eyespot in wheat. Front. Genet. 2021, 12, 699342. [Google Scholar] [CrossRef] [PubMed]
- Różewicz, M.; Wyzińska, M.; Grabiński, J. The most important fungal diseases of cereals—Problems and possible solutions. Agronomy 2021, 11, 714. [Google Scholar] [CrossRef]
- Xu, X.; Nicholson, P. Community ecology of fungal pathogens causing wheat head blight. Annu. Rev. Phytopathol. 2009, 47, 83–103. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Yuan, L.; Ma, L.; Qi, K.; Zhang, Y.; Zhang, B.; Ma, G.; Qi, J. Histone H3 N-terminal lysine acetylation governs fungal growth, conidiation, and pathogenicity through regulating gene expression in Fusarium pseudograminearum. J. Fungi 2024, 10, 379. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Yang, G.; Wang, J.; Song, Y.; Liu, L.; Zhao, K.; Li, Y.; Han, Z. Spatial distribution of root and crown rot fungi associated with winter wheat in the north China plain and its relationship with climate variables. Front. Microbiol. 2018, 9, 1054. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Gardiner, D.M. Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: Recent progress and future prospects. Mol. Plant Pathol. 2018, 19, 1547–1562. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; He, X.; Wang, S.; Ma, Q.; Sun, B.; Ding, S.; Chen, L.; Zhang, M.; Li, H. Diversity of the Fusarium pathogens associated with crown rot in the Huanghuai wheat-growing region of China. Environ. Microbiol. 2019, 21, 2740–2754. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Sun, H.Y.; Shen, C.M.; Li, W.; Yu, H.S.; Chen, H.G. Survey of Fusarium spp. causing wheat crown rot in major winter wheat growing regions of China. Plant Dis. 2015, 99, 1610–1615. [Google Scholar] [CrossRef]
- Ma, G.; Wang, H.; Qi, K.; Ma, L.; Zhang, B.; Zhang, Y.; Jiang, H.; Wu, X.; Qi, J. Isolation, characterization, and pathogenicity of Fusarium species causing crown rot of wheat. Front. Microbiol. 2024, 15, 1405115. [Google Scholar] [CrossRef]
- Xu, F.; Song, Y.L.; Yang, G.Q.; Wang, J.M.; Liu, L.L.; Li, Y.H. First report of Fusarium pseudograminearum from wheat heads with Fusarium head blight in north China plain. Plant Dis. 2015, 99, 156. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ma, L.; Qi, K.; Zhang, Y.; Zhang, B.; Ma, G.; Qi, J. First report of maize seedling blight caused by Fusarium pseudograminearum in China. Plant Dis. 2022, 106, 2519. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Kong, L.; Li, Q.; Wang, L.; Chen, D.; Ma, P. First report of Fusarium pseudograminearum causing Fusarium head blight of wheat in Hebei Province, China. Plant Dis. 2015, 100, 220. [Google Scholar] [CrossRef]
