Exploring the Role of the Environment as a Reservoir of Antimicrobial-Resistant Campylobacter: Insights from Wild Birds and Surface Waters
Abstract
1. Introduction
2. Materials and Methods
2.1. Campylobacter Isolates Collection
2.2. Disk Diffusion Method
2.3. Genomic DNA Extraction and Whole-Genome Sequencing
2.4. Genomic Assembly and Characterisation
2.5. Result Interpretation and Statistical Analyses
3. Results
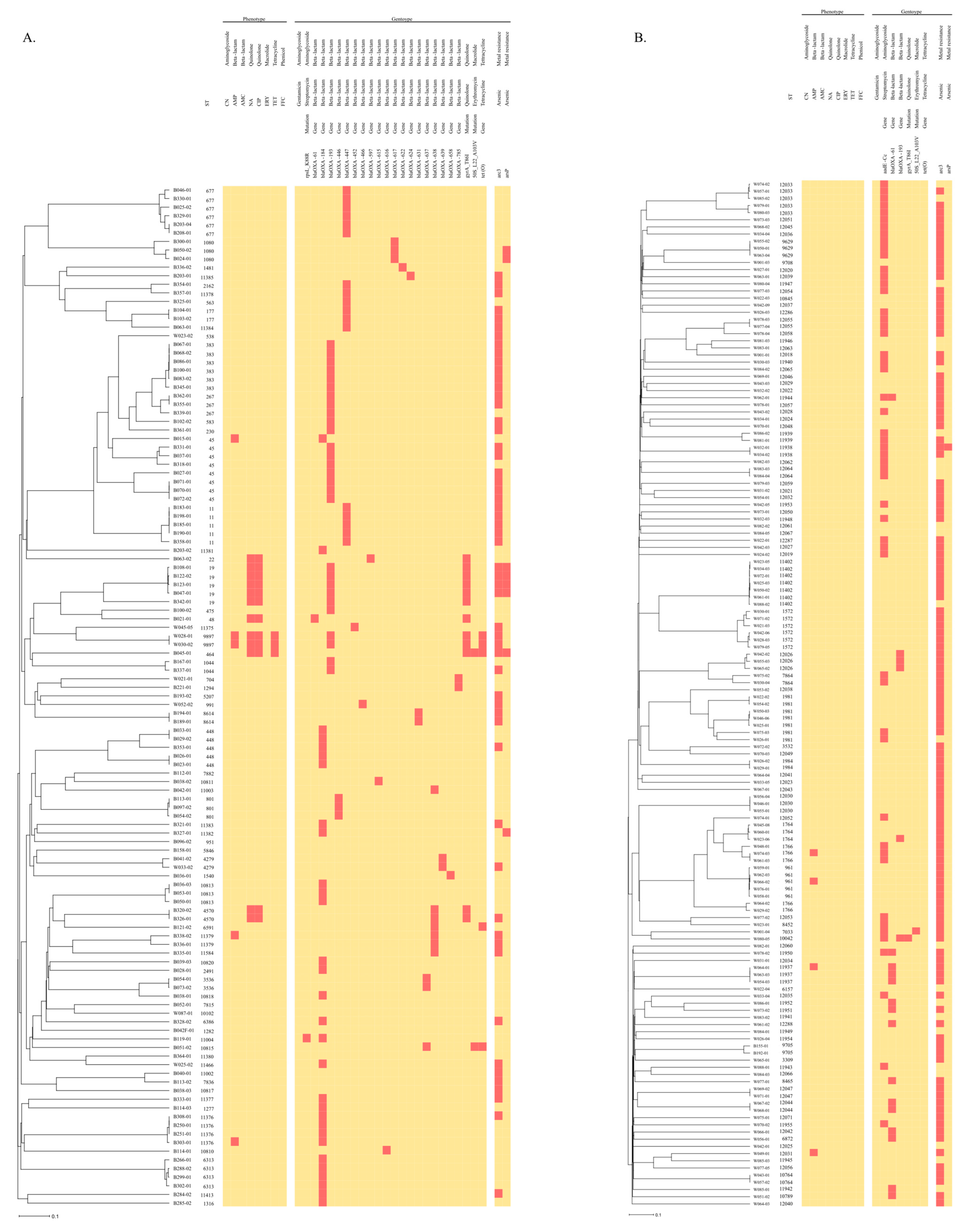
3.1. AMR Phenotypes and Genotypes of Isolates
3.2. Association between AMR, ST, and Heavy Metals
3.3. Virulence Genes of Isolates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistance Infections Globally: Final Report and Recommendations. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 27 January 2023).
