Haloarchaea as Promising Chassis to Green Chemistry
Abstract
:1. Introduction
2. Presentation of Haloarchaea
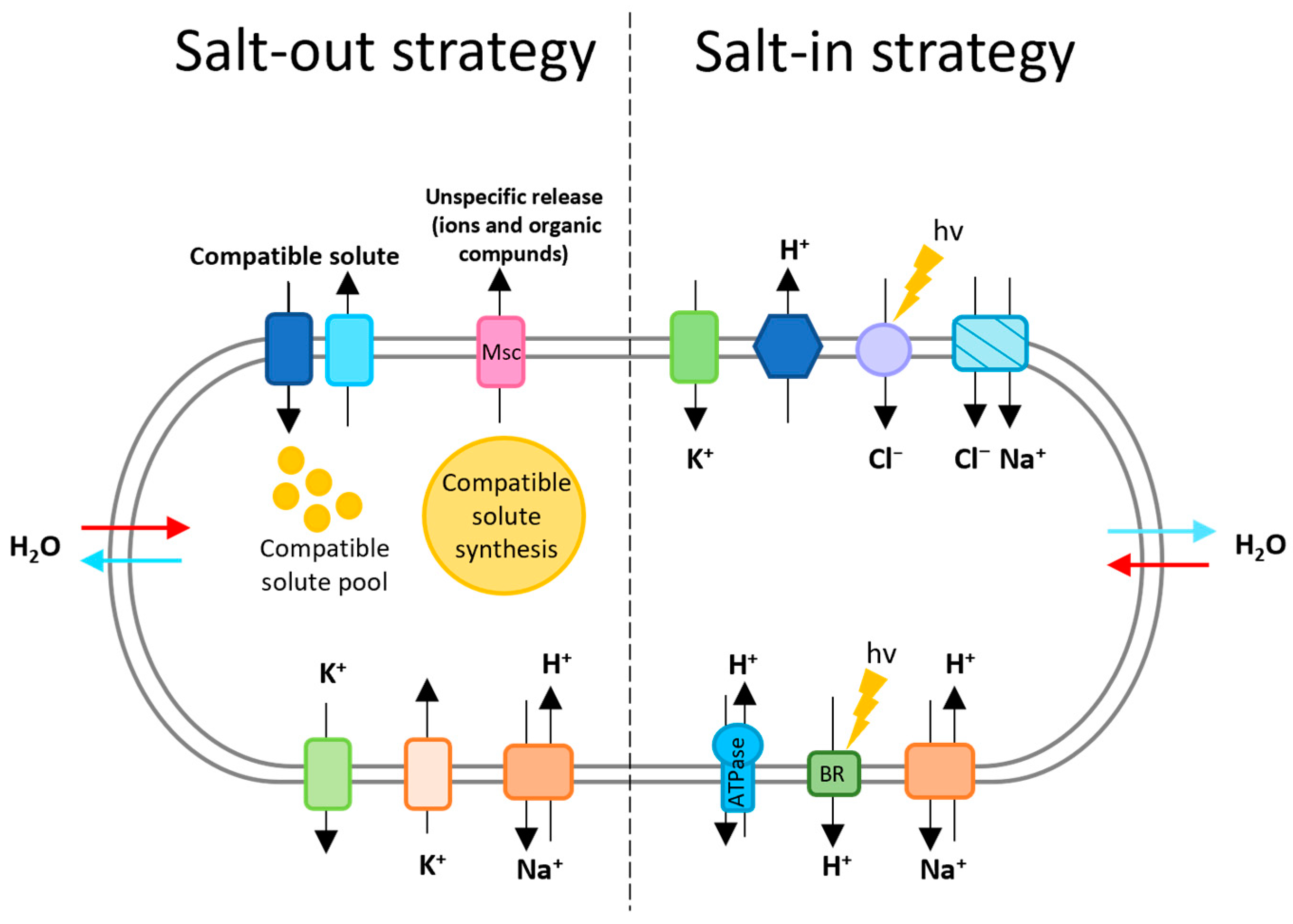
2.1. Osmotic Adaptation
2.2. Haloarchaea: Invaluable Resources of Enzymes for Industry
3. Haloarchaea Chassis: Current State and Challenges
3.1. Specifications for a Haloarchaea Chassis
3.2. The Number of Potential Haloarchaea Chassis Is Limited by the Genetic Tools Available
3.3. Haloarchaea Chassis Construction: Genetic Tools Available
3.3.1. Transformation Methods
3.3.2. Selectable Markers
- Uracil auxotrophy: this is based on the pyrE2 gene of Hfx. volcanii, encoding an oroate phosphoribosyl transferase, based on the pyrF gene of Haloferax mediterranei/Har. hispanica or based on ura3 of Hbt. salinarum, encoding an orotidine-5′-phosphate decarboxylase. This selectable marker allows counter-selection by the addition of 5-fluoro-orotic acid (5-FOA), which inhibits the growth of strains with the wild-type pyrF gene. A loss of growth results from the inhibition of nucleic acid synthesis [4,12,91].