- Fan, X.; Yan, Z.; Yang, M.; Waalwijk, C.; van der Lee, T.; van Diepeningen, A.; Brankovics, B.; Chen, W.; Feng, J.; Zhang, H. Contamination and translocation of deoxynivalenol and its derivatives associated with Fusarium crown rot of wheat in northern China. Plant Dis. 2021, 105, 3397–3406. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Shi, R.; Liu, L.; Li, S.; Wang, J.; Han, Z.; Liu, W.; Wang, H.; Liu, J.; Fan, J.; et al. Fusarium pseudograminearum biomass and toxin accumulation in wheat tissues with and without Fusarium crown rot symptoms. Front. Plant Sci. 2024, 15, 1356723. [Google Scholar] [CrossRef] [PubMed]
- Mawcha, K.T.; Zhang, N.; Zhang, F.; Zhang, L.; Hailu, M.; Tian, J.; Yang, W. Resistance evaluation of 43 Ethiopian wheat cultivars to crown rot caused by Fusarium pseudograminearum. J. Plant Dis. Prot. 2022, 129, 1105–1114. [Google Scholar] [CrossRef]
- Zhang, N.; Xu, Y.; Zhang, Q.; Zhao, L.; Zhu, Y.; Wu, Y.; Li, Z.; Yang, W. Detection of fungicide resistance to fludioxonil and tebuconazole in Fusarium pseudograminearum, the causal agent of Fusarium crown rot in wheat. PeerJ 2023, 11, e14705. [Google Scholar] [CrossRef]
- Yu, F.; Chen, Y.; Huang, X.; Shi, J.; Xu, J.; He, Y. Does straw returning affect the root rot disease of crops in soil? A systematic review and meta-analysis. J. Environ. Manag. 2023, 336, 117673. [Google Scholar] [CrossRef]
- Liu, L.; Wang, B. Protection of halophytes and their uses for cultivation of saline-alkali soil in china. Biology 2021, 10, 353. [Google Scholar] [CrossRef]
- Du, Y.; Liu, X.; Zhang, L.; Zhou, W. Drip irrigation in agricultural saline-alkali land controls soil salinity and improves crop yield: Evidence from a global meta-analysis. Sci. Total Environ. 2023, 880, 163226. [Google Scholar] [CrossRef]
