Surface-Shaving of Staphylococcus aureus Strains and Quantitative Proteomic Analysis Reveal Differences in Protein Abundance of the Surfaceome
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.1.1. Control Conditions: Selection of Buffer, Enzymes, and Digestion Time in a Pilot Step
2.1.2. Control Procedure: Absence of the Proteins Linked to the Knock-Out Genes
2.1.3. Control Strains: Step 1—The Protein Products of Knock-Out Mutant Strains Are Absent
2.1.4. Step 2: Strains of Different Clinical Origin and Pathogenicity
2.2. Cultivation of Bacteria and Preparation of Samples
2.3. RNA Isolation and qPCR
2.4. Lipid-Based Protein Immobilization and Digestion of Proteins into Peptides
2.5. Liquid Chromatography and Tandem Mass Spectrometry (LC-MS/MS)
2.6. Protein Identification and Quantification
2.7. Functional Annotation
2.8. Statistical Evaluation
3. Results
3.1. Pilot Experimient—Surface-Shaving Optimization: PBS Is the Most Suitable Buffer to Perform Surface-Shaving of Intact S. aureus
3.2. Step 1-Mutant Strains Step: The Protein Products of the Knocked-Out Genes Were Not Identified in the Mutant Strains by Surface-Shaving
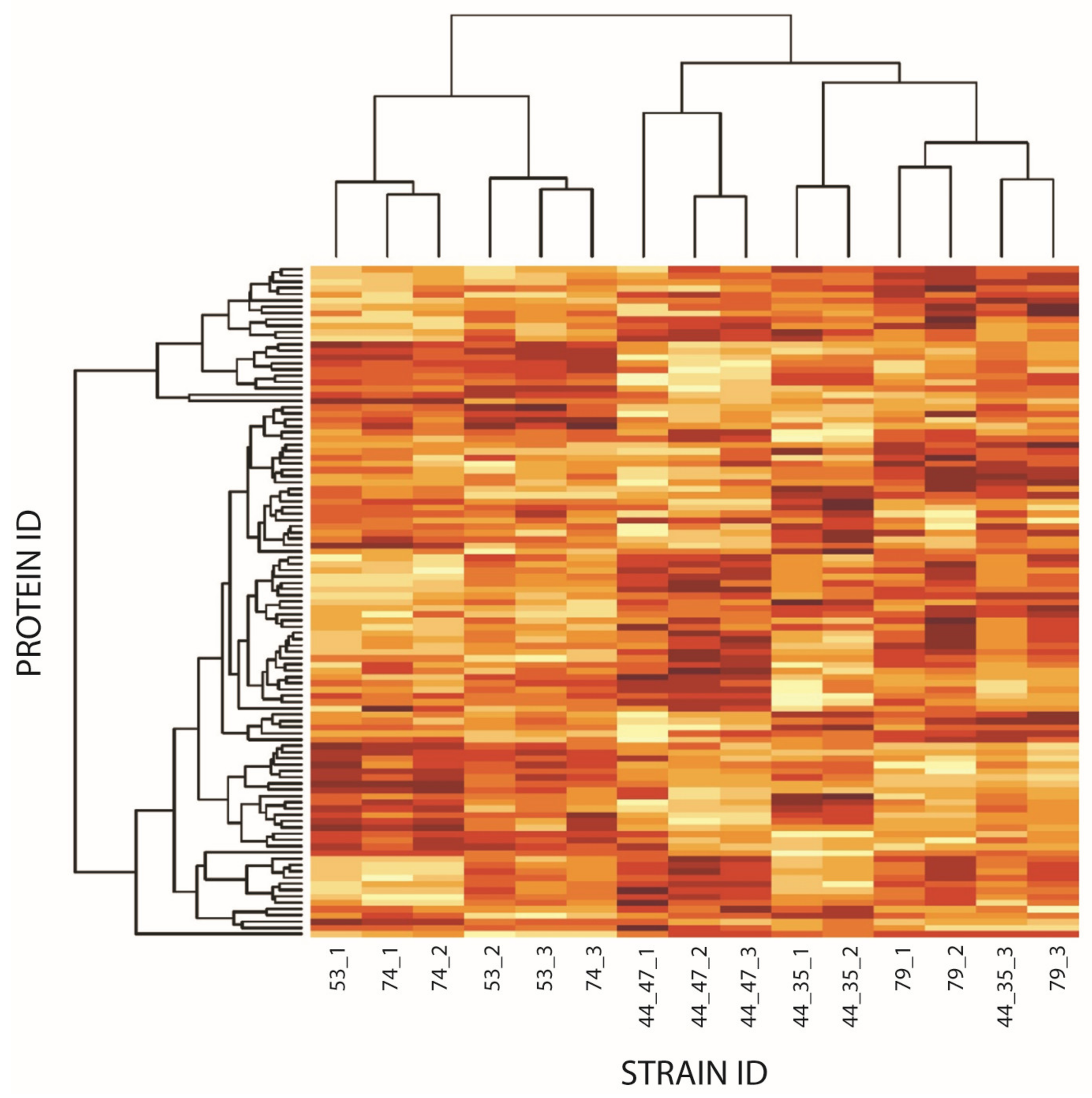
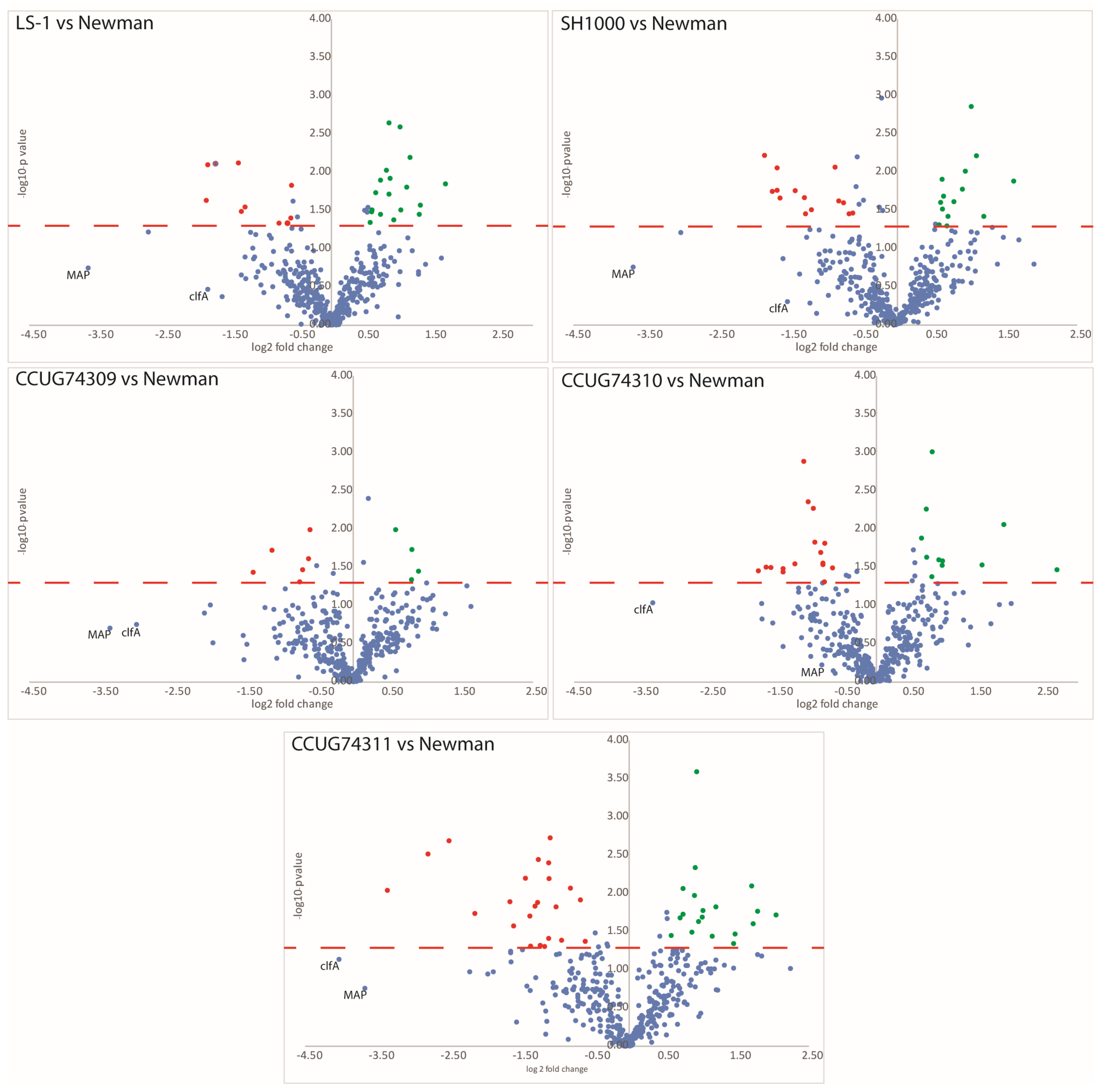
3.3. Step 2—Clinical Strains Step: Differential Protein Expression Is Observed in the Clinical Strains as Compared to the Virulent Newman Strain
3.4. Comparison of Both Steps: Mutant Strains and Clinical Isolates Vs. Newman Strain
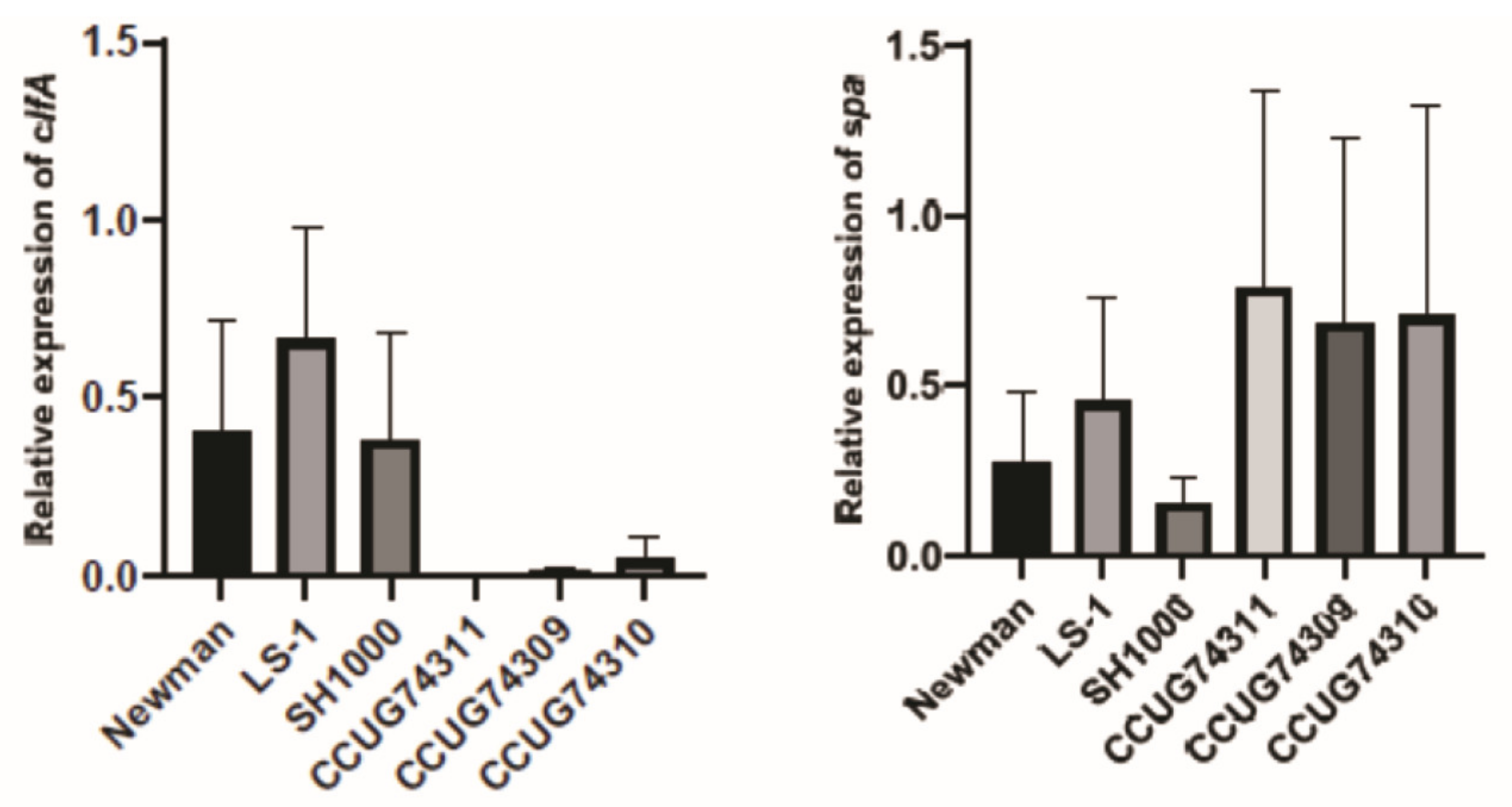
3.5. Clinical Step Results and qPCR—The Expression Trends of Genes and Proteins Are in Accordance within the Clinical Strains
4. Discussion
4.1. Mutant Strains: Plausible Cascade Pattern with Compensational Mechanisms after Deleting Crucial Genes
4.2. Clinical Strains: The Clinical Strains Displayed Higher Protein Expression of SpA, a Key Virulence Factor Involved in S. aureus Immune Evasion
5. Conclusions and Next Steps
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sakr, A.; Brégeon, F.; Mège, J.-L.; Rolain, J.-M.; Blin, O. Staphylococcus aureus Nasal Colonization: An Update on Mechanisms, Epidemiology, Risk Factors, and Subsequent Infections. Front. Microbiol. 2018, 9, 2419. [Google Scholar] [CrossRef]
- Vestergaard, M.; Frees, D.; Ingme, H. Antibiotic Resistance and the MRSA Problem. Microbiol. Spectr. 2019, 7, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A.J.; Lindsay, J.A. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: Implications for vaccine design and host-pathogen interactions. BMC Microbiol. 2010, 10, 173. [Google Scholar] [CrossRef]
- Sause, W.E.; Buckley, P.T.; Strohl, W.R.; Lynch, A.S.; Torres, V.J. Antibody-Based Biologics and Their Promise to Combat Staphylococcus aureus Infections. Trends Pharmacol. Sci. 2016, 37, 231–241. [Google Scholar] [CrossRef]