- Leucine auxotrophy: this is based on the leuB gene of Hfx. volcanii, which encodes 3-isopropylmalate dehydrogenase, essential for leucine biosynthesis [32].
- Tryptophan auxotrophy: this is based on the Hfx. volcanii trpA gene encoding one of the two subunits of tryptophan synthase [32].
- Methionine auxotrophy: this is based on the Hfx. volcanii metX gene encoding a homoserine O-acetyltransferase [12].
- Histidine auxotrophy: this is based on the Hfx. volcanii hisC gene encoding a histidinol-phosphate aminotransferase [12].
3.3.3. Promoters
3.3.4. Vectors
3.3.5. Gene Knock-Out Methods
4. Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chapman, J.; Ismail, A.; Dinu, C. Industrial Applications of Enzymes: Recent Advances, Techniques, and Outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef]
- Espina, G.; Muñoz-Ibacache, S.A.; Cáceres-Moreno, P.; Amenabar, M.J.; Blamey, J.M. From the Discovery of Extremozymes to an Enzymatic Product: Roadmap Based on Their Applications. Front. Bioeng. Biotechnol. 2022, 9, 752281. [Google Scholar] [CrossRef] [PubMed]
- Delgado-García, M.; Valdivia-Urdiales, B.; Aguilar-González, C.N.; Contreras-Esquivel, J.C.; Rodríguez-Herrera, R. Halophilic Hydrolases as a New Tool for the Biotechnological Industries. J. Sci. Food Agric. 2012, 92, 2575–2580. [Google Scholar] [CrossRef]
- Allers, T.; Barak, S.; Liddell, S.; Wardell, K.; Mevarech, M. Improved Strains and Plasmid Vectors for Conditional Overexpression of His-Tagged Proteins in Haloferax volcanii. Appl. Environ. Microbiol. 2010, 76, 1759–1769. [Google Scholar] [CrossRef]
- Munawar, N.; Engel, P.C. Halophilic Enzymes: Characteristics, Structural Adaptation and Potential Applications for Biocatalysis. Curr. Biotechnol. 2013, 2, 334–344. [Google Scholar] [CrossRef]
- Matarredona, L.; Camacho, M.; Zafrilla, B.; Bonete, M.-J.; Esclapez, J. The Role of Stress Proteins in Haloarchaea and Their Adaptive Response to Environmental Shifts. Biomolecules 2020, 10, 1390. [Google Scholar] [CrossRef]
- Moopantakath, J.; Imchen, M.; Anju, V.T.; Busi, S.; Dyavaiah, M.; Martínez-Espinosa, R.M.; Kumavath, R. Bioactive Molecules from Haloarchaea: Scope and Prospects for Industrial and Therapeutic Applications. Front. Microbiol. 2023, 14, 1113540. [Google Scholar] [CrossRef]
- Wood, R.B.; Talling, J.F. Chemical and Algal Relationships in a Salinity Series of Ethiopian Inland Waters. Hydrobiologia 1988, 158, 29–67. [Google Scholar] [CrossRef]
- Oren, A. Life at High Salt Concentrations. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 263–282. ISBN 978-0-387-25492-0. [Google Scholar]
- Becker, E.A.; Seitzer, P.M.; Tritt, A.; Larsen, D.; Krusor, M.; Yao, A.I.; Wu, D.; Madern, D.; Eisen, J.A.; Darling, A.E.; et al. Phylogenetically Driven Sequencing of Extremely Halophilic Archaea Reveals Strategies for Static and Dynamic Osmo-Response. PLoS Genet. 2014, 10, e1004784. [Google Scholar] [CrossRef]
- Thombre, R.S.; Shinde, V.D.; Oke, R.S.; Dhar, S.K.; Shouche, Y.S. Biology and Survival of Extremely Halophilic Archaeon Haloarcula marismortui RR12 Isolated from Mumbai Salterns, India in Response to Salinity Stress. Sci. Rep. 2016, 6, 25642. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.A.; Albers, S.-V.; Atomi, H.; Allers, T. Model Organisms for Genetics in the Domain Archaea: Methanogens, Halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 2011, 35, 577–608. [Google Scholar] [CrossRef]
