Microbial Diversity of Soil in a Mediterranean Biodiversity Hotspot: Parque Nacional La Campana, Chile
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
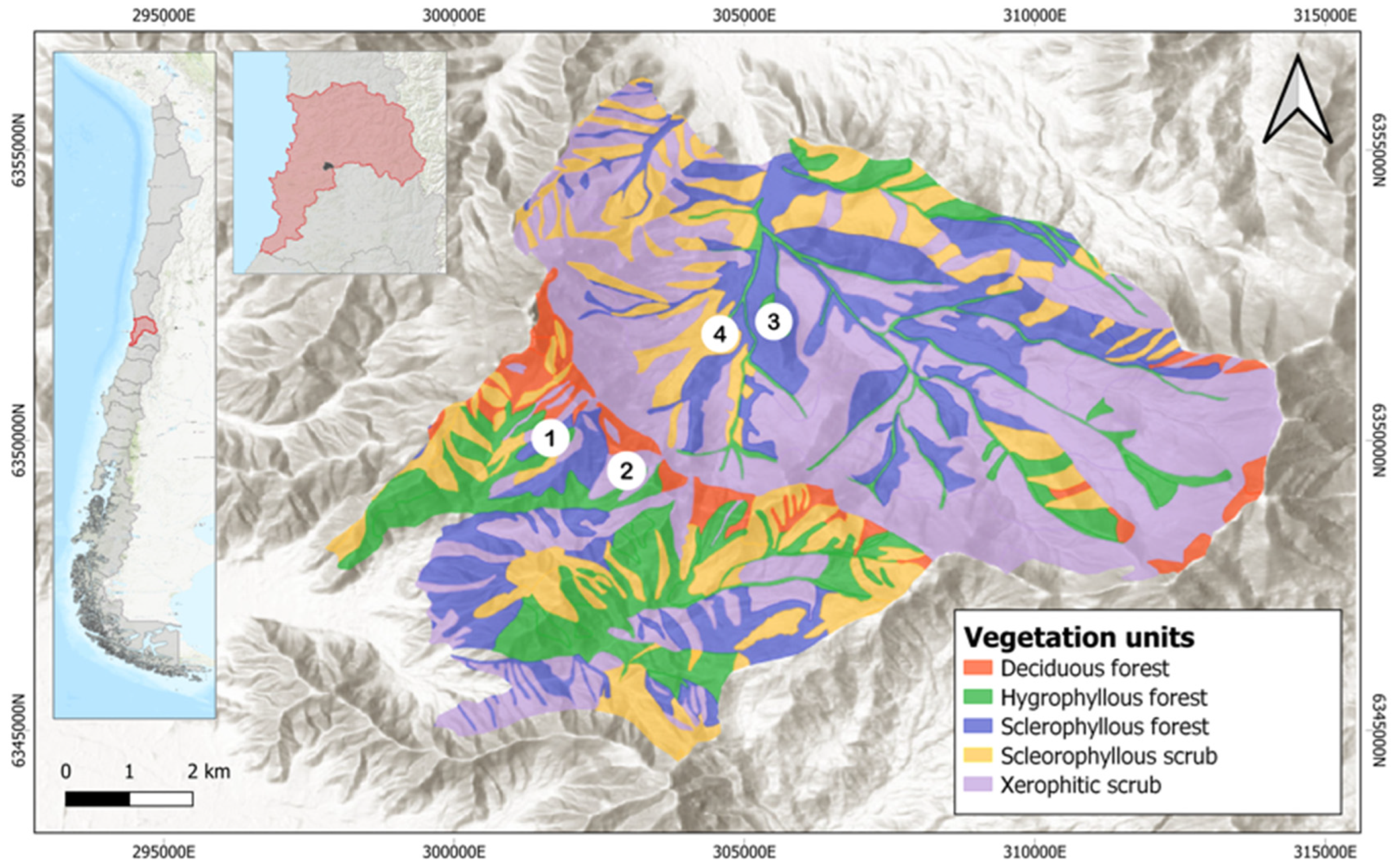
2.2. Study Sites and Sample Collection
2.3. Climate Variables in PNLC
2.4. Soil and Organic Matter Physicochemical Analysis
2.5. DNA Extraction and Sequencing of Prokaryote and Fungal Phylogenetic Markers
2.6. Analysis of Microbial Composition and Other Statistic Data
3. Results
3.1. Environmental and Soil Physicochemical Conditions
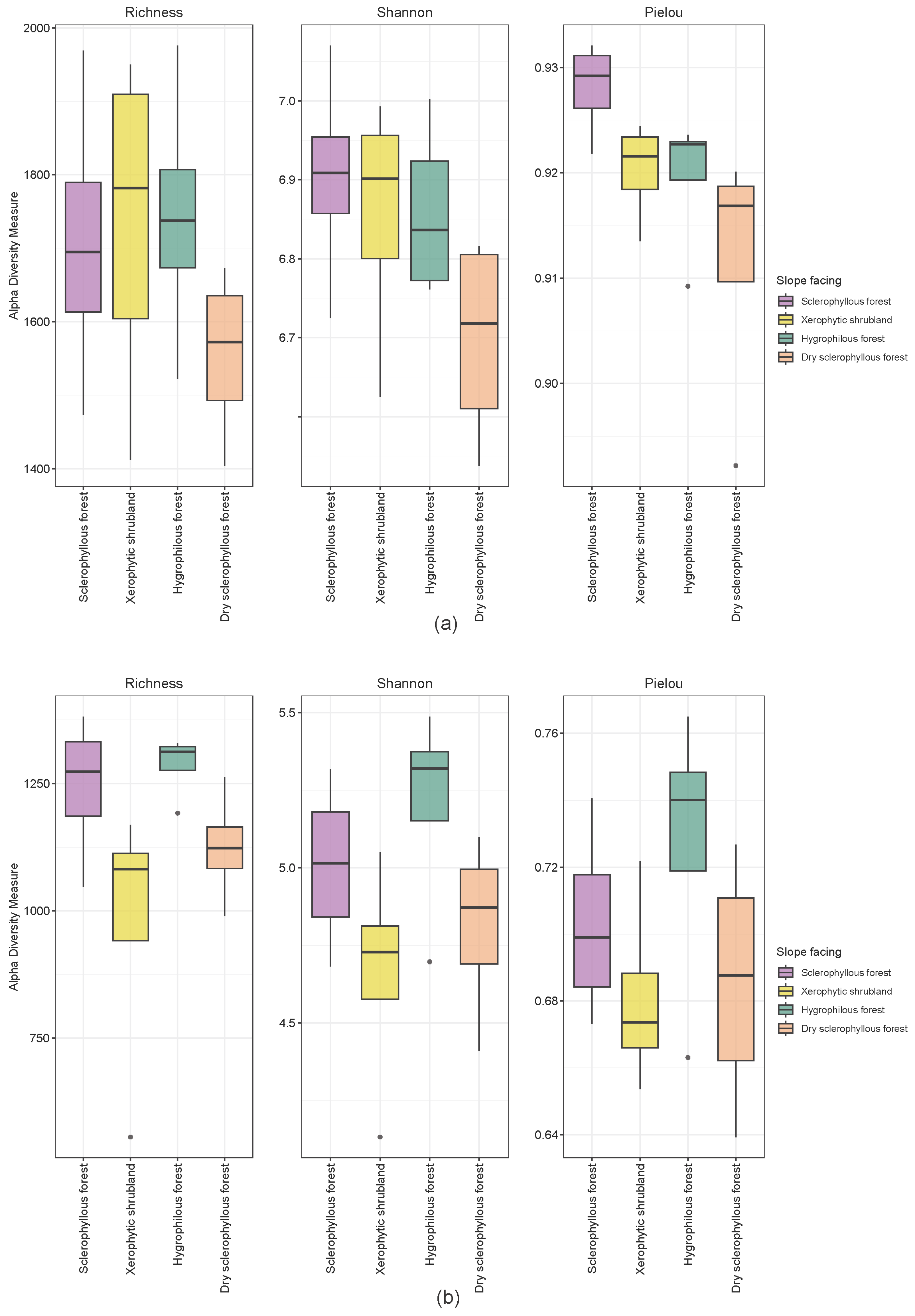
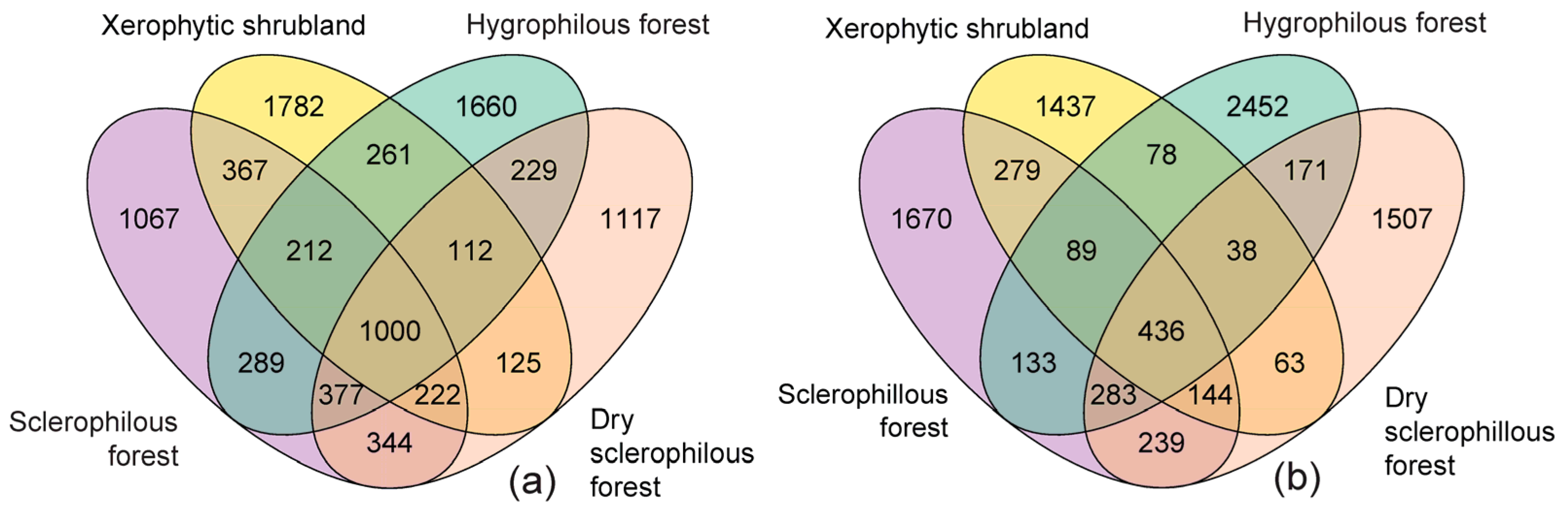
3.2. Sequencing and Structure of the Soil Microbial Community
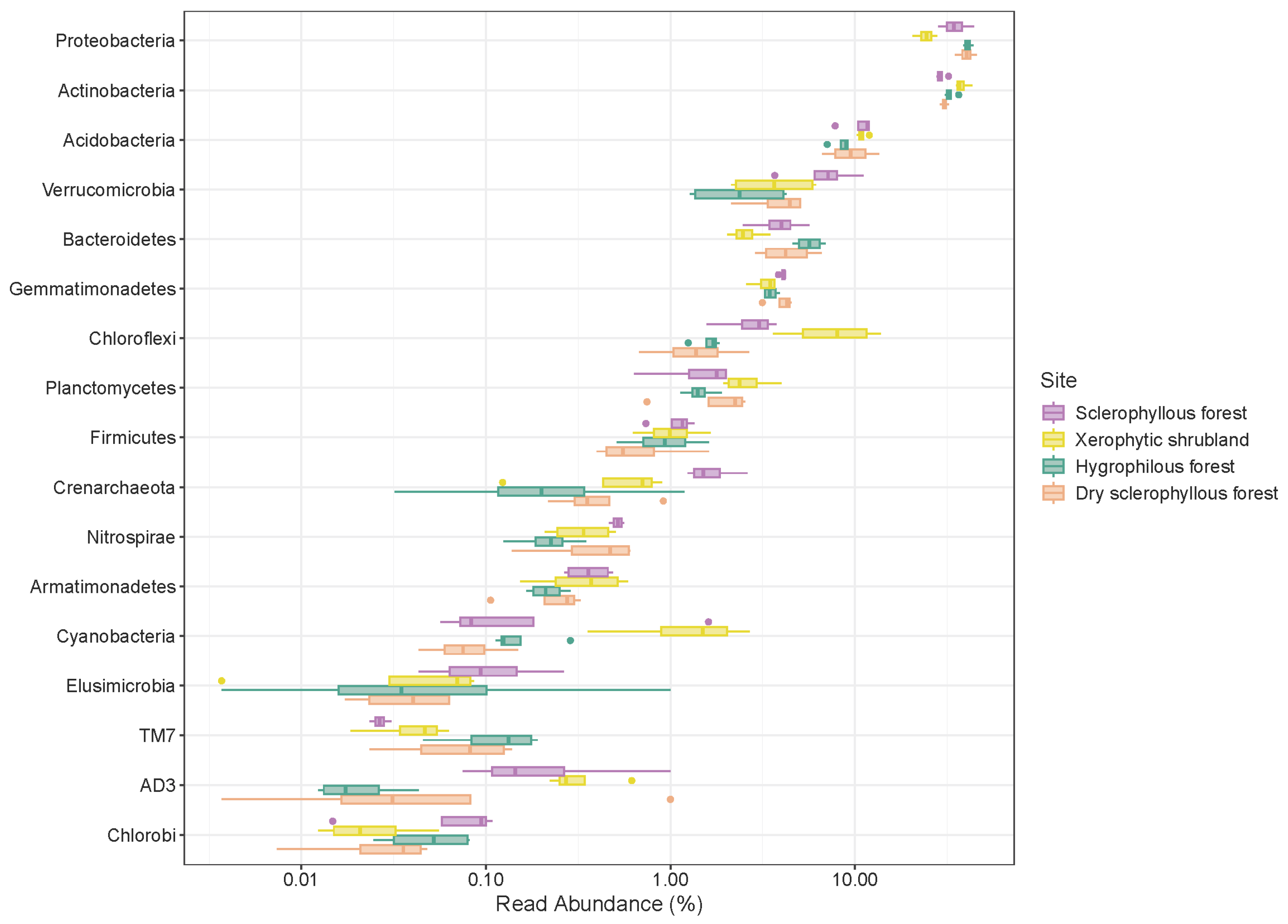
3.3. Microbial Composition in Different Types of Environments
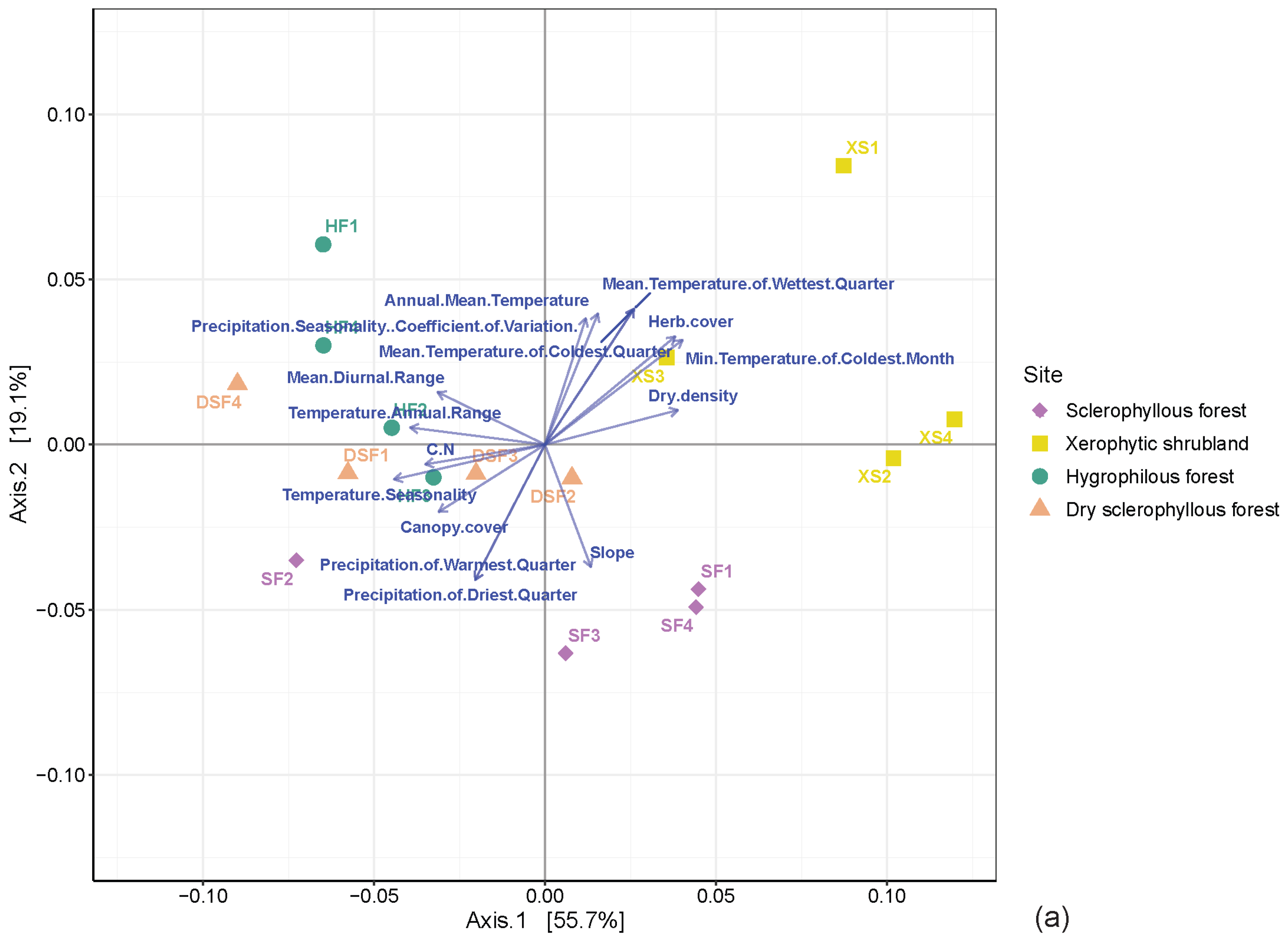
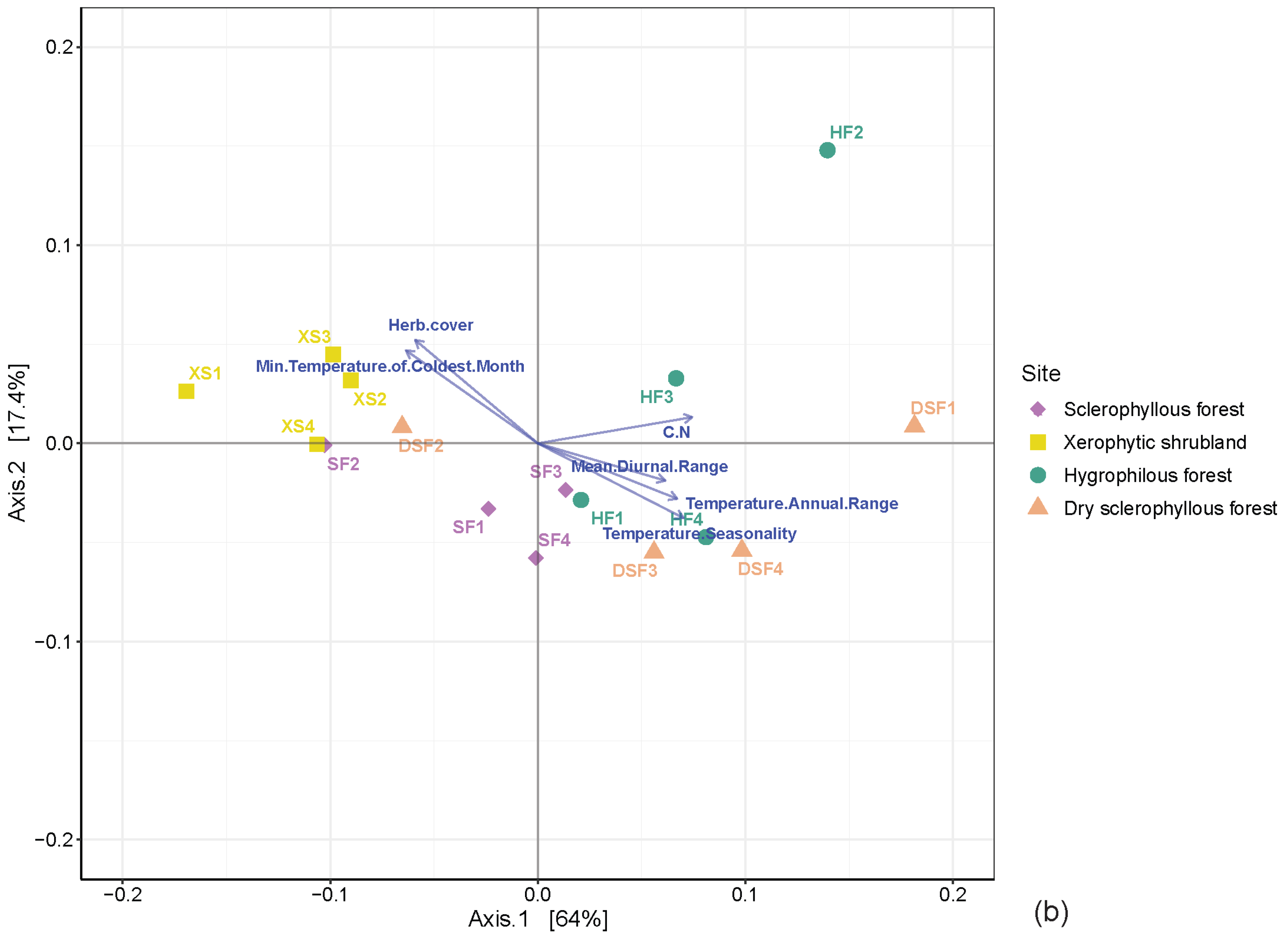
3.4. Distribution of Microbial Communities among the Different Plant Communities
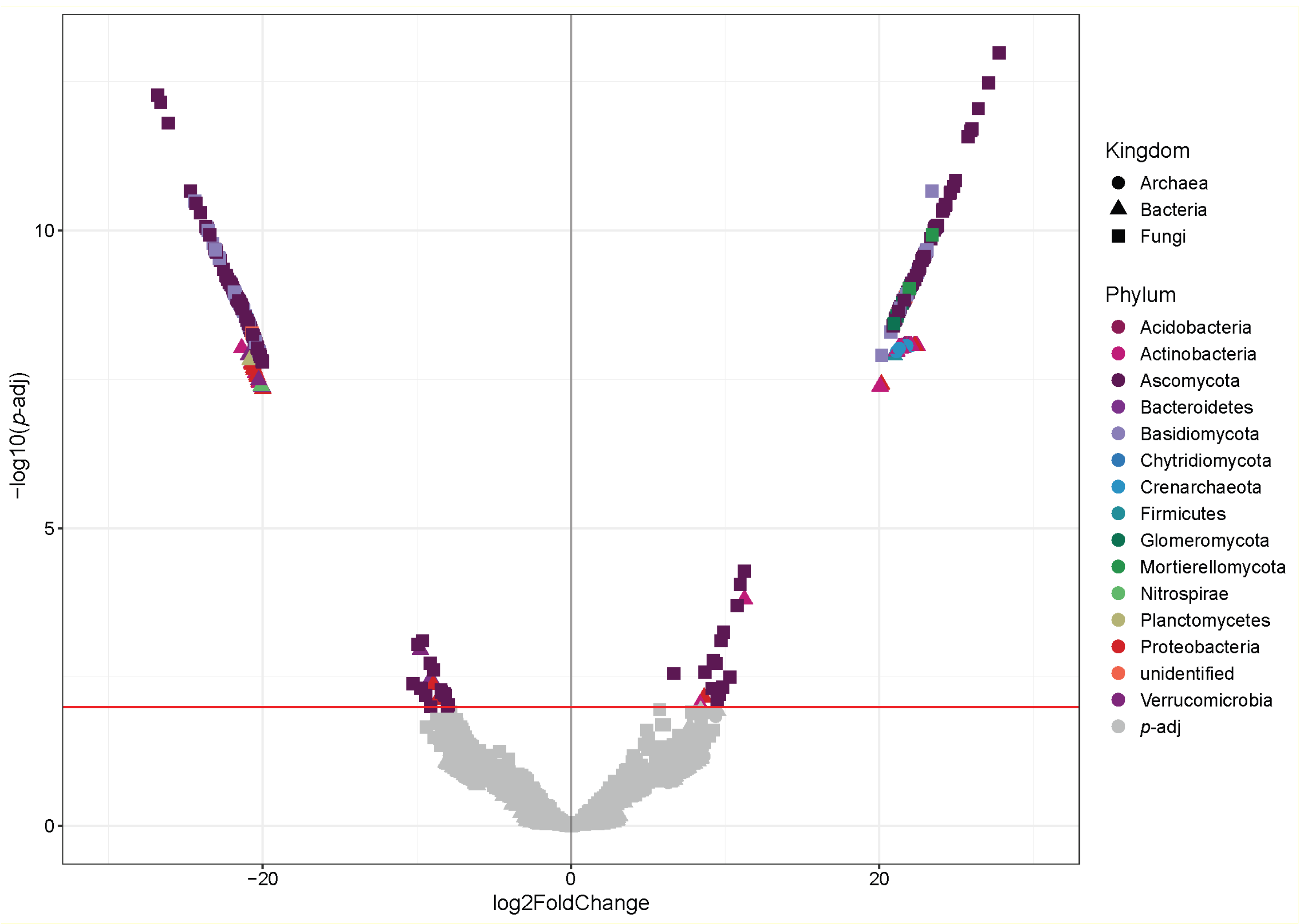
3.5. Analysis of Differential Abundance of Microorganisms
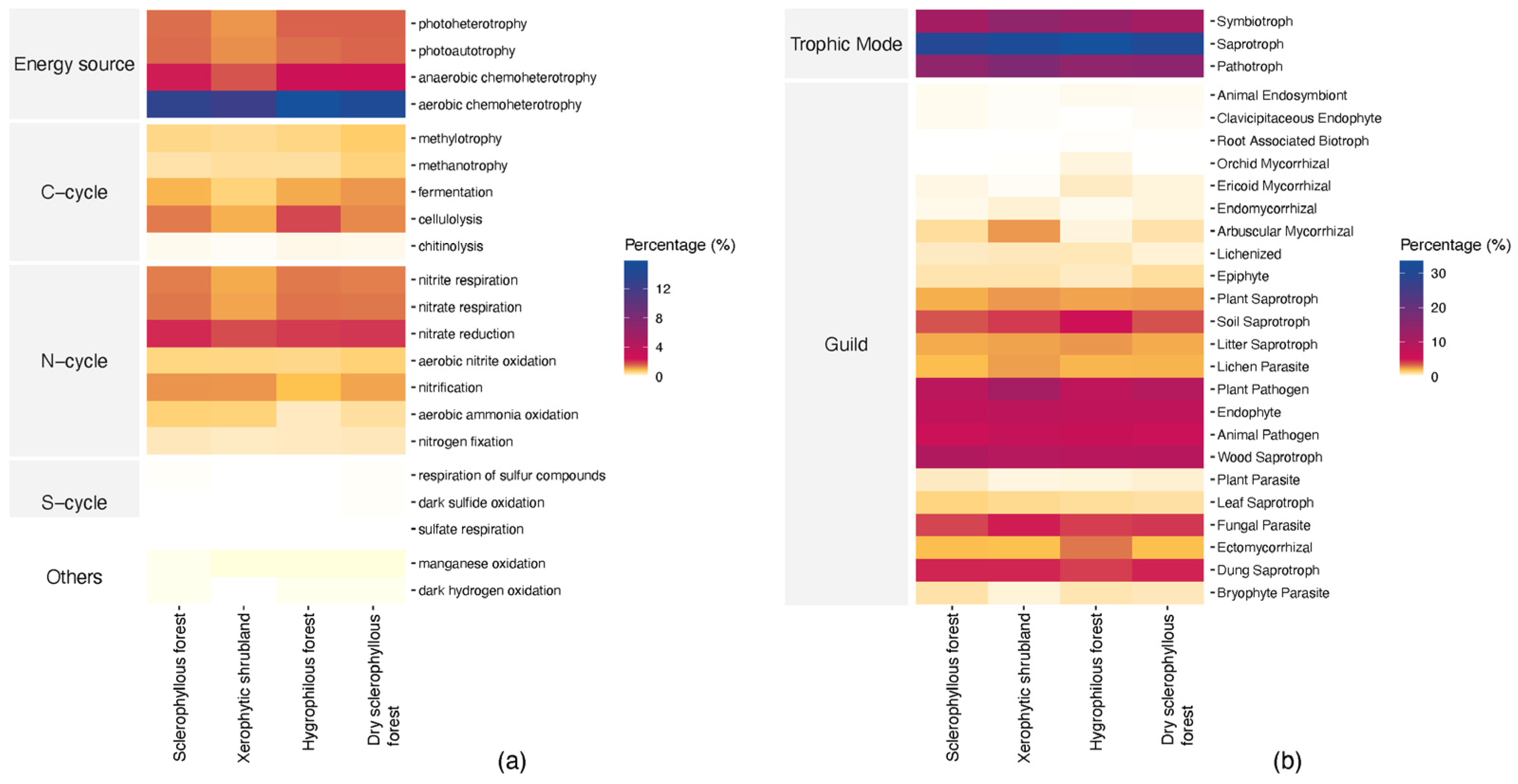
3.6. Predicted Potential Microbial Function
4. Discussion
4.1. Toposequence, Microbial and Plant Communities in PNLC
4.2. Variation in the Microbial Community and Its Association with the Environmental Conditions of the Plant Communities
4.3. Key Microbial Taxa, Potential Functions and Their Relation with the Different Plant Communities of PNLC
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The Importance of the Microbiome of the Plant Holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Bardelli, T.; Gómez-Brandón, M.; Ascher-Jenull, J.; Fornasier, F.; Arfaioli, P.; Francioli, D.; Egli, M.; Sartori, G.; Insam, H.; Pietramellara, G. Effects of Slope Exposure on Soil Physico-Chemical and Microbiological Properties along an Altitudinal Climosequence in the Italian Alps. Sci. Total Environ. 2017, 575, 1041–1055. [Google Scholar] [CrossRef]
- Torsvik, V.; Øvreås, L. Microbial Diversity and Function in Soil: From Genes to Ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going Back to the Roots: The Microbial Ecology of the Rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the Unknown: Disentangling the Complexities of the Soil Microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Cederlund, H.; Wessén, E.; Enwall, K.; Jones, C.M.; Juhanson, J.; Pell, M.; Philippot, L.; Hallin, S. Soil Carbon Quality and Nitrogen Fertilization Structure Bacterial Communities with Predictable Responses of Major Bacterial Phyla. Appl. Soil Ecol. 2014, 84, 62–68. [Google Scholar] [CrossRef]
- Sul, W.J.; Asuming-Brempong, S.; Wang, Q.; Tourlousse, D.M.; Penton, C.R.; Deng, Y.; Rodrigues, J.L.M.; Adiku, S.G.K.; Jones, J.W.; Zhou, J.; et al. Tropical Agricultural Land Management Influences on Soil Microbial Communities through Its Effect on Soil Organic Carbon. Soil Biol. Biochem. 2013, 65, 33–38. [Google Scholar] [CrossRef]
- Oliverio, A.M.; Bradford, M.A.; Fierer, N. Identifying the Microbial Taxa That Consistently Respond to Soil Warming across Time and Space. Glob. Change Biol. 2017, 23, 2117–2129. [Google Scholar] [CrossRef]
- Pett-Ridge, J.; Firestone, M.K. Redox Fluctuation Structures Microbial Communities in a Wet Tropical Soil. Appl. Environ. Microbiol. 2005, 71, 6998–7007. [Google Scholar] [CrossRef]
- Erktan, A.; Or, D.; Scheu, S. The Physical Structure of Soil: Determinant and Consequence of Trophic Interactions. Soil Biol. Biochem. 2020, 148, 107876. [Google Scholar] [CrossRef]
- Parkin, T.B. Soil Microsites as a Source of Denitrification Variability. Soil Sci. Soc. Am. J. 1987, 51, 1194–1199. [Google Scholar] [CrossRef]