- Bonar, E.; Wójcik, I.; Wladyka, B. Proteomics in studies of Staphylococcus aureus virulence. Acta Biochim. Pol. 2015, 62, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Budzyńska, A.; Skowron, K.; Kaczmarek, A.; Wietlicka-Piszcz, M.; Gospodarek-Komkowska, E. Virulence Factor Genes and Antimicrobial Susceptibility of Staphylococcus aureus Strains Isolated from Blood and Chronic Wounds. Toxins 2021, 14, 491. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jin, T.; Mohammad, M.; Pullerits, R.; Ali, A. Bacteria and Host Interplay in Staphylococcus aureus Septic Arthritis and Sepsis. Pathogens 2021, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.; Geoghegan, J.; Ganesh, V.; Höök, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.A. Challenges for a universal Staphylococcus aureus vaccine. Clin. Infect. Dis. 2012, 54, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.A. Is there a future for a Staphylococcus vaccine? Vaccine 2012, 30, 2921–2927. [Google Scholar] [CrossRef]
- Duthie, E.S.; Lorenz, L.L. Staphylococcal coagulase: Mode of action and antigenicity. J. Gen. Microbiol. 1952, 6, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Bae, T.; Schneewind, O.; Takeuchi, F.; Hiramatsu, K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: Polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 2008, 190, 300–310. [Google Scholar] [CrossRef]
- Foster, T.J.; McDevitt, D. Surface-associated proteins of Staphylococcus aureus: Their possible roles in virulence. FEMS Microbiol. Lett. 1994, 118, 199–205. [Google Scholar] [CrossRef]
- Josefsson, E.; Hartford, O.; O’Brien, L.; Patti, J.M.; Foster, T. Protection against experimental Staphylococcus aureus arthritis by vaccination with clumping factor A, a novel virulence determinant. J. Infect. Dis. 2001, 184, 1572–1580. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Palmqvist, N.; Foster, T.; Tarkowski, A.; Josefsson, E. Protein A is a virulence factor in Staphylococcus aureus arthritis and septic death. Microb. Pathog. 2002, 33, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Kwiecinski, J.; Jin, T.; Josefsson, E. Surface proteins of Staphylococcus aureus play an important role in experimental skin infection. APMIS 2014, 122, 1240–1250. [Google Scholar] [CrossRef]
- McDevitt, D.; Francois, P.; Vaudaux, P.; Foster, T.J. Identification of the ligand-binding domain of the surface-located fi-brinogen receptor (clumping factor) of Staphylococcus aureus. Mol. Microbiol. 1995, 16, 895–907. [Google Scholar] [CrossRef]
- Josefsson, E.; Higgins, J.; Foster, T.J.; Tarkowski, A. Fibrinogen binding sites P336 and Y338 of clumping factor A are crucial for Staphylococcus aureus virulence. PLoS ONE 2008, 3, e2206. [Google Scholar] [CrossRef]
- Higgins, J.; Loughman, A.; van Kessel, K.P.; van Strijp, J.A.; Foster, T.J. Clumping factor A of Staphylococcus aureus inhibits phagocytosis by human polymorphonuclear leucocytes. FEMS Microbiol. Lett. 2006, 258, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.D.; DeLeo, F.R. Staphylococcus aureus protein A promotes immune suppression. mBio 2013, 4, e00764-13. [Google Scholar] [CrossRef]