- Hermann, L.; Mais, C.-N.; Czech, L.; Smits, S.H.J.; Bange, G.; Bremer, E. The Ups and Downs of Ectoine: Structural Enzymology of a Major Microbial Stress Protectant and Versatile Nutrient. Biol. Chem. 2020, 401, 1443–1468. [Google Scholar] [CrossRef] [PubMed]
- Orellana, R.; Macaya, C.; Bravo, G.; Dorochesi, F.; Cumsille, A.; Valencia, R.; Rojas, C.; Seeger, M. Living at the Frontiers of Life: Extremophiles in Chile and Their Potential for Bioremediation. Front. Microbiol. 2018, 9, 2309. [Google Scholar] [CrossRef]
- Kokoeva, M.V. A Novel Mode of Sensory Transduction in Archaea: Binding Protein-Mediated Chemotaxis towards Osmoprotectants and Amino Acids. EMBO J. 2002, 21, 2312–2322. [Google Scholar] [CrossRef]
- Youssef, N.H.; Savage-Ashlock, K.N.; McCully, A.L.; Luedtke, B.; Shaw, E.I.; Hoff, W.D.; Elshahed, M.S. Trehalose/2-Sulfotrehalose Biosynthesis and Glycine-Betaine Uptake Are Widely Spread Mechanisms for Osmoadaptation in the Halobacteriales. ISME J. 2014, 8, 636–649. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, C.D. Potential for Industrial Products from the Halophilic Archaea. J. Ind. Microbiol. Biotechnol. 2011, 38, 1635–1647. [Google Scholar] [CrossRef] [PubMed]
- DasSarma, P.; Zamora, R.C.; Müller, J.A.; DasSarma, S. Genome-Wide Responses of the Model Archaeon Halobacterium Sp. Strain NRC-1 to Oxygen Limitation. J. Bacteriol. 2012, 194, 5530–5537. [Google Scholar] [CrossRef] [PubMed]
- Weerasooriya, R.R.; Liyanage, L.P.K.; Rathnappriya, R.H.K.; Bandara, W.B.M.A.C.; Perera, T.A.N.T.; Gunarathna, M.H.J.P.; Jayasinghe, G.Y. Industrial Water Conservation by Water Footprint and Sustainable Development Goals: A Review. Environ. Dev. Sustain. 2021, 23, 12661–12709. [Google Scholar] [CrossRef]
- Cho, E.-S.; Cha, I.-T.; Roh, S.W.; Seo, M.-J. Haloferax litoreum Sp. Nov., Haloferax marinisediminis Sp. Nov., and Haloferax marinum Sp. Nov., Low Salt-Tolerant Haloarchaea Isolated from Seawater and Sediment. Antonie van Leeuwenhoek 2021, 114, 2065–2082. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, Y.; Chen, G.-Q. Halophiles as Chassis for Bioproduction. Adv. Biosyst. 2018, 2, 1800088. [Google Scholar] [CrossRef]
- Díaz, S.; Pérez-Pomares, F.; Pire, C.; Ferrer, J.; Bonete, M.-J. Gene Cloning, Heterologous Overexpression and Optimized Refolding of the NAD-Glutamate Dehydrogenase from Haloferax Mediterranei. Extremophiles 2006, 10, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Calero, P.; Nikel, P.I. Chasing Bacterial Chassis for Metabolic Engineering: A Perspective Review from Classical to Non-Traditional Microorganisms. Microb. Biotechnol. 2019, 12, 98–124. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Choe, D.; Lee, D.-H.; Cho, B.-K. Engineering Biology to Construct Microbial Chassis for the Production of Difficult-to-Express Proteins. Int. J. Mol. Sci. 2020, 21, 990. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. The Family Halobacteriaceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 41–121. ISBN 978-3-642-38953-5. [Google Scholar]
- National Center for Biotechnology Information (NCBI). Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 13 July 2024).