- Petersen, S.O.; Nielsen, T.H.; Frostegård, Å.; Olesen, T. O2 Uptake, C Metabolism and Denitrification Associated with Manure Hot-Spots. Soil Biol. Biochem. 1996, 28, 341–349. [Google Scholar] [CrossRef]
- Lynch, J.M.; Whipps, J.M. Substrate Flow in the Rhizosphere. Plant Soil 1990, 129, 1–10. [Google Scholar] [CrossRef]
- Aguilera, P.; Cornejo, P.; Borie, F.; Barea, J.M.; von Baer, E.; Oehl, F. Diversity of Arbuscular Mycorrhizal Fungi Associated with Triticum aestivum L. Plants Growing in an Andosol with High Aluminum Level. Agric. Ecosyst. Environ. 2014, 186, 178–184. [Google Scholar] [CrossRef]
- Benedetti, S.; Balocchi, F.; Hormazábal, M. Hongos Micorrícicos Arbusculares (HMA) Asociados a Poblaciones Naturales de Peumus Boldus En Chile Central Arbuscular Mycorrhizal Fungi (AMF) Linked to Peumus Boldus Natural Formation in Central. Gayana Bot. 2018, 75, 431–437. [Google Scholar] [CrossRef]
- Castillo, C.G.; Borie, F.; Oehl, F.; Sieverding, E. Arbuscular Mycorrhizal Fungi Biodiversity: Prospecting in Southern-Central Zone of Chile. A Review. J. Soil Sci. Plant Nutr. 2016, 16, 400–422. [Google Scholar] [CrossRef]
- Bull, A.T.; Asenjo, J.A. Microbiology of Hyper-Arid Environments: Recent Insights from the Atacama Desert, Chile. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2013, 103, 1173–1179. [Google Scholar] [CrossRef]
- Okoro, C.K.; Brown, R.; Jones, A.L.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M.; Bull, A.T. Diversity of Culturable Actinomycetes in Hyper-Arid Soils of the Atacama Desert, Chile. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2009, 95, 121–133. [Google Scholar] [CrossRef]
- Schulze-Makuch, D.; Wagner, D.; Kounaves, S.P.; Mangelsdorf, K.; Devine, K.G.; de Vera, J.-P.; Schmitt-Kopplin, P.; Grossart, H.-P.; Parro, V.; Kaupenjohann, M.; et al. Transitory Microbial Habitat in the Hyperarid Atacama Desert. Proc. Natl. Acad. Sci. USA 2018, 115, 2670–2675. [Google Scholar] [CrossRef]
- Zhang, Q.; Acuña, J.J.; Inostroza, N.G.; Mora, M.L.; Radic, S.; Sadowsky, M.J.; Jorquera, M.A. Endophytic Bacterial Communities Associated with Roots and Leaves of Plants Growing in Chilean Extreme Environments. Sci. Rep. 2019, 9, 4950. [Google Scholar] [CrossRef]
- Oeser, R.A.; Stroncik, N.; Moskwa, L.M.; Bernhard, N.; Schaller, M.; Canessa, R.; van den Brink, L.; Köster, M.; Brucker, E.; Stock, S.; et al. Chemistry and Microbiology of the Critical Zone along a Steep Climate and Vegetation Gradient in the Chilean Coastal Cordillera. Catena 2018, 170, 183–203. [Google Scholar] [CrossRef]
- Bernhard, N.; Moskwa, L.M.; Schmidt, K.; Oeser, R.A.; Aburto, F.; Bader, M.Y.; Baumann, K.; von Blanckenburg, F.; Boy, J.; van den Brink, L.; et al. Pedogenic and Microbial Interrelations to Regional Climate and Local Topography: New Insights from a Climate Gradient (Arid to Humid) along the Coastal Cordillera of Chile. Catena 2018, 170, 335–355. [Google Scholar] [CrossRef]
- Rodriguez, V.; Moskwa, L.M.; Oses, R.; Kühn, P.; Riveras-Muñoz, N.; Seguel, O.; Scholten, T.; Wagner, D. Impact of Climate and Slope Aspects on the Composition of Soil Bacterial Communities Involved in Pedogenetic Processes along the Chilean Coastal Cordillera. Microorganisms 2022, 10, 847. [Google Scholar] [CrossRef] [PubMed]
- Vanderplank, S.E.; Moreira-Muñoz, A.A.; Hobohm, C.; Pils, G.; Noroozi, J.; Clark, V.R. Endemism in Vascular Plants; Hobohm, C., Ed.; Springer: Dordrecht, The Netherlands, 2014; Volume 9, ISBN 978-94-007-6912-0. [Google Scholar]
- Mittermeier, R.A.; Turner, W.R.; Larsen, F.W.; Brooks, T.M.; Gascon, C. Global biodiversity conservation: The critical role of hotspots. In Biodiversity Hotspots; Springer: Berlin/Heidelberg, Germany, 2011; pp. 3–22. [Google Scholar]