- Verdrengh, M.; Tarkowski, A. Role of neutrophils in experimental septicemia and septic arthritis induced by Staphylococcus aureus. Infect. Immun. 1997, 65, 2517–2521. [Google Scholar] [CrossRef]
- Kim, H.K.; Kim, H.Y.; Schneewind, O.; Missiakas, D. Identifying protective antigens of Staphylococcus aureus, a pathogen that suppresses host immune responses. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2011, 10, 3605–3612. [Google Scholar] [CrossRef]
- McDevitt, D.; Francois, P.; Vaudaux, P.; Foster, T.J. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 1994, 11, 237–248. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Ton-That, H.; Su, K.; Schneewind, O. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 2293–2298. [Google Scholar] [CrossRef]
- Jonsson, I.M.; Mazmanian, S.K.; Scheewind, O.; Bremell, T.; Tarkowski, A. The role of Staphylococcus aureus sortase A and sortase B in murine arthritis. Microbes Infect. 2003, 5, 775–780. [Google Scholar] [CrossRef]
- Marchetti, M.; De Bei, O.; Bettati, S.; Campanini, B.; Kovachka, S.; Gianquinto, E.; Spyrakis, F.; Ronda, L. Iron Metabolism at the Interface between Host and Pathogen: From Nutritional Immunity to Antibacterial Development. Int. J. Mol. Sci. 2020, 21, 2145. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Liu, G.; Jensen, E.R.; Lenoy, E.; Schneewind, O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA 2000, 97, 5510–5515. [Google Scholar] [CrossRef]
- Na, M.; Hu, Z.; Mohammad, M.; Stroparo, M.; Ali, A.; Fei, Y.; Jarnesborn, A.; Verhamme, P.; Schneewind, O.; Missiakas, D.; et al. The Expression of von Willebrand Factor-Binding Protein Determines Joint-Invading Capacity of Staphylococcus aureus, a Core Mechanism of Septic Arthritis. mBio 2020, 11, e02472-20. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Stewart, G.C. Regulatory elements of the Staphylococcus aureus protein A (Spa) promoter. J. Bacteriol. 2004, 186, 3738–3748. [Google Scholar] [CrossRef] [PubMed]
- Ventura, C.L.; Malachowa, N.; Hammer, C.H.; Nardone, G.A.; Robinson, M.A.; Kobayashi, S.D.; DeLeo, F.R. Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS ONE 2010, 5, e11634. [Google Scholar] [CrossRef]
- Eliuk, S.; Makarov, A. Evolution of Orbitrap mass spectrometry instrumentation. Annu. Rev. Anal. Chem. 2015, 8, 61–80. [Google Scholar] [CrossRef]
- Heil, L.R.; Damoc, E.; Arrey, T.N.; Pashkova, A.; Denisov, E.; Petzoldt, J.; Peterson, A.C.; Hsu, C.; Searle, B.C.; Shulman, N.; et al. Evaluating the Performance of the Astral Mass Analyzer for Quantitative Proteomics Using Data-Independent Acquisition. J. Proteome Res. 2023, 22, 3290–3300. [Google Scholar] [CrossRef]
