Bacillus licheniformisYB06: A Rhizosphere–Genome-Wide Analysis and Plant Growth-Promoting Analysis of a Plant Growth-Promoting Rhizobacterium Isolated from Codonopsis pilosula
Abstract
1. Introduction
2. Materials and Methods
2.1. Species Identification and Biological Characteristics Determination of B. licheniformis YB06
2.1.1. Strain Material
2.1.2. Morphological and Molecular Biological Identification of B. licheniformis YB06
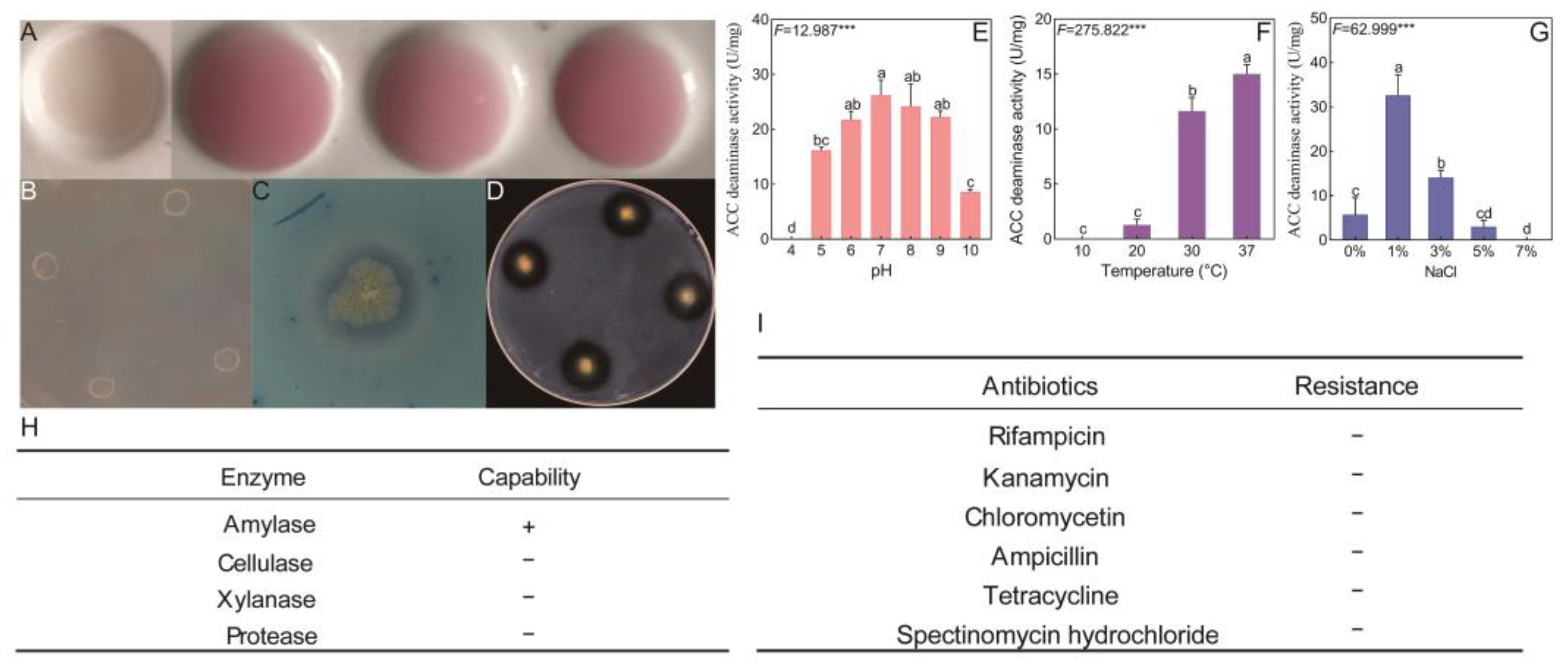
2.1.3. Determination of Biological Characteristics of B. licheniformis YB06
2.1.4. Determination of Plant Growth Promotion (PGP) of B. licheniformis YB06 with C. pilosula Seeds
2.2. Comprehensive Whole-Genome Sequencing of B. licheniformis YB06
2.2.1. Preparation, Sequencing, and Assembly of Genomic DNA
2.2.2. Gene Prediction, Functional Annotation, and Analysis of Plant Growth-Promoting Information