- Manríquez, H.; Mansilla, P.; Figueroa-Sterquel, R.; Moreira-Muñoz, A. Geodiversity Meets Biodiversity: A Landscape Approach for Biogeocultural Conservation and Governance in Mediterranean Central Chile. J. Prot. Mt. Areas Res. 2019, 11, 43–48. [Google Scholar] [CrossRef]
- Leguia-Cruz, M.; Tejer-Nos, C.; Cubillos, N.O.; Mansilla-Quiñones, P.; Moreira-Muñoz, A. Biocultural Resistance and Re-Existence through a Dialogue of Knowledges and Citizen Art in a Threatened Biosphere Reserve. J. Prot. Mt. Areas Res. 2021, 13, 102–107. [Google Scholar] [CrossRef]
- Moreira-Muñoz, A.; del Río, C.; Leguia-Cruz, M.; Mansilla-Quiñones, P. Spatial Dynamics in the Urban-Rural-Natural Interface within a Social-Ecological Hotspot. Appl. Geogr. 2023, 159, 103060. [Google Scholar] [CrossRef]
- Mittermeier, R.; Myers, N.; Gill, P.; Mittermeier, C. Hotspots: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions. J. Mammal. 2002, 83, 630–633. [Google Scholar] [CrossRef]
- Elórtegui, S.; Moreira-Muñoz, A. (Eds.) El Paisaje: Formas, Texturas y Colores. In Parque Nacional La Campana: Origen de Una Reserva de La Biósfera En Chile Central, 2nd ed.; Taller la Era: Viña del Mar, Chile, 2009. [Google Scholar]
- Casale, J.-F.; Borsdorf, A.; Moreira-Muñoz, Á. Reservas de la biosfera como laboratorios para la sustentabilidad: Paisajes de conservación y ordenamiento territorial. In Reservas de la Biosfera de Chile: Laboratorios para la Sustentabilidad; Serie Geolibros; Academia de Ciencias Austriaca-Pontifiia Universidad Católica de Chile-Instituto de Geografía: Santiago, Chile, 2014; Volume 17, pp. 272–293. [Google Scholar]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Da Fonseca, G.A.B.; Kent, J. Biodiversity Hotspots for Conservation Priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Quintanilla, V.G.; Cadiñanos, J.A.; Latasa, I.; Lozano, P.J. Aproximación Biogeográfica a Los Bosques de La Zona Mediterránea de Chile: Caracterización e Inventario. Boletín Asoc. Geógrafos Españoles 2012, 2012, 91–114. [Google Scholar] [CrossRef]
- Quintanilla, V. Situación Postincendio En Ecosistemas de Un Macizo Montañoso de Gran Valor Geobotánico, En La Cordillera Meditarránea de Chile. Cad. Geografia. Coimbra FLUC 2009, 28, 149–150. [Google Scholar]
- Gajardo, R. La Vegetación Natural de Chile: Clasificación y Distribución Geográfica; Universitaria: Santiago, Chile, 1995. [Google Scholar]
- Villaseñor, R. Guía para el Reconocimiento de las Especies Arbóreas y Arbustivas del Parque Nacional La Campana; CONAF V Región: Valparaíso, Chile, 1998. [Google Scholar]
- Hauck, L.; Moreira-Muñoz, A.; Nezadal, W. La Flora Exótica Ruderal Del Parque Nacional La Campana, Región de Valparaíso, Chile Central. Gayana Bot. 2016, 73, 206–219. [Google Scholar] [CrossRef]
- Luebert, F.; Muñoz Schick, M.; Moreira-Muñoz, A. Vegetación y flora. In Parque Nacional La Campana: Origen de una Reserva de la Biósfera en Chile Central; Elórtegui, S., Moreira-Muñoz, A., Eds.; Taller La Era: Viña del Mar, Chile, 2009. [Google Scholar]
- Luebert, F.; Pliscoff, P. Sinopsis Bioclimática y Vegetacional de Chile; Editorial Universitaria: Santiago, Chile, 2006. [Google Scholar]
- Dierschke, H. Pflanzensoziologie: Grundlagen und Methoden, 1st ed.; UTB: Stuttgart, Germany, 1994. [Google Scholar]
- Knapp, R. Sampling Methods and Taxon Analysis in Vegetation Science; Springer: Dordrecht, The Netherland, 1984; Volume 4. [Google Scholar]