- Wild, A.C.; Moinova, H.R.; Mulcahy, R.T. Regulation of γ-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 1999, 274, 33627–33636. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial resistance. Bull. World Health Organ. 2015, 93, 363–364. [Google Scholar]
- Góchez, D.; Raicek, M.; Ferreira, J.P.; Jeannin, M.; Moulin, G.; Erlacher-Vindel, E. OIE annual report on antimicrobial agents intended for use in animals: Methods used. Front. Vet. Sci. 2019, 6, 317. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018, 42, 68–80. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2021, 20, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Garofolo, G.; Di Pasquale, A.; Cammà, C. Monitoring AMR in Campylobacter jejuni from Italy in the last 10 years (2011–2021): Microbiological and WGS data risk assessment. EFSA J. 2022, 20, 200406. [Google Scholar] [CrossRef] [PubMed]
- EFSA; ECDC. The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, 8442. [Google Scholar] [CrossRef]
- Hansson, I.; Sandberg, M.; Habib, I.; Lowman, R.; Engvall, E.O. Knowledge gaps in control of Campylobacter for prevention of campylobacteriosis. Transbound. Emerg. Dis. 2018, 65, 30–48. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of campylobacter infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef] [PubMed]
- Wysok, B.; Sołtysiuk, M.; Stenzel, T. Wildlife Waterfowl as a Source of Pathogenic Campylobacter Strains. Pathogens 2022, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, S.K.; Dallas, J.F.; Strachan, N.J.C.; MacRae, M.; McCarthy, N.D.; Wilson, D.J.; Gormley, F.J.; Falush, D.; Ogden, L.D.; Maiden, M.C.J.; et al. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 2009, 48, 1072–1078. [Google Scholar] [CrossRef] [PubMed]
- Hock, L.; Herold, M.; Walczak, C.; Schoos, A.; Penny, C.; Cauchie, H.M.; Ragimbeau, C. Environmental dynamics of Campylobacter jejuni genotypes circulating in Luxembourg: What is the role of wild birds? Microb. Genom. 2023, 9, 001031. [Google Scholar] [CrossRef]
- Société Française de Microbiologie. CASFM/EUCAST; Société Française de Microbiologie: Paris, France, 2020. [Google Scholar]
- Tang, Y.; Sahin, O.; Pavlovic, N.; Lejeune, J.; Carlson, J.; Wu, Z.; Dai, L.; Zhang, Q. Rising fluoroquinolone resistance in Campylobacter isolated from feedlot cattle in the United States. Sci. Rep. 2017, 7, 494. [Google Scholar] [CrossRef]
- Jünemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; Von Haeseler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013, 31, 294–296. [Google Scholar] [CrossRef]
- Dingle, K.E.; Colles, F.M.; Wareing, D.R.A.; Ure, R.; Fox, A.J.; Bolton, F.E.; Bootsma, H.J.; Willems, R.J.L.; Urwin, R.; Maiden, M.C.J. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 2001, 39, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFINder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159. [Google Scholar] [CrossRef]