- Rahman, M.M.; Mostofa, M.G.; Keya, S.S.; Siddiqui, M.N.; Ansary, M.M.U.; Das, A.K.; Rahman, M.A.; Tran, L.S. Adaptive mechanisms of halophytes and their potential in improving salinity tolerance in plants. Int. J. Mol. Sci. 2021, 22, 10733. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Yan, R.; Wang, Q.; Zhou, B.; Zhu, H.; Liu, G.; Guo, X.; Zhang, Z. Integrated microbiological and metabolomics analyses to understand the mechanism that allows modified biochar to affect the alkalinity of saline soil and winter wheat growth. Sci. Total Environ. 2023, 866, 161330. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ma, H.; Feng, Z.; Yan, Z.; Song, B.; Wang, J.; Zheng, Y.; Hao, W.; Zhang, W.; Yao, M.; et al. Integrated organic and inorganic fertilization and reduced irrigation altered prokaryotic microbial community and diversity in different compartments of wheat root zone contributing to improved nitrogen uptake and wheat yield. Sci. Total Environ. 2022, 842, 156952. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.; Haddix, M.L.; Byrne, P.F.; Cotrufo, M.F.; Schipanski, M.; Kallenbach, C.M.; Wallenstein, M.D.; Fonte, S.J. Divergent belowground carbon allocation patterns of winter wheat shape rhizosphere microbial communities and nitrogen cycling activities. Soil. Biol. Biochem. 2022, 165, 108518. [Google Scholar] [CrossRef]
- Effah, Z.; Li, L.; Xie, J.; Liu, C.; Xu, A.; Karikari, B.; Anwar, S.; Zeng, M. Regulation of nitrogen metabolism, photosynthetic activity, and yield attributes of spring wheat by nitrogen fertilizer in the semi-arid loess plateau region. J. Plant Growth Regul. 2023, 42, 1120–1133. [Google Scholar] [CrossRef]
- Bhattacharyya, S.S.; Ros, G.H.; Furtak, K.; Iqbal, H.M.N.; Parra-Saldívar, R. Soil carbon sequestration—An interplay between soil microbial community and soil organic matter dynamics. Sci. Total Environ. 2022, 815, 152928. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Sharifi, R.; Khan, M.S.S.; Islam, F.; Bhat, J.A.; Kui, L.; Majeed, A. Wheat microbiome: Structure, dynamics, and role in improving performance under stress environments. Front. Microbiol. 2021, 12, 821546. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wang, W.; Wei, J.; Li, H.; Li, X.; Liu, G.; Chen, Y.; Ye, S. Evolution and prediction of drought-flood abrupt alternation events in Huang-Huai-Hai River Basin, China. Sci. Total Environ. 2023, 869, 161707. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kunz, I.G.Z.; Reed, K.J.; Metcalf, J.L.; Hassel, D.M.; Coleman, R.J.; Hess, T.M.; Coleman, S.J. Equine fecal microbiota changes associated with anthelmintic administration. J. Equine Vet. Sci. 2019, 77, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Fricke, W. Energy costs of salinity tolerance in crop plants: Night-time transpiration and growth. New Phytol. 2020, 225, 1152–1165. [Google Scholar] [CrossRef]