- San Martín, C.; Ramírez, C.; San Martín, J.; Villaseñor, R. Flora Y Vegetacion Del Estero Reñaca (V Region, Chile). Gayana Bot. 2001, 58, 31–46. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km Spatial Resolution Climate Surfaces for Global Land Areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Boisier, J.P. CR2MET: A High-Resolution Precipitation and Temperature Dataset for the Period 1960–2021 in Continental Chile. (v2.5) [Data Set]; Zenodo: Genève, Switzerland, 2023. [Google Scholar]
- Ball, D.F. Loss-on-Ignition as an Estimate of Organic Matter and Organic Carbon in Non-Calcareous Soils. J. Soil Sci. 1964, 15, 84–92. [Google Scholar] [CrossRef]
- Sadzawka, R.A.; Carrasco, R.M.A.; Grez, Z.R.; Mora, G.M.d.l.L.; Flores, P.H.; Neaman, A. Métodos de Análisis Recomendados para los Suelos de Chile. Revisión 2006; Instituto de Investigaciones Agropecuarias: Santiago, Chile, 2006; Volume 34. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Costello, E.K.; Berg-Lyons, D.; Gonzalez, A.; Stombaugh, J.; Knights, D.; Gajer, P.; Ravel, J.; Fierer, N.; et al. Moving Pictures of the Human Microbiome. Genome Biol. 2011, 12, R50. [Google Scholar] [CrossRef] [PubMed]
- Degnan, P.H.; Ochman, H. Illumina-Based Analysis of Microbial Community Diversity. ISME J. 2012, 6, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Maignien, L.; Sul, W.J.; Murphy, L.G.; Grim, S.L.; Morrison, H.G.; Sogin, M.L. Oligotyping: Differentiating between Closely Related Microbial Taxa Using 16S rRNA Gene Data. Methods Ecol. Evol. 2013, 4, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S RRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE Database for Molecular Identification of Fungi—Recent Updates and Future Perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE QIIME Release for Fungi 2; Version 10.05.2021; UNITE Community: Greenville, SC, USA, 2021. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhan, L.; Tang, W.; Wang, Q.; Dai, Z.; Zhou, L.; Feng, T.; Chen, M.; Wu, T.; Hu, E.; et al. MicrobiotaProcess: A Comprehensive R Package for Deep Mining Microbiome. Innovation 2023, 4, 100388. [Google Scholar] [CrossRef]
- Andersen, K.S.; Kirkegaard, R.H.; Karst, S.M.; Albertsen, M. Ampvis2: An R Package to Analyse and Visualise 16S RRNA Amplicon Data. bioRxiv 2018, 299537. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. Microeco: An R Package for Data Mining in Microbial Community Ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling Function and Taxonomy in the Global Ocean Microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An Open Annotation Tool for Parsing Fungal Community Datasets by Ecological Guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Oksanen, J. Design Decisions and Implementation Details in Vegan; Vignette of the Package Vegan. R Package Version 2016; R Foundation: Vienna, Austria, 2016; pp. 2–4. [Google Scholar]
- Egli, M.; Sartori, G.; Mirabella, A.; Favilli, F.; Giaccai, D.; Delbos, E. Effect of North and South Exposure on Organic Matter in High Alpine Soils. Geoderma 2009, 149, 124–136. [Google Scholar] [CrossRef]
- Carletti, P.; Vendramin, E.; Pizzeghello, D.; Concheri, G.; Zanella, A.; Nardi, S.; Squartini, A. Soil Humic Compounds and Microbial Communities in Six Spruce Forests as Function of Parent Material, Slope Aspect and Stand Age. Plant Soil 2009, 315, 47–65. [Google Scholar] [CrossRef]
- Barbosa, W.R.; Romero, R.E.; Júnior, V.S.d.S.; Cooper, M.; Sartor, L.R.; Partiti, C.S.d.M.; Jorge, F.d.O.; Cohen, R.; de Jesus, S.L.; Ferreira, T.O. Effects of Slope Orientation on Pedogenesis of Altimontane Soils from the Brazilian Semi-Arid Region (Baturité Massif, Ceará). Environ. Earth Sci. 2015, 73, 3731–3743. [Google Scholar] [CrossRef]
- Egli, M.; Mirabella, A.; Sartori, G.; Zanelli, R.; Bischof, S. Effect of North and South Exposure on Weathering Rates and Clay Mineral Formation in Alpine Soils. Catena 2006, 67, 155–174. [Google Scholar] [CrossRef]