- El-Adawy, H.; Hotzel, H.; García-Soto, S.; Tomaso, H.; Hafez, H.M.; Schwarz, S.; Neubauer, H.; Linde, J. Genomic insight into Campylobacter jejuni isolated from commercial turkey flocks in Germany using whole-genome sequencing analysis. Front. Vet. Sci. 2023, 10, 1092179. [Google Scholar] [CrossRef] [PubMed]
- Ghielmetti, G.; Seth-Smith, H.M.B.; Roloff, T.; Cernela, N.; Biggel, M.; Stephan, R.; Egli, A. Whole-genome-based characterization of Campylobacter jejuni from human patients with gastroenteritis collected over an 18 year period reveals increasing prevalence of antimicrobial resistance. Microb. Genom. 2023, 9, 000941. [Google Scholar] [CrossRef]
- Waldenström, J.; Axelsson-Olsson, D.; Olsen, B.; Hasselquist, D.; Griekspoor, P.; Jansson, L.; Teneberg, S.; Svensson, L.; Ellström, P. Campylobacter jejuni colonization in wild birds: Results from an infection experiment. PLoS ONE 2010, 5, e9082. [Google Scholar] [CrossRef]
- Mencía-Gutiérrez, A.; Martín-Maldonado, B.; Pastor-Tiburón, N.; Moraleda, V.; González, F.; García-Peña, F.J.; Pérez-Cobo, I.; Revuelta, L.; Marín, M. Prevalence and antimicrobial resistance of Campylobacter from wild birds of prey in Spain. Comp. Immunol. Microbiol. Infect. Dis. 2021, 79, 147–9571. [Google Scholar] [CrossRef]
- Batista, R.; Saraiva, M.; Lopes, T.; Silveira, L.; Coelho, A.; Furtado, R.; Castro, R.; Correia, C.B.; Rodrigues, D.; Henriques, P.; et al. Genotypic and Phenotypic Characterization of Pathogenic Escherichia coli, Salmonella spp., and Campylobacter spp., in Free-Living Birds in Mainland Portugal. Int. J. Environ. Res. Public Health 2023, 20, 223. [Google Scholar] [CrossRef]
- Wieczorek, K.; Osek, J. Antimicrobial resistance mechanisms among Campylobacter. BioMed Res. Int. 2013, 2013, 340605. [Google Scholar] [CrossRef]
- Szczepanska, B.; Andrzejewska, M.; Spica, D.; Klawe, J.J. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from children and environmental sources in urban and suburban areas. BMC Microbiol. 2017, 17, 80. [Google Scholar] [CrossRef]
- Garcia-Fernandez, A.; Janowicz, A.; Marotta, F.; Napoleoni, M.; Arena, S.; Primavilla, S.; Pitti, M.; Romantini, R.; Tomei, F.; Garofolo, G.; et al. Antibiotic resistance, plasmids, and virulence-associated markers in human strains of Campylobacter jejuni and Campylobacter coli isolated in Italy. Front. Microbiol. 2023, 14, 1293666. [Google Scholar] [CrossRef]
- Ocejo, M.; Oporto, B.; Lavín, J.L.; Hurtado, A. Whole genome-based characterisation of antimicrobial resistance and genetic diversity in Campylobacter jejuni and Campylobacter coli from ruminants. Sci. Rep. 2021, 11, 8998. [Google Scholar] [CrossRef] [PubMed]