- Armengaud, J. Microbiology and proteomics, getting the best of both worlds! Environ. Microbiol. 2013, 15, 12–23. [Google Scholar] [CrossRef]
- Dumas, T.; Martinez Pinna, R.; Lozano, C.; Radau, S.; Pible, O.; Grenga, L.; Armengaud, J. The astounding exhaustiveness and speed of the Astral mass analyzer for highly complex samples is a quantum leap in the functional analysis of microbiomes. Microbiome 2024, 12, 46. [Google Scholar] [CrossRef]
- Rodríguez-Ortega, M.J.; Norais, N.; Bensi, G.; Liberatori, S.; Capo, S.; Mora, M.; Scarselli, M.; Doro, F.; Ferrari, G.; Garaguso, I.; et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat. Biotechnol. 2006, 24, 191–197. [Google Scholar] [CrossRef]
- Glowalla, E.; Tosetti, B.; Krönke, M.; Krut, O. Proteomics-based identification of anchorless cell wall proteins as vaccine candidates against Staphylococcus aureus. Infect. Immun. 2009, 77, 2719–2729. [Google Scholar] [CrossRef] [PubMed]
- Dreisbach, A.; Hempel, K.; Buist, G.; Hecker, M.; Becher, D.; van Dijl, J.M. Profiling the surfacome of Staphylococcus aureus. Proteomics 2010, 10, 3082–3096. [Google Scholar] [CrossRef]
- Solis, N.; Larsen, M.R.; Cordwell, S.J. Improved accuracy of cell surface shaving proteomics in Staphylococcus aureus using a false-positive control. Proteomics 2010, 10, 2037–2049. [Google Scholar] [CrossRef] [PubMed]
- Dreisbach, A.; van Dijl, J.M.; Buist, G. The cell surface proteome of Staphylococcus aureus. Proteomics 2011, 11, 3154–3168. [Google Scholar] [CrossRef]
- Dreisbach, A.; van der Kooi-Pol, M.; Otto, A.; Gronau, K.; Bonarius, H.P.; Westra, H.; Groen, H.; Becher, D.; Hecker, M.; van Dijl, J.M. Surface shaving as a versatile tool to profile global interactions between human serum proteins and the Staphylococcus aureus cell surface. Proteomics 2011, 11, 2921–2930. [Google Scholar] [CrossRef] [PubMed]
- Hempel, K.; Herbst, F.A.; Moche, M.; Hecke, M.; Becher, D. Quantitative proteomic view on secreted, cell surface-associated and cytoplasmic proteins of the methicillin-resistant human pathogen Staphylococcus aureus under iron-limited conditions. J. Proteome. Res. 2011, 10, 1657–1666. [Google Scholar] [CrossRef]
- Karlsson, R.; Davidson, M.; Svensson-Stadler, L.; Karlsson, A.; Olesen, K.; Carlsohn, E.; Moore, E.R.B. Strain-level typing and identification of bacteria using mass spectrometry-based proteomics. J. Proteome Res. 2012, 11, 2710–2720. [Google Scholar] [CrossRef]
- Olaya-Abril, A.; Jiménez-Munguía, I.; Gómez-Gascón, L.; Rodríguez-Ortega, M.J. Surfomics: Shaving live organisms for a fast proteomic identification of surface proteins. J. Proteom. 2014, 97, 164–176. [Google Scholar] [CrossRef]
- Olaya-Abril, A.; González-Reyes, J.A.; Rodríguez-Ortega, M.J. Approaching In Vivo Models of Pneumococcus-Host Interaction: Insights into Surface Proteins, Capsule Production, and Extracellular Vesicles. Pathogens 2021, 10, 1098. [Google Scholar] [CrossRef]