- Zhang, B.; Liang, C.; He, H.; Zhang, X. Variations in Soil Microbial Communities and Residues Along an Altitude Gradient on the Northern Slope of Changbai Mountain, China. PLoS ONE 2013, 8, e66184. [Google Scholar] [CrossRef] [PubMed]
- Djukic, I.; Zehetner, F.; Tatzber, M.; Gerzabek, M.H. Soil Organic-matter Stocks and Characteristics along an Alpine Elevation Gradient. J. Plant Nutr. Soil Sci. 2010, 173, 30–38. [Google Scholar] [CrossRef]
- Samolov, E.; Baumann, K.; Büdel, B.; Jung, P.; Leinweber, P.; Mikhailyuk, T.; Karsten, U.; Glaser, K. Biodiversity of Algae and Cyanobacteria in Biological Soil Crusts Collected along a Climatic Gradient in Chile Using an Integrative Approach. Microorganisms 2020, 8, 1047. [Google Scholar] [CrossRef] [PubMed]
- Rivas, Y.; Aponte, H.; Rivera-Salazar, D.; Matus, F.; Martínez, O.; Encina, C.; Retamal-Salgado, J. Microbial Community and Enzyme Activity of Forest Plantation, Natural Forests, and Agricultural Land in Chilean Coastal Cordillera Soils. Forests 2023, 14, 938. [Google Scholar] [CrossRef]
- Almonacid-Muñoz, L.; Herrera, H.; Fuentes-Ramírez, A.; Vargas-Gaete, R.; Larama, G.; Jara, R.; Fernández-Urrutia, C.; da Silva Valadares, R.B. Tree Cover Species Modify the Diversity of Rhizosphere-Associated Microorganisms in Nothofagus Obliqua (Mirb.) Oerst Temperate Forests in South-Central Chile. Forests 2022, 13, 756. [Google Scholar] [CrossRef]
- Villaseñor, R. Unidades Fisionómicas y Florísticas Del Parque Nacional La Campana. Chloris Chil. Rev. Chil. Flora Veg. 2021, 24, 128–134. [Google Scholar]
- Siles, J.A.; Rachid, C.T.C.C.; Sampedro, I.; García-Romera, I.; Tiedje, J.M. Microbial Diversity of a Mediterranean Soil and Its Changes after Biotransformed Dry Olive Residue Amendment. PLoS ONE 2014, 9, e103035. [Google Scholar] [CrossRef]
- Frenk, S.; Hadar, Y.; Minz, D. Resilience of Soil Bacterial Community to Irrigation with Water of Different Qualities under M Editerranean Climate. Environ. Microbiol. 2014, 16, 559–569. [Google Scholar] [CrossRef]
- Pii, Y.; Borruso, L.; Brusetti, L.; Crecchio, C.; Cesco, S.; Mimmo, T. The Interaction between Iron Nutrition, Plant Species and Soil Type Shapes the Rhizosphere Microbiome. Plant Physiol. Biochem. 2016, 99, 39–48. [Google Scholar] [CrossRef]
- Shen, C.; Ge, Y.; Yang, T.; Chu, H. Verrucomicrobial Elevational Distribution Was Strongly Influenced by Soil PH and Carbon/Nitrogen Ratio. J. Soils Sediments 2017, 17, 2449–2456. [Google Scholar] [CrossRef]
- Brewer, T.E.; Handley, K.M.; Carini, P.; Gilbert, J.A.; Fierer, N. Genome Reduction in an Abundant and Ubiquitous Soil Bacterium ‘Candidatus Udaeobacter Copiosus’. Nat. Microbiol. 2016, 2, 16198. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Khan, S.T. Microbial Communities and Their Predictive Functional Profiles in the Arid Soil of Saudi Arabia. Soil 2020, 6, 513–521. [Google Scholar] [CrossRef]
- Buckley, D.H.; Huangyutitham, V.; Hsu, S.-F.; Nelson, T.A. Stable Isotope Probing with 15N2 Reveals Novel Noncultivated Diazotrophs in Soil. Appl. Environ. Microbiol. 2007, 73, 3196–3204. [Google Scholar] [CrossRef]
- Catania, V.; Bueno, R.S.; Alduina, R.; Grilli, E.; La Mantia, T.; Castaldi, S.; Quatrini, P. Soil Microbial Biomass and Bacterial Diversity in Southern European Regions Vulnerable to Desertification. Ecol. Indic. 2022, 145, 109725. [Google Scholar] [CrossRef]
- Vohník, M.; Mrnka, L.; Lukešová, T.; Bruzone, M.C.; Kohout, P.; Fehrer, J. The Cultivable Endophytic Community of Norway Spruce Ectomycorrhizas from Microhabitats Lacking Ericaceous Hosts Is Dominated by Ericoid Mycorrhizal Meliniomyces Variabilis. Fungal Ecol. 2013, 6, 281–292. [Google Scholar] [CrossRef]