- Hodges, L.M.; Taboada, E.N.; Koziol, A.; Mutschall, S.; Blais, B.W.; Inglis, G.D.; Leclair, D.; Carrillo, C.D. Systematic Evaluation of Whole-Genome Sequencing Based Prediction of Antimicrobial Resistance in Campylobacter jejuni and C. coli. Front. Microbiol. 2021, 12, 776967. [Google Scholar] [CrossRef]
- Papadopoulos, D.; Petridou, E.; Papageorgiou, K.; Giantsis, I.A.; Delis, G.; Economou, V.; Frydas, I.; Papadopoulos, G.; Hatzistylianou, M.; Kritas, S.K. Phenotypic and molecular patterns of resistance among Campylobacter coli and Campylobacter jejuni isolates, from pig farms. Animals 2021, 11, 2394. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO List of Medically Important Antimicrobials: A Risk Management Tool for Mitigating; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 2021, 19, e06651. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2021–2022. EFSA J. 2024, 10, 2598. [Google Scholar] [CrossRef]
- Andrzejewska, M.; Grudlewska-Buda, K.; Śica, D.; Skowron, K.; Ćwiklińska-Jurkowska, M.; Szady-Grad, M.; Indykiewicz, P.; Wiktorczyk-Kapischke, N.; Klawe, J.J.; King, L.; et al. Genetic relatedness, virulence, and drug susceptibility of Campylobacter isolated from water and wild birds. Front. Cell. Infect. Microbiol. 2022, 12, 1005085. [Google Scholar] [CrossRef]
- Casalino, G.; D’Amico, F.; Dinardo, F.R.; Bozzo, G.; Napoletano, V.; Camarda, A.; Bove, A.; Lombardi, R.; D’Onghia, F.P.; Circella, E. Prevalence and Antimicrobial Resistance of Campylobacter jejuni and Campylobacter coli in Wild Birds from a Wildlife Rescue Centre. Animals 2022, 12, 2889. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; Penny, C.; Ragimbeau, C.; Schets, F.M.; Blaak, H.; Duim, B.; Wagenaar, J.A.; de Boer, A.; Cauchie, H.M.; Mossong, J.; et al. Quantifying potential sources of surface water contamination with Campylobacter jejuni and Campylobacter coli. Water Res. 2016, 101, 36–45. [Google Scholar] [CrossRef]
- Painset, A.; Day, M.; Doumith, M.; Rigby, J.; Jenkins, C.; Grant, K.; Dallman, T.J.; Godbole, G.; Swift, C. Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of Campylobacter jejuni and Campylobacter coli isolated from cases of diarrhoeal disease in England and Wales, 2015–16. J. Antimicrob. Chemother. 2020, 75, 883–889. [Google Scholar] [CrossRef]
- Hormeño, L.; Campos, M.J.; Vadillo, S.; Quesada, A. Occurrence of tet(O/M/O) mosaic gene in tetracycline-resistant Campylobacter. Microorganisms 2020, 8, 710. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Elhadidy, M.; Ali, M.M.; El-Shibiny, A.; Miller, W.G.; Elkhatib, W.F.; Botteldoorn, N.; Dierick, K. Antimicrobial resistance patterns and molecular resistance markers of Campylobacter jejuni isolates from human diarrheal cases. PLoS ONE 2020, 15, e227833. [Google Scholar] [CrossRef] [PubMed]