- Rengel, Z. Availability of Mn, Zn and Fe in the rhizosphere. J. Soil. Sci. Plant Nutr. 2014, 15, 397–409. [Google Scholar] [CrossRef]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Shams, M.; Khadivi, A. Mechanisms of salinity tolerance and their possible application in the breeding of vegetables. BMC Plant Biol. 2023, 23, 139. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, R.P.; Taylor, N.L.; Millar, A.H. The role of mitochondrial respiration in salinity tolerance. Trends Plant Sci. 2011, 16, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Mansour, E.; Moustafa, E.S.A.; Desoky, E.M.; Ali, M.M.A.; Yasin, M.A.T.; Attia, A.; Alsuhaibani, N.; Tahir, M.U.; El-Hendawy, S. Multidimensional evaluation for detecting salt tolerance of bread wheat genotypes under actual saline field growing conditions. Plants 2020, 9, 1324. [Google Scholar] [CrossRef] [PubMed]
- Saddiq, M.S.; Iqbal, S.; Hafeez, M.B.; Ibrahim, A.M.H.; Raza, A.; Fatima, E.M.; Baloch, H.; Jahanzaib; Woodrow, P.; Ciarmiello, L.F. Effect of salinity stress on physiological changes in winter and spring wheat. Agronomy 2021, 11, 1193. [Google Scholar] [CrossRef]
- Deliopoulos, T.; Kettlewell, P.S.; Hare, M.C. Fungal disease suppression by inorganic salts: A review. Crop Protect. 2010, 29, 1059–1075. [Google Scholar] [CrossRef]
- Berlanas, C.; Ojeda, S.; López-Manzanares, B.; Andrés-Sodupe, M.; Bujanda, R.; Del Pilar Martínez-Diz, M.; Díaz-Losada, E.; Gramaje, D. Occurrence and diversity of black-foot disease fungi in symptomless grapevine nursery stock in Spain. Plant Dis. 2020, 104, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, M.; Chen, H.; Chen, Y.; Wang, L.; Wang, R. Organic Amendments promote saline-alkali soil desalinization and enhance maize growth. Front. Plant Sci. 2023, 14, 1177209. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Yan, L.; Tong, K.; Yu, H.; Lu, M.; Wang, L.; Niu, Y. The potential and prospects of modified biochar for comprehensive management of salt-affected soils and plants: A critical review. Sci. Total Environ. 2024, 912, 169618. [Google Scholar] [CrossRef]