- Wolden, R.; Pain, M.; Karlsson, R.; Karlsson, A.; Fredheim, E.G.A.; Cavanagh, J.P. Identification of surface proteins in a clinical Staphylococcus haemolyticus isolate by bacterial surface shaving. BMC Microbiol. 2020, 20, 80. [Google Scholar]
- Thompson, A.; Schafer, J.; Kuhn, K.; Kienle, S.; Schwarz, J.; Schmidt, G.; Neumann, T.; Hamon, C. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003, 75, 1895–1904. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, R.; Thorell, K.; Hosseini, S.; Kenny, D.; Sihlbom, C.; Sjöling, Å.; Karlsson, A.; Nookaew, I. Comparative Analysis of Two Helicobacter pylori Strains using Genomics and Mass Spectrometry-Based Proteomics. Front. Microbiol. 2016, 7, 1757. [Google Scholar] [CrossRef]
- Gonzales-Siles, L.; Karlsson, R.; Kenny, D.; Karlsson, A.; Sjöling, Å. Proteomic analysis of enterotoxigenic Escherichia coli (ETEC) in neutral and alkaline conditions. BMC Microbiol. 2017, 17, 11. [Google Scholar] [CrossRef]
- Bremell, T.; Lange, S.; Holmdahl, R.; Rydén, C.; Hansson, G.K.; Tarkowski, A. Immunopathological features of rat arthritis. Infect. Immun. 1994, 62, 2334–2344. [Google Scholar]
- Horsburgh, M.J.; Aish, J.L.; White, I.J.; Shaw, L.; Lithgow, J.K.; Foster, S.J. σB modulates virulence determinant expression and stress resistance—Characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 2002, 184, 5457–5467. [Google Scholar] [CrossRef]
- O’Neill, A.J. Staphylococcus aureus SH1000 and 8325-4: Comparative genome sequences of key laboratory strains in staphylococcal research. Lett. Appl. Microbiol. 2010, 51, 358–361. [Google Scholar] [CrossRef]
- Jacobsson, G.; Dashti, S.; Wahlberg, T.; Andersson, R. The epidemiology of and risk factors for invasive Staphylococcus aureus infections in western Sweden. Scand. J. Infect. Dis. 2007, 39, 6–13. [Google Scholar] [CrossRef]
- Ren, Z.; Yu, J.; Du, J.; Zhang, Y.; Hamushan, M.; Jiang, F.; Zhang, F.; Wang, B.; Tang, J.; Shen, H.; et al. A General Map of Transcriptional Expression of Virulence, Metabolism, and Biofilm Formation Adaptive Changes of Staphylococcus aureus when Exposed to Different Antimicrobials. Front. Microbiol. 2022, 13, 825041. [Google Scholar] [CrossRef]
- Xiao, Y.; Hsiao, T.H.; Suresh, U.; Chen, H.I.; Wu, X.; Wolf, S.E.; Chen, Y. A novel significance score for gene selection and ranking. Bioinformatics 2012, 30, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar]
- Hemmadi, V.; Biswas, M. An overview of moonlighting proteins in Staphylococcus aureus infection. Arch. Microbiol. 2021, 203, 481–498. [Google Scholar] [CrossRef]
- Harvey, K.L.; Jarocki, V.M.; Charles, I.G.; Djordjevic, S.P. The Diverse Functional Roles of Elongation Factor Tu (EF-Tu) in Microbial Pathogenesis. Front. Microbiol. 2019, 10, 2351. [Google Scholar] [CrossRef]