- Griggs, D.J.; Peake, L.; Johnson, M.M.; Ghori, S.; Mott, A.; Piddock, L.J.V. β-lactamase-mediated β-lactam resistance in Campylobacter species: Prevalence of Cj0299 (blaOXA-61) and evidence for a novel β-lactamase in C. jejuni. Antimicrob. Agents Chemother. 2009, 53, 3357–3364. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, Y.; Zhang, Q.; Shen, J. Antimicrobial Resistance in Campylobacter spp. Microbiol. Spectr. 2018, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- EFSA; ECDC. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, 6490. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 1–20. [Google Scholar] [CrossRef]
- Smith, O.M.; Snyder, W.E.; Owen, J.P. Are we overestimating risk of enteric pathogen spillover from wild birds to humans? Biol. Rev. 2020, 95, 652–679. [Google Scholar] [CrossRef] [PubMed]
- Bravo, V.; Katz, A.; Porte, L.; Weitzel, T.; Varela, C.; Gonzalez-Escalona, N.; Blondel, C.J. Genomic analysis of the diversity, antimicrobial resistance and virulence potential of clinical Campylobacter jejuni and Campylobacter coli strains from chile. PLoS Negl. Trop. Dis. 2021, 15, e0009207. [Google Scholar] [CrossRef]
- Hull, D.M.; Harrel, E.; Harden, L.; Thakur, S. Detection of resistance and virulence plasmids in Campylobacter coli and Campylobacter jejuni isolated from North Carolina food animal production, 2018–2019. Food Microbiol. 2023, 116, 104348. [Google Scholar] [CrossRef]
- Lopes, G.V.; Ramires, T.; Kleinubing, N.R.; Scheik, L.K.; Fiorentini, Â.M.; Padilha da Silva, W. Virulence factors of foodborne pathogen Campylobacter jejuni. Microb. Pathog. 2021, 161, 105265. [Google Scholar] [CrossRef] [PubMed]
- Edet, U.O.; Bassey, I.U.; Joseph, A.P. Heavy metal co-resistance with antibiotics amongst bacteria isolates from an open dumpsite soil. Heliyon 2023, 9, e13457. [Google Scholar] [CrossRef] [PubMed]
- Mencía-Ares, O.; Borowiak, M.; Argüello, H.; Cobo-Díaz, J.F.; Malorny, B.; Álvarez-Ordóñez, A.; Carvajal, A.; Deneke, C. Genomic Insights into the Mobilome and Resistome of Sentinel Microorganisms Originating from Farms of Two Different Swine Production Systems. Microbiol. Spectr. 2022, 10, e02896-22. [Google Scholar] [CrossRef] [PubMed]
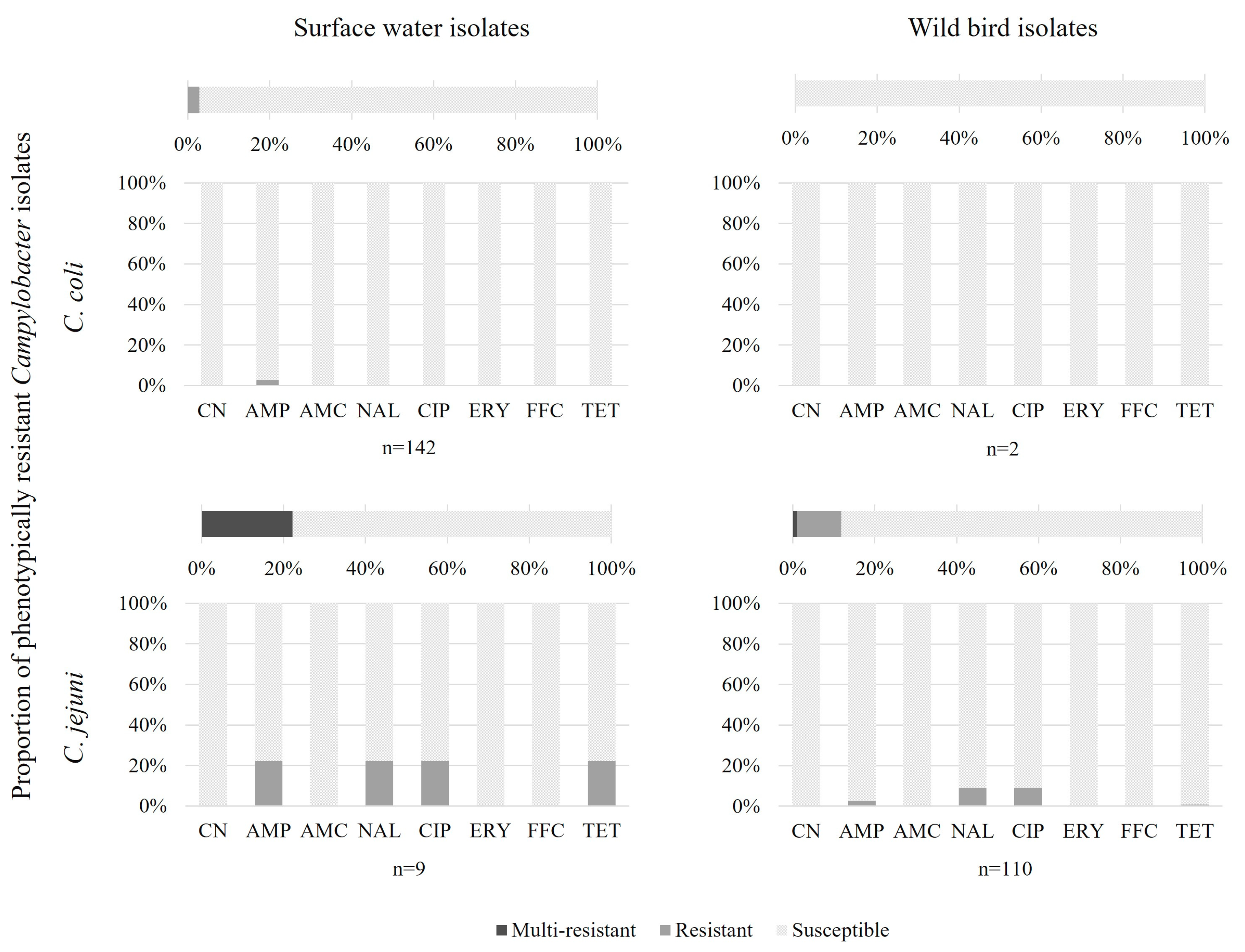


| C. coli | C. jejuni | |||
|---|---|---|---|---|
| AMR Profiles | Bird (n = 2) | Water (n = 142) | Bird (n = 110) | Water (n = 9) |
| Susceptible | 2 (100%) | 138 (97.2%, CI95: 93.0–98.9%) | 97 (88.2%, CI95: 80.8–93.0%) | 7 (78%) |
| AMP | 4 (2.8%, CI95: 1.1–7.0%) | 3 (2.7%, CI95: 0.9–7.7%) | ||
| NAL-CIP | 9 (8.2%, CI95: 4.4–14.8%) | |||
| NAL-CIP-TET | 1 (0.9%, CI95: 0.2–5.0%) | |||
| AMP-NAL-CIP-TET | 2 (22%) | |||
| C. coli | C. jejuni | |||
|---|---|---|---|---|
| AMR Profiles | Bird (n = 2) | Water (n = 142) | Bird (n = 110) | Water (n = 9) |
| Susceptible | 2 (100%) | 74 (52.1%, CI95: 43.9–60.2%) | 10 (9.1%, CI95: 5.0–15.9%) | 1 (11.1%) |
| Aminoglycoside | 48 (33.8%, CI95: 26.5–41.9%) | |||
| Beta-lactam | 16 (11.3%, CI95: 7.1–17.5%) | 87 (79.1%, CI95: 70.6–85.6%) | 6 (66.7%) | |