- Roh, S.W.; Nam, Y.-D.; Chang, H.-W.; Sung, Y.; Kim, K.-H.; Oh, H.-M.; Bae, J.-W. Halalkalicoccus jeotgali Sp. Nov., a Halophilic Archaeon from Shrimp Jeotgal, a Traditional Korean Fermented Seafood. Int. J. Syst. Evol. Microbiol. 2007, 57, 2296–2298. [Google Scholar] [CrossRef]
- Xue, Y.; Fan, H.; Ventosa, A.; Grant, W.D.; Jones, B.E.; Cowan, D.A.; Ma, Y. Halalkalicoccus tibetensis Gen. Nov., Sp. Nov., Representing a Novel Genus of Haloalkaliphilic Archaea. Int. J. Syst. Evol. Microbiol. 2005, 55, 2501–2505. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-B.; Narsing Rao, M.P.; Yin, X.-Q.; Li, X.; Salam, N.; Zhang, Y.; Alkhalifah, D.H.M.; Hozzein, W.N.; Li, W.-J. Description of Halegenticoccus soli Gen. Nov., Sp. Nov., a Halophilic Archaeon Isolated from a Soil Sample of Ebi Lake. Extremophiles 2019, 23, 521–528. [Google Scholar] [CrossRef]
- Liu, B.-B.; Salam, N.; Cheng, S.; Zhang, W.; Zhou, Y.; Guo, S.; Li, W.-J. Halegenticoccus tardaugens Sp. Nov., an Extremely Halophilic Archaeon Isolated from a Saline Soil. Extremophiles 2021, 25, 483–492. [Google Scholar] [CrossRef]
- Cline, S.W.; Doolittle, W.F. Transformation of Members of the Genus Haloarcula with Shuttle Vectors Based on Halobacterium halobium and Haloferax volcanii Plasmid Replicons. J. Bacteriol. 1992, 174, 1076–1080. [Google Scholar] [CrossRef]
- Atomi, H.; Imanaka, T.; Fukui, T. Overview of the Genetic Tools in the Archaea. Front. Microbiol. 2012, 3, 00337. [Google Scholar] [CrossRef]
- Juez, G.; Rodriguez-Valera, F.; Ventosa, A.; Kushner, D.J. Haloarcula hispanica Spec. Nov. and Haloferax gibbonsii Spec, Nov., Two New Species of Extremely Halophilic Archaebacteria. Syst. Appl. Microbiol. 1986, 8, 75–79. [Google Scholar] [CrossRef]
- Nakamura, S.; Nakasone, K.; Takashina, T. Genetics and Genomics of Triangular Disc-Shaped Halophilic Archaeon Haloarcula japonica Strain TR-1. In Extremophiles Handbook; Horikoshi, K., Ed.; Springer Japan: Tokyo, Japan, 2011; pp. 363–381. ISBN 978-4-431-53897-4. [Google Scholar]
- Baliga, N.S.; Bonneau, R.; Facciotti, M.T.; Pan, M.; Glusman, G.; Deutsch, E.W.; Shannon, P.; Chiu, Y.; Weng, R.S.; Gan, R.R.; et al. Genome Sequence of Haloarcula marismortui: A Halophilic Archaeon from the Dead Sea. Genome Res. 2004, 14, 2221–2234. [Google Scholar] [CrossRef]
- Kixmüller, D.; Greie, J.-C. Construction and Characterization of a Gradually Inducible Expression Vector for Halobacterium Salinarum, Based on the Kdp Promoter. Appl. Environ. Microbiol. 2012, 78, 2100–2105. [Google Scholar] [CrossRef] [PubMed]
- Haloarchaea. Available online: https://haloarchaea.com/halohandbook/ (accessed on 13 July 2024).
- Patenge, N.; Haase, A.; Bolhuis, H.; Oesterhelt, D. The Gene for a Halophilic β-Galactosidase (bgaH) of Haloferax alicantei as a Reporter Gene for Promoter Analyses in Halobacterium Salinarum. Mol. Microbiol. 2000, 36, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Shimoshige, H.; Yamada, T.; Minegishi, H.; Echigo, A.; Shimane, Y.; Kamekura, M.; Itoh, T.; Usami, R. Halobaculum magnesiiphilum Sp. Nov., a Magnesium-Dependent Haloarchaeon Isolated from Commercial Salt. Int. J. Syst. Evol. Microbiol. 2013, 63, 861–866. [Google Scholar] [CrossRef]