- Radke, E.E.; Brown, S.M.; Pelzek, A.J.; Fulmer, Y.; Hernandez, D.N.; Torres, V.J.; Thomsen, I.P.; Chiang, W.K.; Miller, A.O.; Shopsin, B.; et al. Hierarchy of human IgG recognition within the Staphylococcus aureus immunome. Sci. Rep. 2018, 8, 13296. [Google Scholar] [CrossRef]
- Geraci, J.; Neubauer, S.; Pöllath, C.; Hansen, U.; Rizzo, F.; Krafft, C.; Westermann, M.; Hussain, M.; Peters, G.; Pletz, M.W. The Staphylococcus aureus extracellular matrix protein (Emp) has a fibrous structure and binds to different extracellular matrices. Sci. Rep. 2017, 7, 13665. [Google Scholar] [CrossRef]
- Smith, E.J.; Visai, L.; Kerrigan, S.W.; Speziale, P.; Foster, T.J. The Sbi Protein Is a Multifunctional Immune Evasion Factor of Staphylococcus aureus. Infect. Immun. 2011, 79, 3801–3809. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.Y.; Miyamoto, Y.J.; McIntyre, B.W.; Höök, M.; McCrea, K.W.; McDevitt, D.; Brown, E.L. The Staphylococcus aureus Map protein is an immunomodulator that interferes with T cell-mediated responses. J. Clin. Investig. 2002, 110, 1461–1471. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kreikemeyer, B.; McDevitt, D.; Podbielski, A. The role of the map protein in Staphylococcus aureus matrix protein and eukaryotic cell adherence. Int. J. Med. Microbiol. 2002, 292, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
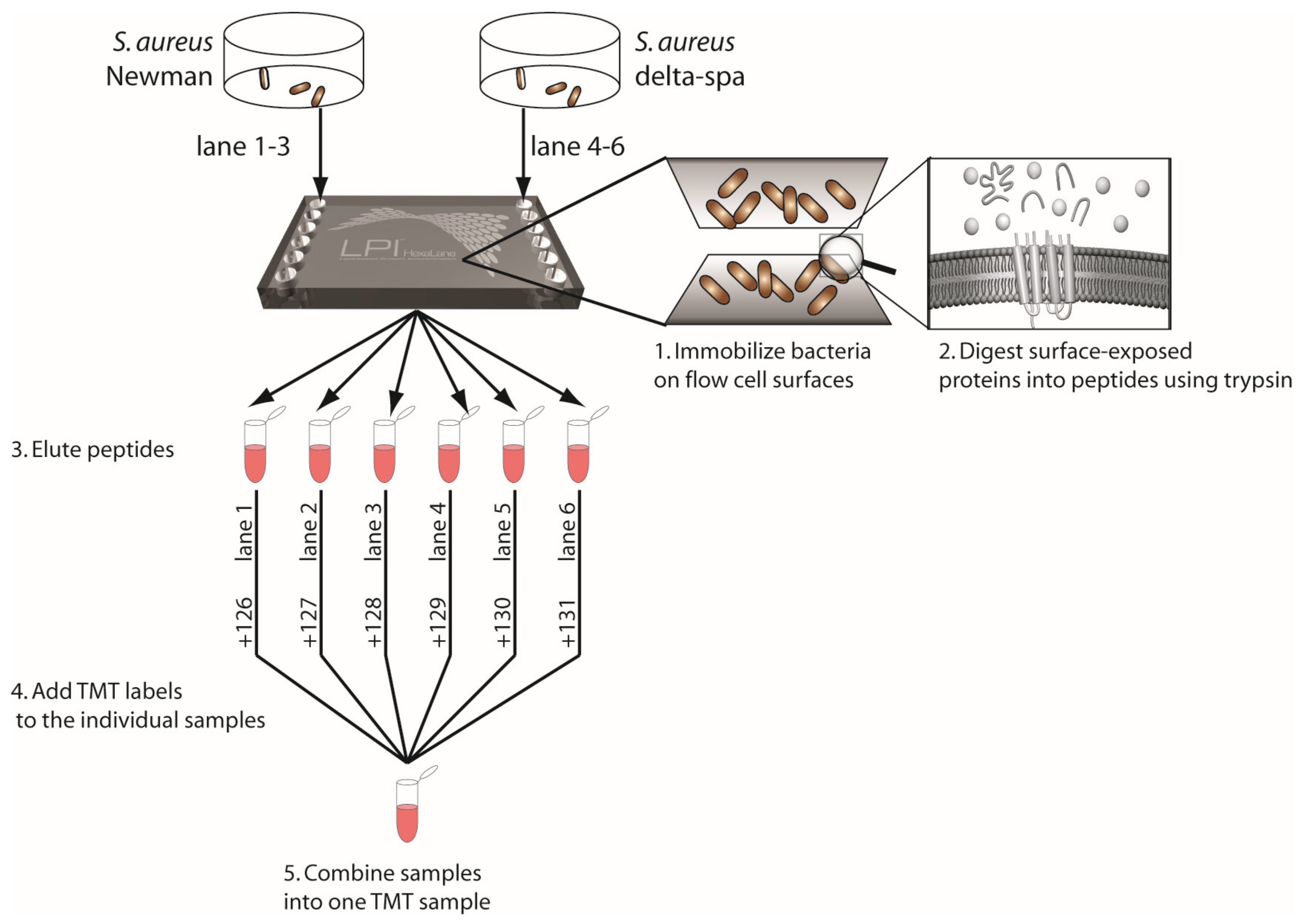
| Step 1—Mutant Strains | Step 2—Clinical Strains | ||
|---|---|---|---|
| Strain | Description | Strain | Description |
| CCUG 74308 “Newman” | Newman strain (Foster lab), Duthie and Lorenz 1952; Baba et al., 2008 [11,12] | CCUG 74308 “Newman” | Newman strain (Foster lab), Duthie and Lorenz 1952; Baba et al., 2008 [11,12] |
| CCUG 74306 “∆spa” | Newman Δspa DU5873 Newman strain (Foster lab) knock-out mutant of Staphylococcal protein A, Kim et al., 2011; Kobayashi and DeLeo 2013 [20,22] | CCUG 74304 “LS-1” | LS-1 Bremell et al., 1994; Verdrengh and Tarkowski 1997 [21,49] |
| CCUG 74305 “∆clfA” | Newman ΔclfA DU5876 Newman strain (Foster lab) knock-out mutant of Clumping factor A, McDewitt et al., 1994, 1995; Higgins et al., 2006 [17,19,23] | CCUG 74303 “SH1000” | SH1000 Horsburgh et al., 2002; O’Neill 2010 [50,51] |
| CCUG 74307 “ΔsrtAsrtB” | Newman ΔsrtAsrtB SKM14 Newman strain Schneewind lab knock-out mutant of Sortase A and B, Mazmanian et al., 2000; Jonsson et al., 2003 [25,27] | CCUG 74309 | Invasive strain, 44_35 Jacobsson et al., 2007 [52] |
| CCUG 74310 | Invasive strain, 44_47 Jacobsson et al., 2007 [52] | ||
| CCUG 74311 | Clinical strain from mild skin and soft tissue infection, 79 Kwiecinski et al., 2014 [16] | ||
| Accession NCBI | Protein Names from S. aureus Newman (NCBI Name) | Gene Names from S. aureus Newman | Subcellular Location (PSORTb) | NM PBS | Δspa PBS | NM 10% TEAB | Δspa 10% TEAB | NM 25% TEAB | Δspa 25% TEAB | NM 100% TEAB | Δspa 100% TEAB |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WP_000728763.1 | Staphylococcal protein A (Spa) | spa NWMN_0055 | Cellwall | 8 | 0 | 12 | 0 | 11 | 0 | 14 | 0 |