| Aminoglycoside–Macrolide | 1 (0.9%, CI95:0.2–3.9%) | |||
| Aminoglycoside–Beta-lactam | 2 (1.2%, CI95: 0.4–5%) | 1 (0.9%, CI95: 0.2–5%) | ||
| Beta-lactam–Quinolone | 9 (8.2%, CI95: 4.4–14.8%) | |||
| Beta-lactam–Tetracycline | 1 (0.9%, CI95: 0.2–5%) | |||
| Aminoglycoside–Beta-lactam–Quinolone | 1 (0.9%, CI95: 0.2–3.9%) | |||
| Beta-lactam–Macrolide–Tetracycline | 1 (0.9%, CI95: 0.2–5%) | |||
| Beta-lactam–Quinolone–Tetracycline | 2 (22%) | |||
| Macrolide–Quinolone–Tetracycline | 1 (0.9%, CI95: 0.2–5%) | |||
| C. coli | C. jejuni | ||||
|---|---|---|---|---|---|
| Bird (n = 2) | Water (n = 142) | Bird (n = 110) | Water (n = 9) | ||
| Aminoglycoside | No. of isolates with R phenotype | 0 | 0 | 0 | 0 |
| No. of isolates with R genotype | 0 | 52 | 1 | 0 | |
| Concordance (%) | 100 | 63 | 99 | 100 | |
| Beta-lactam | No. of isolates with R phenotype | 0 | 4 | 3 | 2 |
| No. of isolates with R genotype | 0 | 19 | 99 | 8 | |
| Concordance (%) | 100 | 89 | 13 | 33 | |
| Quinolone | No. of isolates with R phenotype | 0 | 0 | 10 | 2 |
| No. of isolates with R genotype | 0 | 1 | 10 | 2 | |
| Concordance (%) | 100 | 99 | 100 | 100 | |
| Macrolide | No. of isolates with R phenotype | 0 | 0 | 0 | 0 |
| No. of isolates with R genotype | 0 | 1 | 2 | 0 | |
| Concordance (%) | 100 | 99 | 98 | 100 | |
| Tetracycline | No. of isolates with R phenotype | 0 | 0 | 1 | 2 |
| No. of isolates with R genotype | 0 | 0 | 3 | 2 | |
| Concordance (%) | 100 | 100 | 98 | 100 | |
| Phenotype: Susceptible | Phenotype: Resistant | |||||||
|---|---|---|---|---|---|---|---|---|
| Antimicrobial | Genotype: Susceptible | Genotype: Resistant | Genotype: Resistant | Genotype: Susceptible | Cohen’s Kappa Coefficient | 95% CI | Interpretation | |
| C. coli | Aminoglycoside | 92 | 52 | 0 | 0 | - | - | |
| Beta-lactam | 122 | 18 | 1 | 3 | 0.04 | −0.11–0.2 | Slight | |
| Quinolone | 143 | 1 | 0 | 0 | - | - | ||
| Macrolide | 143 | 1 | 0 | 0 | - | - | ||
| Tetracylcine | 144 | 0 | 0 | 0 | - | - | ||
| C. jejuni | Aminoglycoside | 118 | 1 | 0 | 0 | - | - | |
| Beta-lactam | 12 | 102 | 5 | 0 | 0.01 | 0–0.02 | Slight | |
| Quinolone | 107 | 0 | 12 | 0 | 1 | 1 | Almost perfect | |
| Macrolide | 117 | 2 | 0 | 0 | - | - | ||
| Tetracylcine | 114 | 2 | 3 | 0 | 0.74 | 0.40–1 | Substantial | |
| C. coli | C. jejuni | ||||
|---|---|---|---|---|---|
| Virulence Gene | Bird (n = 2) | Water (n = 142) | Bird (n = 110) | Water (n = 9) | |
| Flagellin | flaC | 2 | 142 | 110 | 9 |
| Flagellar proteins | flgB, flgC, flgF, flgG, flgI | 2 | 142 | 110 | 9 |
| Flagellar biosynthesis protein | flhA | 2 | 142 | 110 | 9 |
| Flagellar protein | fliE, fliF, fliG, fliI, fliL, fliM, fliS, fliW | 2 | 142 | 110 | 9 |
| Lipooligosaccharide (LOS) synthesis | hldD | 2 | 142 | 110 | 9 |
| Pseudaminic acid synthesis (Flagellin) | pseB | 2 | 142 | 110 | 9 |