- Ding, J.; Li, B.; Sun, M.; Li, X. Different cropping patterns to restore saline-alkali soils in northeast China affect the abundance of functional genes in the soil nitrogen cycle. Sustainability 2023, 15, 6592. [Google Scholar] [CrossRef]
- Irshad, A.; Ahmed, R.I.; Ur Rehman, S.; Sun, G.; Ahmad, F.; Sher, M.A.; Aslam, M.Z.; Hassan, M.M.; Qari, S.H.; Aziz, M.K.; et al. Characterization of salt tolerant wheat genotypes by using morpho-physiological, biochemical, and molecular analysis. Front. Plant Sci. 2022, 13, 956298. [Google Scholar] [CrossRef] [PubMed]
- Lethin, J.; Shakil, S.S.M.; Hassan, S.; Sirijovski, N.; Töpel, M.; Olsson, O.; Aronsson, H. Development and characterization of an EMS-mutagenized wheat population and identification of salt-tolerant wheat lines. BMC Plant Biol. 2020, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, J.; Carvalhais, L.C.; Percy, C.D.; Prakash Verma, J.; Schenk, P.M.; Singh, B.K. Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogens. New Phytol. 2021, 229, 2873–2885. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Babalola, O.O. The rhizosphere microbial complex in plant health: A review of interaction dynamics. J. Integr. Agric. 2022, 21, 2168–2182. [Google Scholar] [CrossRef]

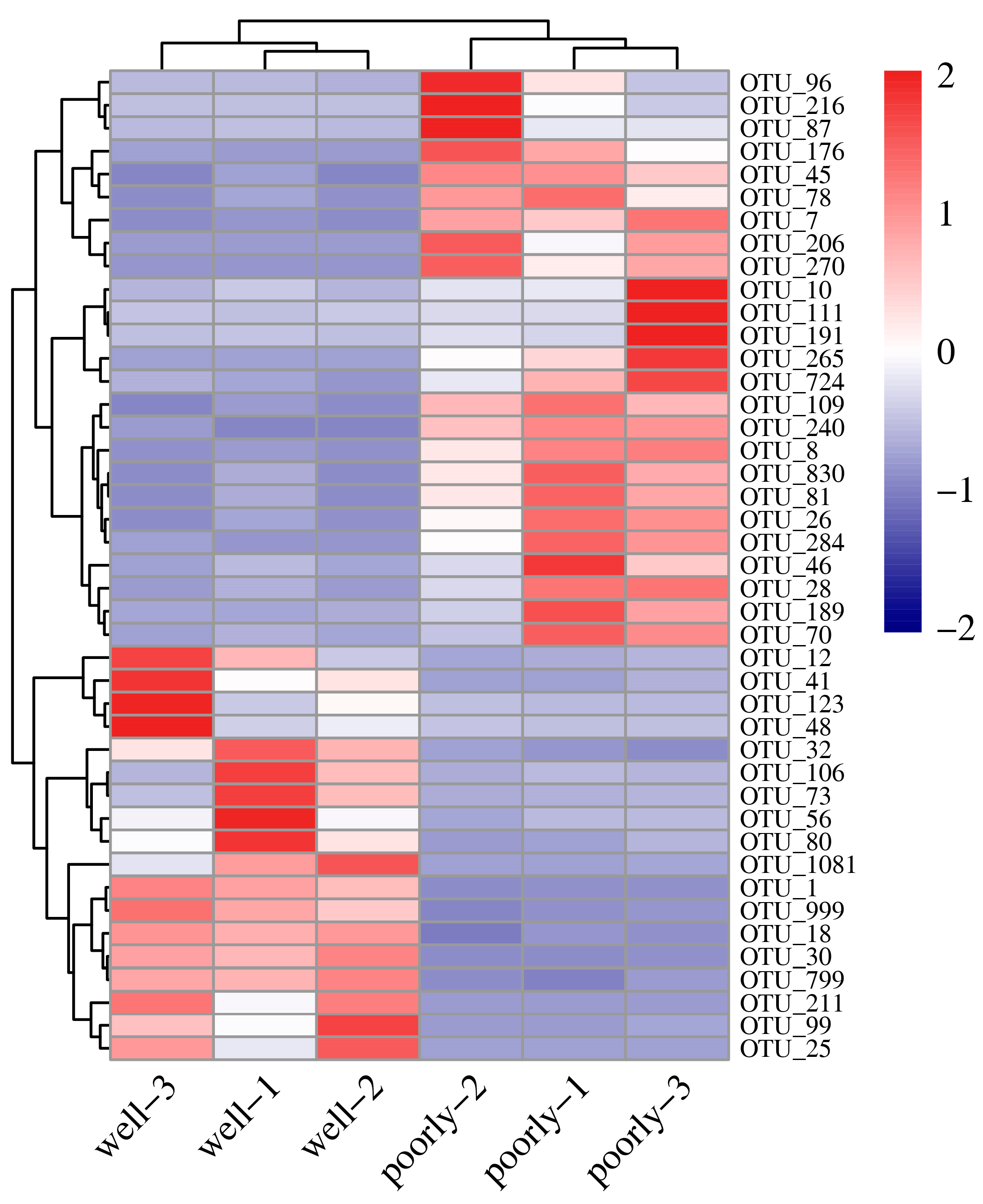

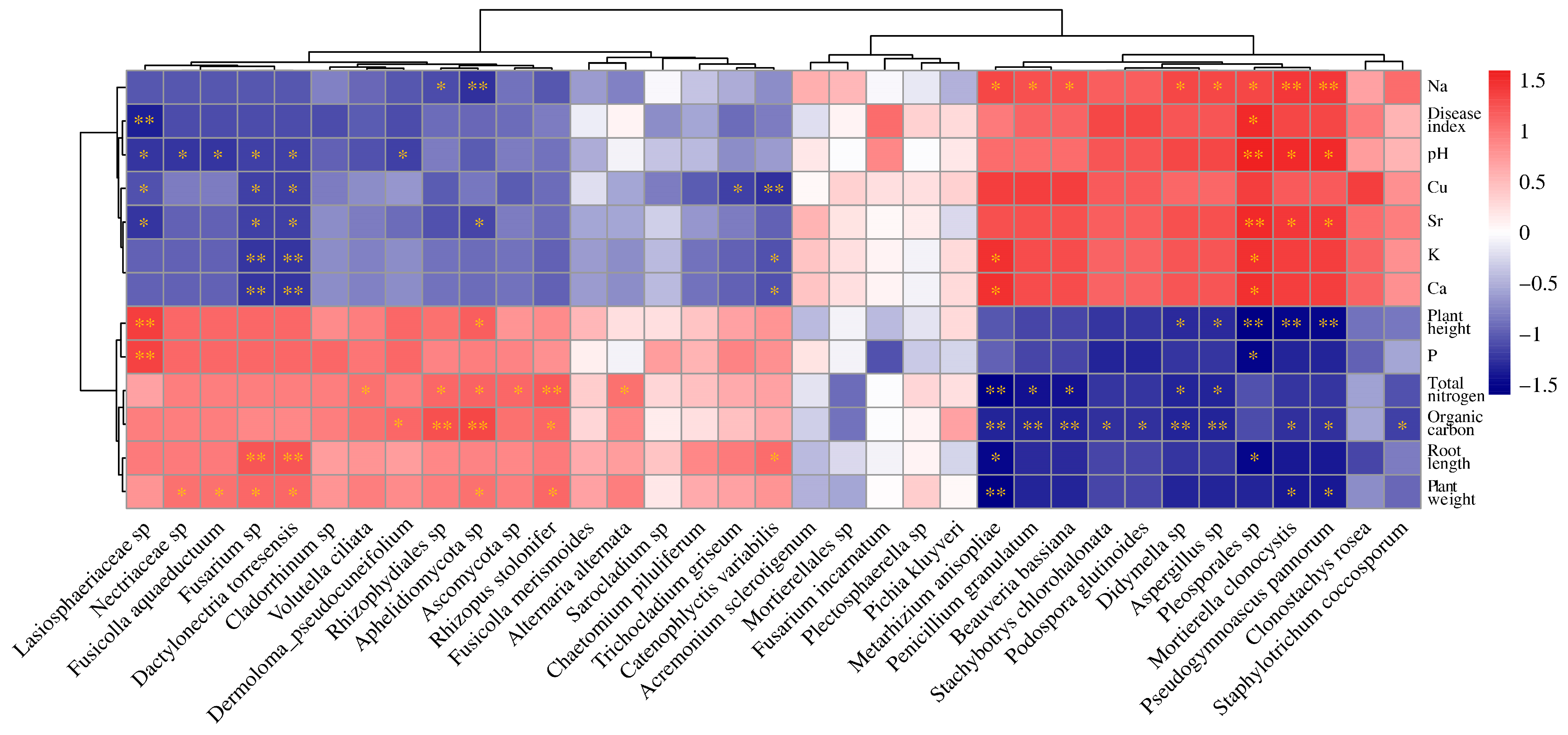
| Sample Name | OTU Richness | Shannon | Simpson | Phylogenetic Diversity |
|---|---|---|---|---|
| Well-growing wheat | 518.67 ± 34.69 a # | 4.82 ± 0.16 a | 0.89 ± 0.01 a | 173.16 ± 10.42 a |
| Poorly growing wheat | 606.67 ± 56.67 a | 5.30 ± 0.18 a | 0.93 ± 0.02 a | 211.67 ± 22.45 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, H.; Ma, L.; Gao, P.; Zhang, Y.; Zhang, B.; Ma, G.; Qi, K.; Qi, J. Relationships between Wheat Development, Soil Properties, and Rhizosphere Mycobiota. Microorganisms 2024, 12, 1516. https://doi.org/10.3390/microorganisms12081516
Jiang H, Ma L, Gao P, Zhang Y, Zhang B, Ma G, Qi K, Qi J. Relationships between Wheat Development, Soil Properties, and Rhizosphere Mycobiota. Microorganisms. 2024; 12(8):1516. https://doi.org/10.3390/microorganisms12081516
Chicago/Turabian StyleJiang, Hang, Liguo Ma, Peixin Gao, Yueli Zhang, Bo Zhang, Guoping Ma, Kai Qi, and Junshan Qi. 2024. "Relationships between Wheat Development, Soil Properties, and Rhizosphere Mycobiota" Microorganisms 12, no. 8: 1516. https://doi.org/10.3390/microorganisms12081516