| WP_001074508.1 | Bifunctional autolysin | atl NWMN_0922 | Extracellular | 7 | 12 | 18 | 12 | 13 | 11 | 27 | 25 |
| WP_000215236.1 | Asp23/Gls24 family envelope stress response protein | NWMN_2086 | Unknown | 5 | 7 | 6 | 4 | 7 | 7 | 7 | 6 |
| WP_001056178.1 | MSCRAMM family adhesin clumping factor A ClfA | clfA NWMN_0756 | Cellwall | 4 | 4 | 5 | 5 | 5 | 4 | 7 | 7 |
| WP_001040568.1 | Elongation factor Tu (EF-Tu) | tuf NWMN_0510 | Cytoplasmic * | 3 | 4 | 5 | 3 | 4 | 4 | 7 | 8 |
| WP_001041586.1 | Heme uptake protein IsdB | isdB frpB sasJ sirH NWMN_1040 | Cellwall | 3 | 5 | 6 | 3 | 4 | 3 | 11 | 10 |
| WP_001549158.1 | Extracellular adherence protein Eap/Map | NWMN_1872 | Cytoplasmic membrane | 3 | 2 | 9 | 3 | 6 | 4 | 13 | 10 |
| WP_000383814.1 | DUF948 domain-containing protein | NWMN_1632 | Cytoplasmic | 2 | 3 | 3 | 1 | 4 | 3 | 7 | 5 |
| WP_000745871.1 | MSCRAMM family adhesin clumping factor B ClfB | clfB NWMN_2529 | Cellwall | 2 | 2 | 3 | 1 | 3 | 2 | 5 | 3 |
| WP_000160859.1 | LPXTG-anchored heme-scavanging protein IsdA | isdA frpA stbA NWMN_1041 | Cellwall | 2 | 3 | 3 | 3 | 2 | 3 | 4 | 3 |
| Σ Proteins > 2 peptides | 10 | 13 | 26 | 11 | 28 | 23 | 89 | 96 |
| Accession Number | NCBI Protein Name | Protein Names from S. aureus Newman | Subcellular Location | Gene Name from S. aureus Newman | Fold Change (FC) vs. Newman | ||
|---|---|---|---|---|---|---|---|
| ∆clfA | ΔsrtAsrtB | ∆spa | |||||
| WP_000728763.1 | Staphylococcal protein A | Immunoglobulin G-binding protein A (IgG-binding protein A) (Staphylococcal protein A) (Spa) | extracellular region; cell wall; membrane | spa NWMN_0055 | - | −11 | −25 |
| WP_001056178.1 | MSCRAMM family adhesin clumping factor ClfA | Clumping factor A (Fibrinogen receptor A) (Fibrinogen-binding protein A) | extracellular region; cell wall; membrane | clfA NWMN_0756 | −24 | −10 | - |
| WP_001549158.1 | Extracellular adherence protein Eap/Map | 65 kDa membrane protein (map-ND2C) | plasma membrane | NWMN_1872 | - | −8 | - |
| WP_000745871.1 | MSCRAMM family adhesin clumping factor ClfB | Clumping factor B (Fibrinogen receptor B) (Fibrinogen-binding protein B) | extracellular region; cell wall; integral component of membrane | clfB NWMN_2529 | - | −3 | 2 |
| Accession Number | NCBI Protein Name | Protein Name from S. aureus Newman | Subcellular Location | Gene Name from S. aureus Newman WT | Fold Change (FC) vs. Newman | ||||
|---|---|---|---|---|---|---|---|---|---|
| LS-1 | SH1000 | CCUG 74310 | CCUG 74309 | CCUG 74311 | |||||
| WP_000728763.1 | Staphylococcal protein A | Immunoglobulin G-binding protein A (IgG-binding protein A) (Staphylococcal protein A) (Spa) | extracellular region; cell wall; membrane | spa NWMN_0055 | −4 | −2 | 2 | 2 | 3 |