| Chemotaxis protein | cheW | 2 | 141 | 110 | 9 |
| Flagellar protein | flgH, flgJ, flgQ | 2 | 141 | 110 | 9 |
| Flagellar motor protein | motA | 2 | 141 | 110 | 9 |
| Chemotaxis protein | cheV | 2 | 142 | 109 | 9 |
| Flagellar biosynthesis protein | fliR | 2 | 142 | 109 | 9 |
| Chemotaxis protein | cheY | 2 | 140 | 110 | 9 |
| Flagellar protein | flhG | 0 | 142 | 110 | 9 |
| Flagellar protein | flgK | 2 | 141 | 109 | 9 |
| Lipooligosaccharide (LOS) synthesis | gmhA | 0 | 139 | 109 | 8 |
| Pseudaminic acid synthesis (flagellin) | pseC | 0 | 109 | 110 | 9 |
| LOS synthesis | hldE | 2 | 96 | 109 | 9 |
| RNA polymerase factor sigma-54 (Pse) | rpoN | 2 | 87 | 110 | 9 |
| Pseudaminic acid synthesis (flagellin) | pseI | 2 | 65 | 110 | 9 |
| Flagellar protein | flgE | 2 | 43 | 110 | 9 |
| Flagellar protein | flaD | 2 | 94 | 50 | 6 |
| Pseudaminic acid synthesis (flagellin) | pseA | 2 | 40 | 92 | 7 |
| Capsular polysaccharide | kpsT | 2 | 15 | 110 | 9 |
| Chemotaxis protein | cheA | 0 | 9 | 109 | 8 |
| Flagellar motor protein | fliN | 0 | 6 | 110 | 9 |
| Flagellar protein | flgM | 0 | 5 | 110 | 9 |
| Pseudaminic acid synthesis (flagellin) | pseF | 0 | 3 | 110 | 9 |
| Pseudaminic acid synthesis (flagellin) | pseG | 0 | 2 | 110 | 9 |
| Flagellar protein | flaG | 0 | 1 | 110 | 9 |
| Flagellar protein | flgQ | 0 | 1 | 110 | 9 |
| Outer membrane fibronectin-binding protein | cadF | 0 | 0 | 110 | 9 |
| Invasion antigen | ciaB, ciaC | 0 | 0 | 110 | 9 |
| Adherence | eptC | 0 | 0 | 110 | 9 |
| Flagellar protein | flgA, flgP, flgR, flgS | 0 | 0 | 110 | 9 |
| Flagellar biosynthesis protein | flhB, flhF | 0 | 0 | 110 | 9 |
| Flagellar biosynthesis protein | fliA, fliH, fliP, fliY | 0 | 0 | 110 | 9 |
| Lipooligosaccharide (LOS) synthesis | gmhB | 0 | 0 | 110 | 9 |
| Adhesin | jlpA | 0 | 0 | 110 | 9 |
| Flagella motor protein | motB | 0 | 0 | 110 | 9 |
| Adhesin | pebA | 0 | 0 | 110 | 9 |
| Flagellar protein | pflA | 0 | 0 | 110 | 9 |
| Lipooligosaccharide (LOS) synthesis | waaC | 0 | 0 | 110 | 9 |
| Capsular polysaccharide | kpsS | 0 | 0 | 109 | 9 |
| Capsular polysaccharide | kpsD, kpsE | 0 | 1 | 109 | 7 |
| Capsular polysaccharide | kpsM | 0 | 2 | 108 | 7 |
| Capsular synthesis | Cj1419c | 2 | 20 | 84 | 7 |
| Cytolethal distending toxin (CDT) | cdtC | 0 | 0 | 99 | 9 |
| Capsular synthesis | Cj1420c | 2 | 21 | 77 | 6 |
| Capsule protein | kpsC | 0 | 0 | 87 | 6 |
| Capsule biosynthesis and transport | Cj1417c | 0 | 0 | 85 | 7 |
| Lipooligosaccharide (LOS) synthesis | gmhA2 | 0 | 4 | 80 | 4 |
| Capsule synthesis | hddA | 0 | 5 | 78 | 4 |
| Flagella protein | pseH | 0 | 0 | 72 | 7 |
| Major outer membrane protein | porA | 0 | 1 | 57 | 1 |
| Cytolethal distending toxin (CDT) | cdtA | 0 | 0 | 54 | 3 |
| Cytolethal distending toxin (CDT) | cdtB | 0 | 0 | 53 | 3 |
| Lipooligosaccharide (LOS) synthesis | waaF | 0 | 1 | 50 | 4 |