- Minegishi, H.; Echigo, A.; Kuwahara, A.; Shimane, Y.; Kamekura, M.; Itoh, T.; Ohkuma, M.; Usami, R. Halocalculus aciditolerans Gen. Nov., Sp. Nov., an Acid-Tolerant Haloarchaeon Isolated from Commercial Salt. Int. J. Syst. Evol. Microbiol. 2015, 65, 1640–1645. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Pal, Y.; Kumar, P.; Krishnamurthi, S. Halocatena pleomorpha Gen. Nov. Sp. Nov., an Extremely Halophilic Archaeon of Family Halobacteriaceae Isolated from Saltpan Soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 3693–3700. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y.; Messina, E.; Smedile, F.; Roman, P.; Damsté, J.S.S.; Ciordia, S.; Mena, M.C.; Ferrer, M.; Golyshin, P.N.; Kublanov, I.V.; et al. Discovery of Anaerobic Lithoheterotrophic Haloarchaea, Ubiquitous in Hypersaline Habitats. ISME J. 2017, 11, 1245–1260. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-W.; Wu, Y.-H.; Wang, C.-S.; Oren, A.; Zhou, P.-J.; Wu, M. Haloferax larsenii Sp. Nov., an Extremely Halophilic Archaeon from a Solar Saltern. Int. J. Syst. Evol. Microbiol. 2007, 57, 717–720. [Google Scholar] [CrossRef]
- Haque, R.U.; Paradisi, F.; Allers, T. Haloferax volcanii for Biotechnology Applications: Challenges, Current State and Perspectives. Appl. Microbiol. Biot. 2020, 104, 1371–1382. [Google Scholar] [CrossRef]
- Cline, S.W.; Schalkwyk, L.C.; Doolittle, W.F. Transformation of the Archaebacterium Halobacterium Volcanii with Genomic DNA. J. Bacteriol. 1989, 171, 4987–4991. [Google Scholar] [CrossRef]
- Ortenberg, R.; Rozenblatt-Rosen, O.; Mevarech, M. The Extremely Halophilic Archaeon Haloferax volcanii Has Two Very Different Dihydrofolate Reductases: Haloferax volcanii Dihydrofolate Reductases. Mol. Microbiol. 2002, 35, 1493–1505. [Google Scholar] [CrossRef]
- Large, A.; Stamme, C.; Lange, C.; Duan, Z.; Allers, T.; Soppa, J.; Lund, P.A. Characterization of a Tightly Controlled Promoter of the Halophilic Archaeon Haloferax volcanii and Its Use in the Analysis of the Essential Cct1 Gene. Mol. Microbiol. 2007, 66, 1092–1106. [Google Scholar] [CrossRef] [PubMed]
- Haque, R.U.; Paradisi, F.; Allers, T. Haloferax volcanii as Immobilised Whole Cell Biocatalyst: New Applications for Halophilic Systems. Appl. Microbiol. Biotechnol. 2019, 103, 3807–3817. [Google Scholar] [CrossRef]
- Lam, W.L.; Doolittle, W.F. Shuttle Vectors for the Archaebacterium Halobacterium volcanii. Proc. Natl. Acad. Sci. USA 1989, 86, 5478–5482. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Soppa, J. Dihydrofolate Reductase (DHFR) Reporter Enzyme Assay for Haloferax volcanii; Research Square: Durham, NC, USA, 2019. [Google Scholar] [CrossRef]
- Reuter, C.J.; Maupin-Furlow, J.A. Analysis of Proteasome-Dependent Proteolysis in Haloferax volcanii Cells, Using Short-Lived Green Fluorescent Proteins. Appl. Environ. Microbiol. 2004, 70, 7530–7538. [Google Scholar] [CrossRef] [PubMed]
- Savage, K.N.; Krumholz, L.R.; Oren, A.; Elshahed, M.S. Halosarcina pallida Gen. Nov., Sp. Nov., a Halophilic Archaeon from a Low-Salt, Sulfide-Rich Spring. Int. J. Syst. Evol. Microbiol. 2008, 58, 856–860. [Google Scholar] [CrossRef]
- Durán-Viseras, A.; Sánchez-Porro, C.; Ventosa, A. Haloglomus irregulare Gen. Nov., Sp. Nov., a New Halophilic Archaeon Isolated from a Marine Saltern. Microorganisms 2020, 8, 206. [Google Scholar] [CrossRef]
- Mou, Y.-Z.; Qiu, X.-X.; Zhao, M.-L.; Cui, H.-L.; Oh, D.; Dyall-Smith, M.L. Halohasta litorea Gen. Nov. Sp. Nov., and Halohasta litchfieldiae Sp. Nov., Isolated from the Daliang Aquaculture Farm, China and from Deep Lake, Antarctica, Respectively. Extremophiles 2012, 16, 895–901. [Google Scholar] [CrossRef]
- Inoue, K.; Itoh, T.; Ohkuma, M.; Kogure, K. Halomarina oriensis Gen. Nov., Sp. Nov., a Halophilic Archaeon Isolated from a Seawater Aquarium. Int. J. Syst. Evol. Microbiol. 2011, 61, 942–946. [Google Scholar] [CrossRef]