- Bickel, S.; Or, D. The Chosen Few—Variations in Common and Rare Soil Bacteria across Biomes. ISME J. 2021, 15, 3315–3325. [Google Scholar] [CrossRef] [PubMed]
- Egidi, E.; Delgado-Baquerizo, M.; Plett, J.M.; Wang, J.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K. A Few Ascomycota Taxa Dominate Soil Fungal Communities Worldwide. Nat. Commun. 2019, 10, 2369. [Google Scholar] [CrossRef]
- Gleason, F.H.; Letcher, P.M.; Commandeur, Z.; Jeong, C.E.; McGee, P.A. The Growth Response of Some Chytridiomycota to Temperatures Commonly Observed in the Soil. Mycol. Res. 2005, 109, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Hanrahan-Tan, D.G.; Lilje, O.; Henderson, L. Chytrids in Soil Environments: Unique Adaptations and Distributions. Encyclopedia 2023, 3, 642–664. [Google Scholar] [CrossRef]
- Vega, F.E.; Meyling, N.V.; Luangsa-ard, J.J.; Blackwell, M. Fungal entomopathogens. In Insect Pathology; Elsevier: Amsterdam, The Netherlands, 2012; pp. 171–220. [Google Scholar]
- Rungjindamai, N.; Jones, E.B.G. Why Are There So Few Basidiomycota and Basal Fungi as Endophytes? A Review. J. Fungi 2024, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Egidi, E.; Wood, J.L.; Celestina, C.; May, T.W.; Mele, P.; Edwards, J.; Powell, J.; Bissett, A.; Franks, A.E. Delving into the Dark Ecology: A Continent-Wide Assessment of Patterns of Composition in Soil Fungal Communities from Australian Tussock Grasslands. Fungal Ecol. 2019, 39, 356–370. [Google Scholar] [CrossRef]
- Tedersoo, L.; Tooming-Klunderud, A.; Anslan, S. PacBio Metabarcoding of Fungi and Other Eukaryotes: Errors, Biases and Perspectives. New Phytol. 2018, 217, 1370–1385. [Google Scholar] [CrossRef]
- Tedersoo, L.; Mikryukov, V.; Anslan, S.; Bahram, M.; Khalid, A.N.; Corrales, A.; Agan, A.; Vasco-Palacios, A.-M.; Saitta, A.; Antonelli, A.; et al. The Global Soil Mycobiome Consortium Dataset for Boosting Fungal Diversity Research. Fungal Divers. 2021, 111, 573–588. [Google Scholar] [CrossRef]
- Pereira, A.P.A.; Durrer, A.; Gumiere, T.; Gonçalves, J.L.M.; Robin, A.; Bouillet, J.-P.; Wang, J.; Verma, J.P.; Singh, B.K.; Cardoso, E.J.B.N. Mixed Eucalyptus Plantations Induce Changes in Microbial Communities and Increase Biological Functions in the Soil and Litter Layers. For. Ecol. Manag. 2019, 433, 332–342. [Google Scholar] [CrossRef]
- Zhang, H.; Shi, Y.; Dong, Y.; Lapen, D.R.; Liu, J.; Chen, W. Subsoiling and Conversion to Conservation Tillage Enriched Nitrogen Cycling Bacterial Communities in Sandy Soils under Long-Term Maize Monoculture. Soil Tillage Res. 2022, 215, 105197. [Google Scholar] [CrossRef]
- Bates, S.T.; Berg-Lyons, D.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. Examining the Global Distribution of Dominant Archaeal Populations in Soil. ISME J. 2011, 5, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, A.M.; Jaffe, A.L.; Nuccio, E.E.; Taga, M.E.; Firestone, M.K.; Banfield, J.F. Soil Candidate Phyla Radiation Bacteria Encode Components of Aerobic Metabolism and Co-Occur with Nanoarchaea in the Rare Biosphere of Rhizosphere Grassland Communities. mSystems 2021, 6, e0120520. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.M.; Hemp, J.; Shih, P.M.; McGlynn, S.E.; Fischer, W.W. Evolution of Phototrophy in the Chloroflexi Phylum Driven by Horizontal Gene Transfer. Front. Microbiol. 2018, 9, 260. [Google Scholar] [CrossRef]
- Sánchez-Baracaldo, P.; Cardona, T. On the Origin of Oxygenic Photosynthesis and Cyanobacteria. New Phytol. 2020, 225, 1440–1446. [Google Scholar] [CrossRef]