| Lipooligosaccharide (LOS) synthesis | htrB | 0 | 0 | 45 | 7 |
| Capsule protein | cysC | 0 | 0 | 47 | 4 |
| Flagella protein | ptmA, ptmB | 0 | 0 | 42 | 8 |
| Flagella protein | flgD | 0 | 1 | 42 | 6 |
| Lipooligosaccharide (LOS) synthesis | neuC1 | 0 | 35 | 10 | 0 |
| Capsule synthesis | Cj1416c | 0 | 0 | 40 | 4 |
| Flagella protein | fliD | 0 | 1 | 39 | 4 |
| Capsule synthesis | hddC | 0 | 1 | 37 | 1 |
| Motility accessory factor PseE | pseE maf5 | 0 | 0 | 32 | 5 |
| Lipooligosaccharide (LOS) synthesis | Cj1135 | 0 | 0 | 23 | 7 |
| Capsule protein | Cj1427c | 0 | 1 | 24 | 1 |
| Lipooligosaccharide (LOS) synthesis | waaV | 0 | 0 | 24 | 1 |
| Flagellin | flaA, flaB | 0 | 0 | 15 | 4 |
| Motility accessory factor PseD | pseD maf2 | 0 | 0 | 14 | 1 |
| Motility accessory factor | maf4 | 0 | 0 | 9 | 2 |
| Capsule protein | rfbC | 0 | 1 | 9 | 0 |
| Lipooligosaccharide (LOS) synthesis | neuA1, neuB1 | 0 | 0 | 9 | 0 |
| Flagellar protein | fliK | 0 | 0 | 8 | 1 |
| Lipooligosaccharide (LOS) synthesis | Cj1137c, Cj1138 | 0 | 0 | 7 | 0 |
| Lipooligosaccharide (LOS) synthesis | wlaN | 0 | 0 | 6 | 0 |
| Lipooligosaccharide (LOS) synthesis | Cj1136 | 0 | 0 | 5 | 0 |
| Lipooligosaccharide (LOS) synthesis | cstIII | 0 | 0 | 5 | 0 |
| Capsule synthesis | fcl | 0 | 1 | 4 | 0 |
| Capsule protein | Cj1432c, Cj1435c, Cj1436c, Cj1440c | 0 | 1 | 1 | 0 |
| Capsule protein | glf | 0 | 1 | 1 | 0 |
| Capsule protein | kfiD | 0 | 1 | 1 | 0 |
| Capsule protein | Cj1426c | 0 | 0 | 1 | 0 |
| Type IV secretion system protein | Cjp54, virB11, virB4, virB8, virD4 | 0 | 1 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hock, L.; Walczak, C.; Mosser, J.; Ragimbeau, C.; Cauchie, H.-M. Exploring the Role of the Environment as a Reservoir of Antimicrobial-Resistant Campylobacter: Insights from Wild Birds and Surface Waters. Microorganisms 2024, 12, 1621. https://doi.org/10.3390/microorganisms12081621
Hock L, Walczak C, Mosser J, Ragimbeau C, Cauchie H-M. Exploring the Role of the Environment as a Reservoir of Antimicrobial-Resistant Campylobacter: Insights from Wild Birds and Surface Waters. Microorganisms. 2024; 12(8):1621. https://doi.org/10.3390/microorganisms12081621
Chicago/Turabian StyleHock, Louise, Cécile Walczak, Juliette Mosser, Catherine Ragimbeau, and Henry-Michel Cauchie. 2024. "Exploring the Role of the Environment as a Reservoir of Antimicrobial-Resistant Campylobacter: Insights from Wild Birds and Surface Waters" Microorganisms 12, no. 8: 1621. https://doi.org/10.3390/microorganisms12081621
APA StyleHock, L., Walczak, C., Mosser, J., Ragimbeau, C., & Cauchie, H.-M. (2024). Exploring the Role of the Environment as a Reservoir of Antimicrobial-Resistant Campylobacter: Insights from Wild Birds and Surface Waters. Microorganisms, 12(8), 1621. https://doi.org/10.3390/microorganisms12081621