- Echigo, A.; Minegishi, H.; Shimane, Y.; Kamekura, M.; Itoh, T.; Usami, R. Halomicroarcula pellucida Gen. Nov., Sp. Nov., a Non-Pigmented, Transparent-Colony-Forming, Halophilic Archaeon Isolated from Solar Salt. Int. J. Syst. Evol. Microbiol. 2013, 63, 3556–3562. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xu, Y.; Sun, S.; Liu, J.; Chen, F. Halomicrococcus hydrotolerans Gen. Nov., Sp. Nov., an Extremely Halophilic Archaeon Isolated from a Subterranean Salt Deposit. Int. J. Syst. Evol. Microbiol. 2020, 70, 4425–4431. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Minegishi, H.; Echigo, A.; Shimane, Y.; Kamekura, M.; Itoh, T.; Ohkuma, M.; Tanaka, A.; Takahashi-Ando, N.; Fukushima, Y.; et al. Haloparvum alkalitolerans Sp. Nov., Alkali-Tolerant Haloarchaeon Isolated from Commercial Salt. Int. J. Syst. Evol. Microbiol. 2016, 66, 5314–5319. [Google Scholar] [CrossRef]
- Chen, S.; Liu, H.-C.; Zhou, J.; Xiang, H. Haloparvum sedimenti Gen. Nov., Sp. Nov., a Member of the Family Haloferacaceae. Int. J. Syst. Evol. Microbiol. 2016, 66, 2327–2334. [Google Scholar] [CrossRef] [PubMed]
- Song, H.S.; Cha, I.-T.; Yim, K.J.; Lee, H.-W.; Hyun, D.-W.; Lee, S.-J.; Rhee, S.-K.; Kim, K.-N.; Kim, D.; Choi, J.-S.; et al. Halapricum Salinum Gen. Nov., Sp. Nov., an Extremely Halophilic Archaeon Isolated from Non-Purified Solar Salt. Antonie van Leeuwenhoek J. Microb. 2014, 105, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Cui, H.-L.; Meng, F. Haloprofundus halophilus Sp. Nov., Isolated from the Saline Soil of Tarim Basin. Antonie van Leeuwenhoek 2019, 112, 553–559. [Google Scholar] [CrossRef]
- Zhang, G.; Gu, J.; Zhang, R.; Rashid, M.; Haroon, M.F.; Xun, W.; Ruan, Z.; Dong, X.; Stingl, U. Haloprofundus marisrubri Gen. Nov., Sp. Nov., an Extremely Halophilic Archaeon Isolated from a Brine–Seawater Interface. Int. J. Syst. Evol. Microbiol. 2017, 67, 9–16. [Google Scholar] [CrossRef]
- Haloweb. Available online: https://haloweb.org/ (accessed on 13 July 2024).
- Antunes, A.; Taborda, M.; Huber, R.; Moissl, C.; Nobre, M.F.; da Costa, M.S. Halorhabdus tiamatea Sp. Nov., a Non-Pigmented, Extremely Halophilic Archaeon from a Deep-Sea, Hypersaline Anoxic Basin of the Red Sea, and Emended Description of the Genus Halorhabdus. Int. J. Syst. Evol. Microbiol. 2008, 58, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Wainø, M.; Tindall, B.J.; Ingvorsen, K. Halorhabdus utahensis Gen. Nov., Sp. Nov., an Aerobic, Extremely Halophilic Member of the Archaea from Great Salt Lake, Utah. Int. J. Syst. Evol. Microbiol. 2000, 50, 183–190. [Google Scholar] [CrossRef]
- Cui, H.-L.; Mou, Y.-Z.; Yang, X.; Zhou, Y.-G.; Liu, H.-C.; Zhou, P.-J. Halorubellus salinus Gen. Nov., Sp. Nov. and Halorubellus litoreus Sp. Nov., Novel Halophilic Archaea Isolated from a Marine Solar Saltern. Syst. Appl. Microbiol. 2012, 35, 30–34. [Google Scholar] [CrossRef]
- Mayrhofer-Iro, M.; Ladurner, A.; Meissner, C.; Derntl, C.; Reiter, M.; Haider, F.; Dimmel, K.; Rössler, N.; Klein, R.; Baranyi, U.; et al. Utilization of Virus ϕCh1 Elements To Establish a Shuttle Vector System for Halo(Alkali)Philic Archaea via Transformation of Natrialba magadii. Appl. Environ. Microbiol. 2013, 79, 2741–2748. [Google Scholar] [CrossRef]
- Liao, Y.; Williams, T.J.; Walsh, J.C.; Ji, M.; Poljak, A.; Curmi, P.M.G.; Duggin, I.G.; Cavicchioli, R. Developing a Genetic Manipulation System for the Antarctic Archaeon, Halorubrum Lacusprofundi: Investigating Acetamidase Gene Function. Sci. Rep. 2016, 6, 34639. [Google Scholar] [CrossRef]
- Cui, H.-L.; Gao, X.; Yang, X.; Xu, X.-W. Halorussus rarus Gen. Nov., Sp. Nov., a New Member of the Family Halobacteriaceae Isolated from a Marine Solar Saltern. Extremophiles 2010, 14, 493–499. [Google Scholar] [CrossRef]