| WP_000241588.1 | Asp23/Gls24 family envelope stress response protein | Uncharacterized protein | NWMN_1430 | 2 | 2 | 3 | 2 | 3 | |
| WP_000069282.1 | Elastin-binding protein EbpS | Elastin-binding protein EbpS | plasma membrane; integral membrane | ebpS NWMN_1389 | 2 | 2 | 1 | 2 | 2 |
| WP_000728056.1 | Extracellular matrix protein-binding adhesin Emp | Extracellular matrix protein-binding protein Emp | cell surface | ssp NWMN_0758 | 2 | 2 | −2 | −3 | −2 |
| WP_000792564.1 | Immunoglobulin-binding protein Sbi | Immunoglobulin-binding protein Sbi | extracellular region; plasma membrane | sbi NWMN_2317 | 2 | 1 | −1 | −2 | −2 |
| WP_000610306.1 | MSCRAMM family adhesion SdrE | Serine-aspartate repeat-containing protein E | extracellular region; cell wall; integral membrane | sdrE NWMN_0525 | −1 | −2 | −3 | −1 | −3 |
| WP_000739209.1 | Complement convertase inhibitor Ecb | Efb-c domain-containing protein | extracellular space | NWMN_1066 | −3 | −2 | −1 | −4 | −3 |
| WP_001549158.1 | Extracellular adherence protein Eap/Map | 65 kDa membrane protein (map-ND2C) | plasma membrane | NWMN_1872 | −12 | −13 | −2 | −10 | −13 |
| WP_001056178.1 | MSCRAMM family adhesion clumping factor ClfA | Clumping factor A (Fibrinogen receptor A) (Fibrinogen-binding protein A) | extracellular region; cell wall; membrane | clfA NWMN_0756 | −4 | −3 | −10 | −8 | −16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karlsson, A.; Alarcón, L.A.; Piñeiro-Iglesias, B.; Jacobsson, G.; Skovbjerg, S.; Moore, E.R.B.; Kopparapu, P.K.; Jin, T.; Karlsson, R. Surface-Shaving of Staphylococcus aureus Strains and Quantitative Proteomic Analysis Reveal Differences in Protein Abundance of the Surfaceome. Microorganisms 2024, 12, 1725. https://doi.org/10.3390/microorganisms12081725
Karlsson A, Alarcón LA, Piñeiro-Iglesias B, Jacobsson G, Skovbjerg S, Moore ERB, Kopparapu PK, Jin T, Karlsson R. Surface-Shaving of Staphylococcus aureus Strains and Quantitative Proteomic Analysis Reveal Differences in Protein Abundance of the Surfaceome. Microorganisms. 2024; 12(8):1725. https://doi.org/10.3390/microorganisms12081725
Chicago/Turabian StyleKarlsson, Anders, Leonarda Achá Alarcón, Beatriz Piñeiro-Iglesias, Gunnar Jacobsson, Susann Skovbjerg, Edward R. B. Moore, Pradeep Kumar Kopparapu, Tao Jin, and Roger Karlsson. 2024. "Surface-Shaving of Staphylococcus aureus Strains and Quantitative Proteomic Analysis Reveal Differences in Protein Abundance of the Surfaceome" Microorganisms 12, no. 8: 1725. https://doi.org/10.3390/microorganisms12081725
APA StyleKarlsson, A., Alarcón, L. A., Piñeiro-Iglesias, B., Jacobsson, G., Skovbjerg, S., Moore, E. R. B., Kopparapu, P. K., Jin, T., & Karlsson, R. (2024). Surface-Shaving of Staphylococcus aureus Strains and Quantitative Proteomic Analysis Reveal Differences in Protein Abundance of the Surfaceome. Microorganisms, 12(8), 1725. https://doi.org/10.3390/microorganisms12081725