- Genitsaris, S.; Stefanidou, N.; Leontidou, K.; Matsi, T.; Karamanoli, K.; Mellidou, I. Bacterial Communities in the Rhizosphere and Phyllosphere of Halophytes and Drought-Tolerant Plants in Mediterranean Ecosystems. Microorganisms 2020, 8, 1708. [Google Scholar] [CrossRef] [PubMed]
- Velázquez-Becerra, C.; Macías-Rodríguez, L.I.; López-Bucio, J.; Altamirano-Hernández, J.; Flores-Cortez, I.; Valencia-Cantero, E. A Volatile Organic Compound Analysis from Arthrobacter Agilis Identifies Dimethylhexadecylamine, an Amino-Containing Lipid Modulating Bacterial Growth and Medicago Sativa Morphogenesis in Vitro. Plant Soil 2011, 339, 329–340. [Google Scholar] [CrossRef]
- Requena, N.; Jimenez, I.; Toro, M.; Barea, J.M. Interactions between Plant-growth-promoting Rhizobacteria (PGPR), Arbuscular Mycorrhizal Fungi and Rhizobium spp. in the Rhizosphere of Anthyllis cytisoides, a Model Legume for Revegetation in Mediterranean Semi-arid Ecosystems. New Phytol. 1997, 136, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Asplund, J.; Kauserud, H.; Bokhorst, S.; Lie, M.H.; Ohlson, M.; Nybakken, L. Fungal Communities Influence Decomposition Rates of Plant Litter from Two Dominant Tree Species. Fungal Ecol. 2018, 32, 1–8. [Google Scholar] [CrossRef]
- Asplund, J.; Kauserud, H.; Ohlson, M.; Nybakken, L. Spruce and Beech as Local Determinants of Forest Fungal Community Structure in Litter, Humus and Mineral Soil. FEMS Microbiol. Ecol. 2019, 95, fiy232. [Google Scholar] [CrossRef] [PubMed]
- Challacombe, J.F.; Hesse, C.N.; Bramer, L.M.; McCue, L.A.; Lipton, M.; Purvine, S.; Nicora, C.; Gallegos-Graves, L.V.; Porras-Alfaro, A.; Kuske, C.R. Genomes and Secretomes of Ascomycota Fungi Reveal Diverse Functions in Plant Biomass Decomposition and Pathogenesis. BMC Genom. 2019, 20, 976. [Google Scholar] [CrossRef]
- Santonja, M.; Foucault, Q.; Rancon, A.; Gauquelin, T.; Fernandez, C.; Baldy, V.; Mirleau, P. Contrasting Responses of Bacterial and Fungal Communities to Plant Litter Diversity in a Mediterranean Oak Forest. Soil Biol. Biochem. 2018, 125, 27–36. [Google Scholar] [CrossRef]
- Funk, A. Foliar Fungi of Western Trees; Pacific Forest Research Centre: Victoria, BC, Canada, 1985. [Google Scholar]
- Selbmann, L.; de Hoog, G.S.; Mazzaglia, A.; Friedmann, E.I.; Onofri, S. Fungi at the Edge of Life: Cryptoendolithic Black Fungi from Antarctic Desert. Stud. Mycol. 2005, 51, 1–32. [Google Scholar]
- Hart, M.; Gorzelak, M.A.; McAmmond, B.M.; Van Hamme, J.D.; Stevens, J.; Abbott, L.; Whiteley, A.S.; Nevill, P. Fungal Communities Resist Recovery in Sand Mine Restoration. Front. For. Glob. Change 2019, 2, 78. [Google Scholar] [CrossRef]
- Das, K.; Lee, S.-Y.; Jung, H.-Y. Cladophialophora lanosa Sp. Nov., a New Species Isolated from Soil. Mycobiology 2019, 47, 173–179. [Google Scholar] [CrossRef]
- Liu, X.-Z.; Wang, Q.-M.; Göker, M.; Groenewald, M.; Kachalkin, A.V.; Lumbsch, H.T.; Millanes, A.M.; Wedin, M.; Yurkov, A.M.; Boekhout, T.; et al. Towards an Integrated Phylogenetic Classification of the Tremellomycetes. Stud. Mycol. 2015, 81, 85–147. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Pärtel, K.; Jairus, T.; Gates, G.; Põldmaa, K.; Tamm, H. Ascomycetes Associated with Ectomycorrhizas: Molecular Diversity and Ecology with Particular Reference to the Helotiales. Environ. Microbiol. 2009, 11, 3166–3178. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.S.; Phillips, A.J.L. Palm Fungi and Their Key Role in Biodiversity Surveys: A Review. J. Fungi 2023, 9, 1121. [Google Scholar] [CrossRef] [PubMed]
- Ozimek, E.; Hanaka, A. Mortierella Species as the Plant Growth-Promoting Fungi Present in the Agricultural Soils. Agriculture 2020, 11, 7. [Google Scholar] [CrossRef]