- Mehrshad, M.; Amoozegar, M.A.; Makhdoumi, A.; Fazeli, S.A.S.; Farahani, H.; Asadi, B.; Schumann, P.; Ventosa, A. Halosiccatus urmianus Gen. Nov., Sp. Nov., a Haloarchaeon from a Salt Lake. Int. J. Syst. Evol. Microbiol. 2016, 66, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Hong, L.-G.; Xu, Q.; Cui, H.-L. Halostella limicola Sp. Nov., Isolated from Saline Soil Sampled at the Tarim Basin. Int. J. Syst. Evol. Microbiol. 2019, 69, 3299–3304. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Cui, H.-L. Halostella pelagica Sp. Nov. and Halostella litorea Sp. Nov., Isolated from Salted Brown Alga Laminaria. Int. J. Syst. Evol. Microbiol. 2020, 70, 1969–1976. [Google Scholar] [CrossRef]
- Song, H.S.; Cha, I.-T.; Rhee, J.-K.; Yim, K.J.; Kim, A.Y.; Choi, J.-S.; Baek, S.J.; Seo, M.-J.; Park, S.-J.; Nam, Y.-D.; et al. Halostella salina Gen. Nov., Sp. Nov., an Extremely Halophilic Archaeon Isolated from Solar Salt. Int. J. Syst. Evol. Microbiol. 2016, 66, 2740–2746. [Google Scholar] [CrossRef]
- Mehrshad, M.; Amoozegar, M.A.; Makhdoumi, A.; Rasooli, M.; Asadi, B.; Schumann, P.; Ventosa, A. Halovarius luteus Gen. Nov., Sp. Nov., an Extremely Halophilic Archaeon from a Salt Lake. Int. J. Syst. Evol. Microbiol. 2015, 65, 2420–2425. [Google Scholar] [CrossRef]
- Liu, Q.; Ren, M.; Zhang, L.-L. Natribaculum breve Gen. Nov., Sp. Nov. and Natribaculum longum Sp. Nov., Halophilic Archaea Isolated from Saline Soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Romano, I.; Poli, A.; Finore, I.; Huertas, F.J.; Gambacorta, A.; Pelliccione, S.; Nicolaus, G.; Lama, L.; Nicolaus, B. Haloterrigena hispanica Sp. Nov., an Extremely Halophilic Archaeon from Fuente de Piedra, Southern Spain. Int. J. Syst. Evol. Microbiol. 2007, 57, 1499–1503. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Khijniak, T.V.; Kostrikina, N.A.; Elcheninov, A.G.; Toshchakov, S.V.; Bale, N.J.; Damsté, J.S.S.; Kublanov, I.V. Natronobiforma cellulositropha Gen. Nov., Sp. Nov., a Novel Haloalkaliphilic Member of the Family Natrialbaceae (Class Halobacteria) from Hypersaline Alkaline Lakes. Syst. Appl. Microbiol. 2018, 41, 355–362. [Google Scholar] [CrossRef]
- Zhao, B.; Hu, Q.; Guo, X.; Liao, Z.; Sarmiento, F.; Mesbah, N.M.; Yan, Y.; Li, J.; Wiegel, J. Natronolimnobius aegyptiacus Sp. Nov., an Extremely Halophilic Alkalithermophilic Archaeon Isolated from the Athalassohaline Wadi An Natrun, Egypt. Int. J. Syst. Evol. Microbiol. 2018, 68, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Orsini, S.S.; James, K.L.; Reyes, D.J.; Couto-Rodriguez, R.L.; Gulko, M.K.; Witte, A.; Carroll, R.K.; Rice, K.C. Bacterial-like Nitric Oxide Synthase in the Haloalkaliphilic Archaeon Natronomonas pharaonis. MicrobiologyOpen 2020, 9, e1124. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.-L.; Tohty, D.; Liu, H.-C.; Liu, S.-J.; Oren, A.; Zhou, P.-J. Natronorubrum sulfidifaciens Sp. Nov., an Extremely Haloalkaliphilic Archaeon Isolated from Aiding Salt Lake in Xin-Jiang, China. Int. J. Syst. Evol. Microbiol. 2007, 57, 738–740. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Romero, E.; Valenzuela-Encinas, C.; López-Ramírez, M.P.; de los Angeles Coutiño-Coutiño, M.; Marsch, R.; Dendooven, L. Natronorubrum texcoconense Sp. Nov., a Haloalkaliphilic Archaeon Isolated from Soil of the Former Lake Texcoco (Mexico). Arch. Microbiol. 2013, 195, 145–151. [Google Scholar] [CrossRef]
- Cui, H.-L.; Yang, X.; Mou, Y.-Z. Salinarchaeum laminariae Gen. Nov., Sp. Nov.: A New Member of the Family Halobacteriaceae Isolated from Salted Brown Alga Laminaria. Extremophiles 2011, 15, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Cui, H.-L. Salinibaculum litoreum Gen. Nov., Sp. Nov., Isolated from Salted Brown Alga Laminaria. Int. J. Syst. Evol. Microbiol. 2020, 70, 2879–2887. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.-L.; Zhang, W.-J. Salinigranum rubrum Gen. Nov., Sp. Nov., a Member of the Family Halobacteriaceae Isolated from a Marine Solar Saltern. Int. J. Syst. Evol. Microbiol. 2014, 64, 2029–2033. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, J.-Q.; Xu, W.-M.; Li, Y.; Zhou, Y.; Lü, Z.-Z.; Hou, J.; Zhu, L.; Cui, H.-L. Salinigranum salinum Sp. Nov., Isolated from a Marine Solar Saltern. Int. J. Syst. Evol. Microbiol. 2016, 66, 3017–3021. [Google Scholar] [CrossRef]
- Hou, J.; Zhao, Y.-J.; Zhu, L.; Cui, H.-L. Salinirubellus salinus Gen. Nov., Sp. Nov., Isolated from a Marine Solar Saltern. Int. J. Syst. Evol. Microbiol. 2018, 68, 1874–1878. [Google Scholar] [CrossRef]
- Cui, H.-L.; Qiu, X.-X. Salinarubrum litoreum Gen. Nov., Sp. Nov.: A New Member of the Family Halobacteriaceae Isolated from Chinese Marine Solar Salterns. Antonie van Leeuwenhoek 2014, 105, 135–141. [Google Scholar] [CrossRef]
- Cui, H.-L.; Lü, Z.-Z.; Li, Y.; Zhou, Y. Salinirussus salinus Gen. Nov., Sp. Nov., Isolated from a Marine Solar Saltern. Int. J. Syst. Evol. Microbiol. 2017, 67, 3622–3626. [Google Scholar] [CrossRef]
- Yin, X.-Q.; Liu, B.-B.; Chu, X.; Salam, N.; Li, X.; Yang, Z.-W.; Zhang, Y.; Xiao, M.; Li, W.-J. Saliphagus infecundisoli Gen. Nov., Sp. Nov., an Extremely Halophilic Archaeon Isolated from a Saline Soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 4154–4160. [Google Scholar] [CrossRef]
- Cline, S.W.; Lam, W.L.; Charlebois, R.L.; Schalkwyk, L.C.; Doolittle, W.F. Transformation Methods for Halophilic Archaebacteria. Can. J. Microbiol. 1989, 35, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Han, J.; Liu, X.; Zhou, J.; Xiang, H. Development of pyrF-Based Gene Knockout Systems for Genome-Wide Manipulation of the Archaea Haloferax mediterranei and Haloarcula hispanica. J. Genet. Genom. 2011, 38, 261–269. [Google Scholar] [CrossRef]
- Falb, M.; Pfeiffer, F.; Palm, P.; Rodewald, K.; Hickmann, V.; Tittor, J.; Oesterhelt, D. Living with Two Extremes: Conclusions from the Genome Sequence of Natronomonas pharaonis. Genome Res. 2005, 15, 1336–1343. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Han, J.; Zhou, L.; Coker, J.A.; DasSarma, P.; DasSarma, S.; Xiang, H. Dissection of the Regulatory Mechanism of a Heat-Shock Responsive Promoter in Haloarchaea: A New Paradigm for General Transcription Factor Directed Archaeal Gene Regulation. Nucleic Acids Res. 2008, 36, 3031–3042. [Google Scholar] [CrossRef] [PubMed]
- Born, J.; Pfeifer, F. Improved GFP Variants to Study Gene Expression in Haloarchaea. Front. Microbiol. 2019, 10, 1200. [Google Scholar] [CrossRef] [PubMed]
- Derntl, C.; Selb, R.; Klein, R.; Alte, B.; Witte, A. Genomic Manipulations in Alkaliphilic Haloarchaea Demonstrated by a Gene Disruption in Natrialba magadii. FEMS Microbiol. Lett. 2015, 362, fnv179. [Google Scholar] [CrossRef]
- Gophna, U.; Allers, T.; Marchfelder, A. Finally, Archaea Get Their CRISPR-Cas Toolbox. Trends Microbiol. 2017, 25, 430–432. [Google Scholar] [CrossRef] [PubMed]
- Khambhati, K.; Bhattacharjee, G.; Gohil, N.; Braddick, D.; Kulkarni, V.; Singh, V. Exploring the Potential of Cell-Free Protein Synthesis for Extending the Abilities of Biological Systems. Front. Bioeng. Biotechnol. 2019, 7, 248. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonnaud, E.; Oger, P.M.; Ohayon, A.; Louis, Y. Haloarchaea as Promising Chassis to Green Chemistry. Microorganisms 2024, 12, 1738. https://doi.org/10.3390/microorganisms12081738
Bonnaud E, Oger PM, Ohayon A, Louis Y. Haloarchaea as Promising Chassis to Green Chemistry. Microorganisms. 2024; 12(8):1738. https://doi.org/10.3390/microorganisms12081738
Chicago/Turabian StyleBonnaud, Emma, Philippe M. Oger, Avigaël Ohayon, and Yoann Louis. 2024. "Haloarchaea as Promising Chassis to Green Chemistry" Microorganisms 12, no. 8: 1738. https://doi.org/10.3390/microorganisms12081738