- Daghino, S.; Murat, C.; Sizzano, E.; Girlanda, M.; Perotto, S. Fungal Diversity Is Not Determined by Mineral and Chemical Differences in Serpentine Substrates. PLoS ONE 2012, 7, e44233. [Google Scholar] [CrossRef]
- van Schöll, L.; Kuyper, T.W.; Smits, M.M.; Landeweert, R.; Hoffland, E.; Breemen, N. van Rock-Eating Mycorrhizas: Their Role in Plant Nutrition and Biogeochemical Cycles. Plant Soil 2008, 303, 35–47. [Google Scholar] [CrossRef]
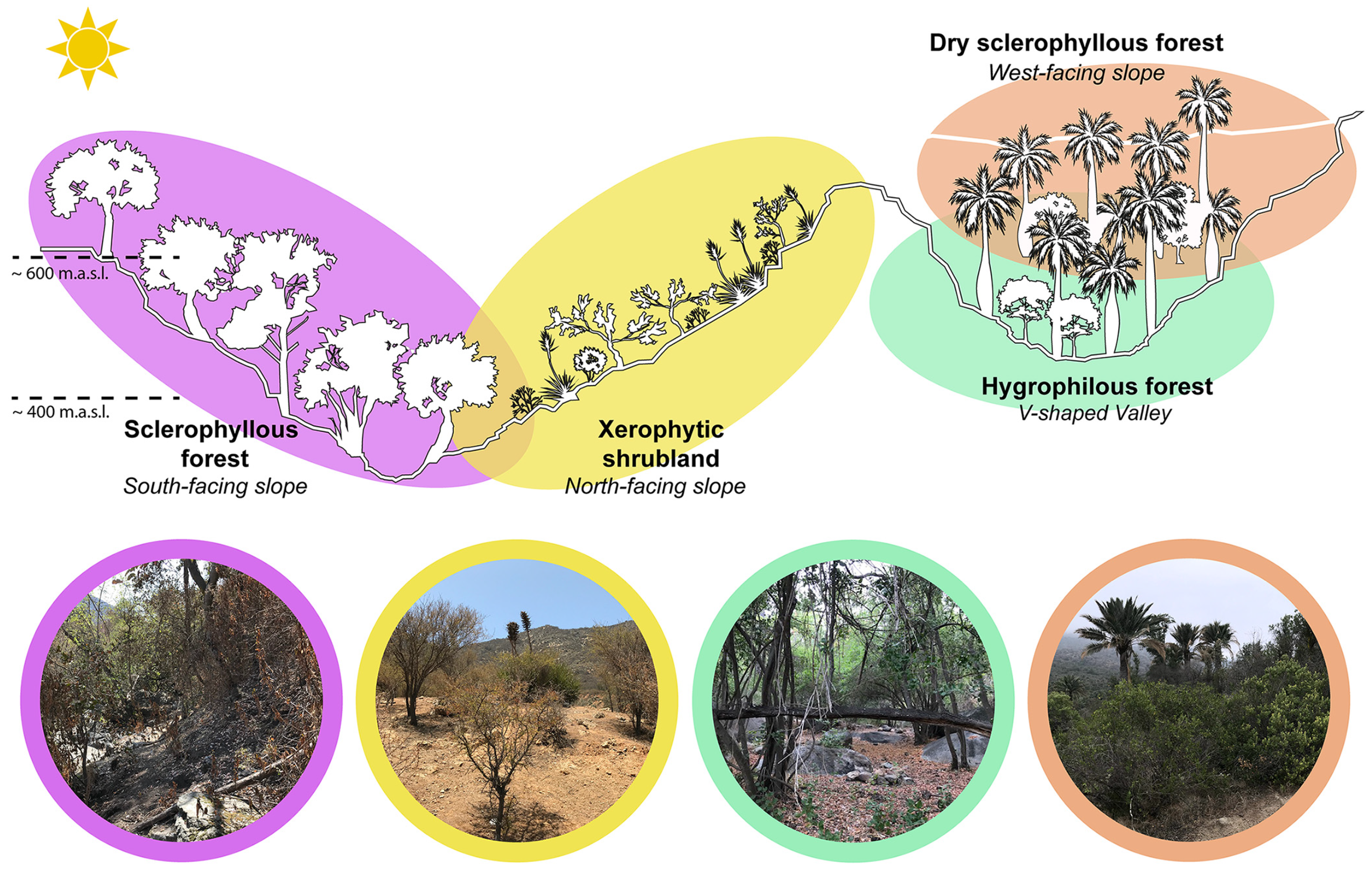
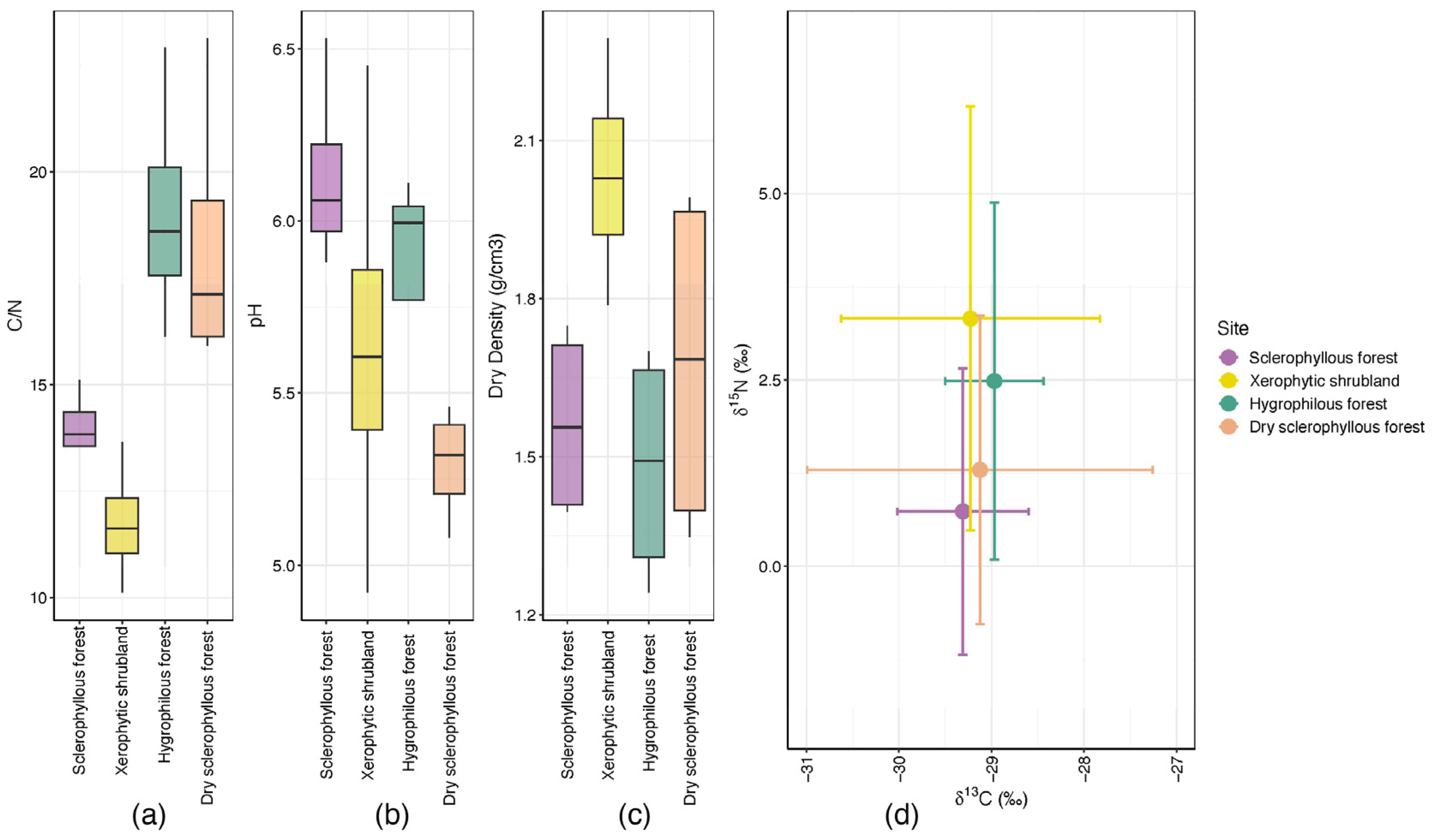
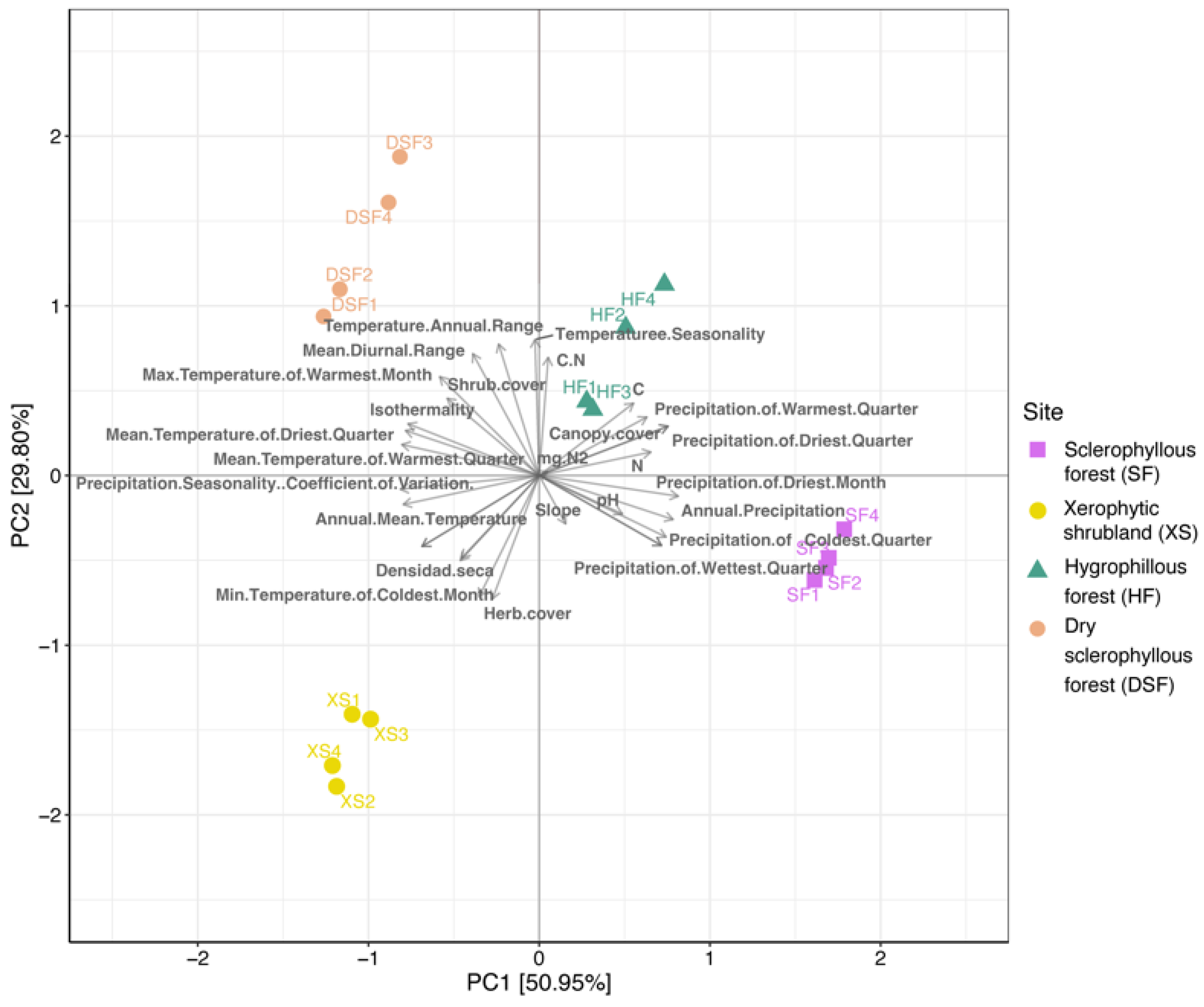




| ID | Site | Slope Facing or Aspect | Geographic Location (Latitude) | Geographic Location (Longitude) | Elevation (masl) | Collection Date |
|---|---|---|---|---|---|---|
| SF1 | Sclerophyllous forest | South | 33°0′12.13″ S | 71°7′24.62″ O | 396 | January 2020 |
| SF2 | Sclerophyllous forest | South | 33°0′12.28″ S | 71°7′25.17″ O | 391 | January 2020 |
| SF3 | Sclerophyllous forest | South | 33°0′12.34″ S | 71°7′26.40″ O | 383 | January 2020 |
| SF4 | Sclerophyllous forest | South | 33°0′12.39″ S | 71°7′25.82″ O | 390 | January 2020 |
| XS1 | Xerophytic shrubland | North | 33°0′16.82″ S | 71°7′28.00″ O | 410 | January 2020 |
| XS2 | Xerophytic shrubland | North | 33°0′16.65″ S | 71°7′27.32″ O | 407 | January 2020 |
| XS3 | Xerophytic shrubland | North | 33°0′16.40″ S | 71°7′27.80″ O | 405 | January 2020 |
| XS4 | Xerophytic shrubland | North | 33°0′17.19″ S | 71°7′27.43″ O | 413 | January 2020 |
| HF1 | Hygrophilous forest | V-shaped valley | 32°57′13.86″ S | 71°5′26.47″ O | 590 | February 2021 |
| HF2 | Hygrophilous forest | V-shaped valley | 32°57′14.28″ S | 71°5′26.84″ O | 587 | February 2021 |
| HF3 | Hygrophilous forest | V-shaped valley | 32°57′14.63″ S | 71°5′25.78″ O | 585 | February 2021 |
| HF4 | Hygrophilous forest | V-shaped valley | 32°57′14.95″ S | 71°5′26.46″ O | 589 | February 2021 |
| DSF1 | Dry sclerophyllous forest | West | 32°57′14.14″ S | 71°5′23.46″ O | 621 | February 2021 |
| DSF2 | Dry sclerophyllous forest | West | 32°57′13.73″ S | 71°5′23.92″ O | 618 | February 2021 |
| DSF3 | Dry sclerophyllous forest | West | 32°57′14.51″ S | 71°5′23.69″ O | 624 | February 2021 |
| DSF4 | Dry sclerophyllous forest | West | 32°57′14.38″ S | 71°5′23.02″ O | 626 | February 2021 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quinteros-Urquieta, C.; Francois, J.-P.; Aguilar-Muñoz, P.; Orellana, R.; Villaseñor, R.; Moreira-Muñoz, A.; Molina, V. Microbial Diversity of Soil in a Mediterranean Biodiversity Hotspot: Parque Nacional La Campana, Chile. Microorganisms 2024, 12, 1569. https://doi.org/10.3390/microorganisms12081569
Quinteros-Urquieta C, Francois J-P, Aguilar-Muñoz P, Orellana R, Villaseñor R, Moreira-Muñoz A, Molina V. Microbial Diversity of Soil in a Mediterranean Biodiversity Hotspot: Parque Nacional La Campana, Chile. Microorganisms. 2024; 12(8):1569. https://doi.org/10.3390/microorganisms12081569
Chicago/Turabian StyleQuinteros-Urquieta, Carolina, Jean-Pierre Francois, Polette Aguilar-Muñoz, Roberto Orellana, Rodrigo Villaseñor, Andres Moreira-Muñoz, and Verónica Molina. 2024. "Microbial Diversity of Soil in a Mediterranean Biodiversity Hotspot: Parque Nacional La Campana, Chile" Microorganisms 12, no. 8: 1569. https://doi.org/10.3390/microorganisms12081569
APA StyleQuinteros-Urquieta, C., Francois, J.-P., Aguilar-Muñoz, P., Orellana, R., Villaseñor, R., Moreira-Muñoz, A., & Molina, V. (2024). Microbial Diversity of Soil in a Mediterranean Biodiversity Hotspot: Parque Nacional La Campana, Chile. Microorganisms, 12(8), 1569. https://doi.org/10.3390/microorganisms12081569