2.3. Analysis of Rhizosphere Bacterial Diversity of C. pilosula under Different Fertilization Treatments
2.3.1. Experimental Fertilizers and Soil Substrates
2.3.2. Determination of Soil Physical and Chemical Properties under Different Fertilization Regimes
2.3.3. Extraction of Soil Bacterial DNA and Illumina MiSeq Sequencing
2.3.4. Data Analysis of Soil Bacterial Diversity
3. Results
3.1. Identification and Biological Characteristics of B. licheniformis YB06
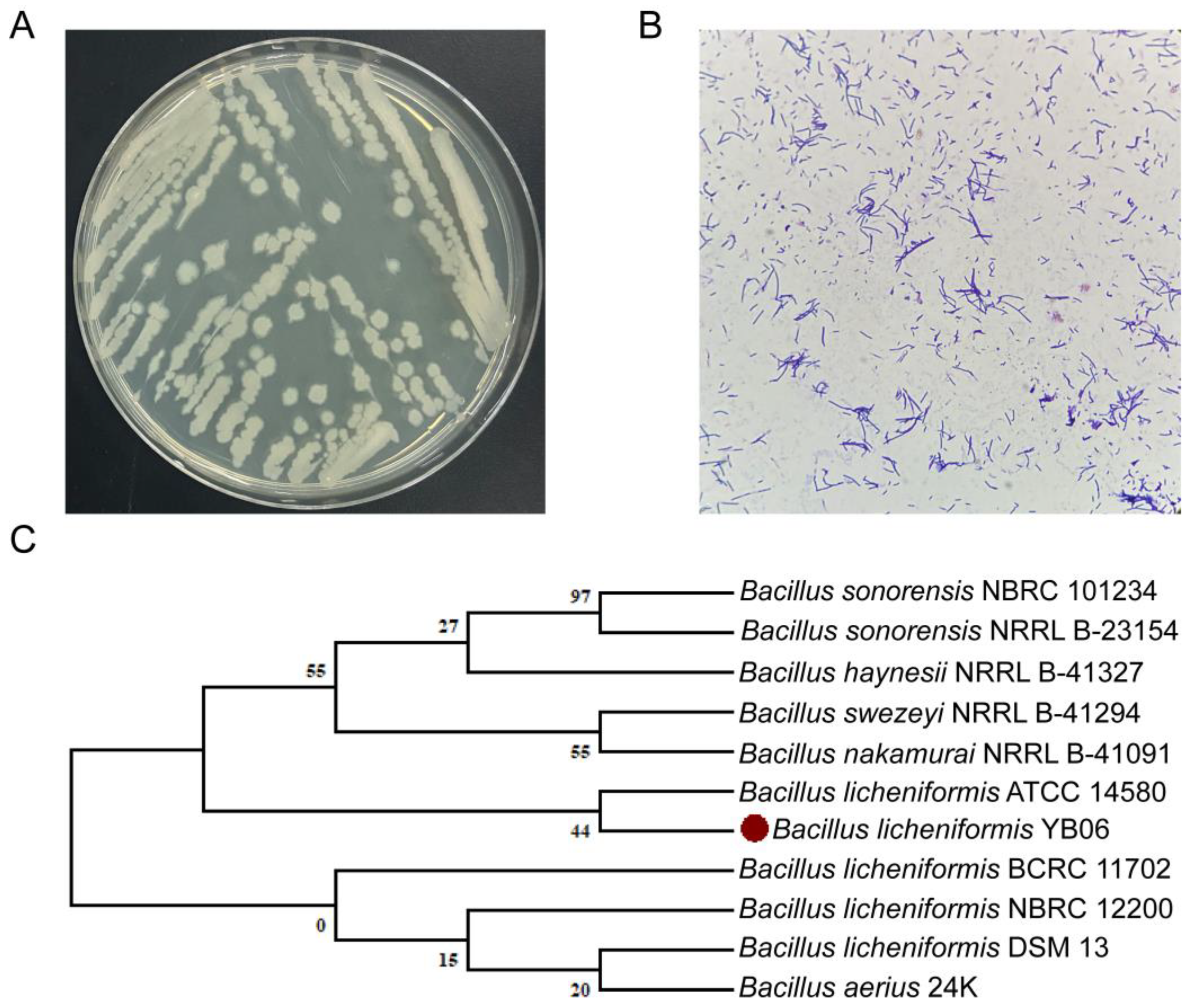
3.1.1. Morphological and Molecular Characterization of B. licheniformis YB06
3.1.2. Determination of the Biological Characteristics of B. licheniformis YB06
3.1.3. Effect of the B. licheniformis YB06 on the Growth of C. pilosula Seedlings
3.2. Whole-Genome Analysis of B. licheniformis YB06
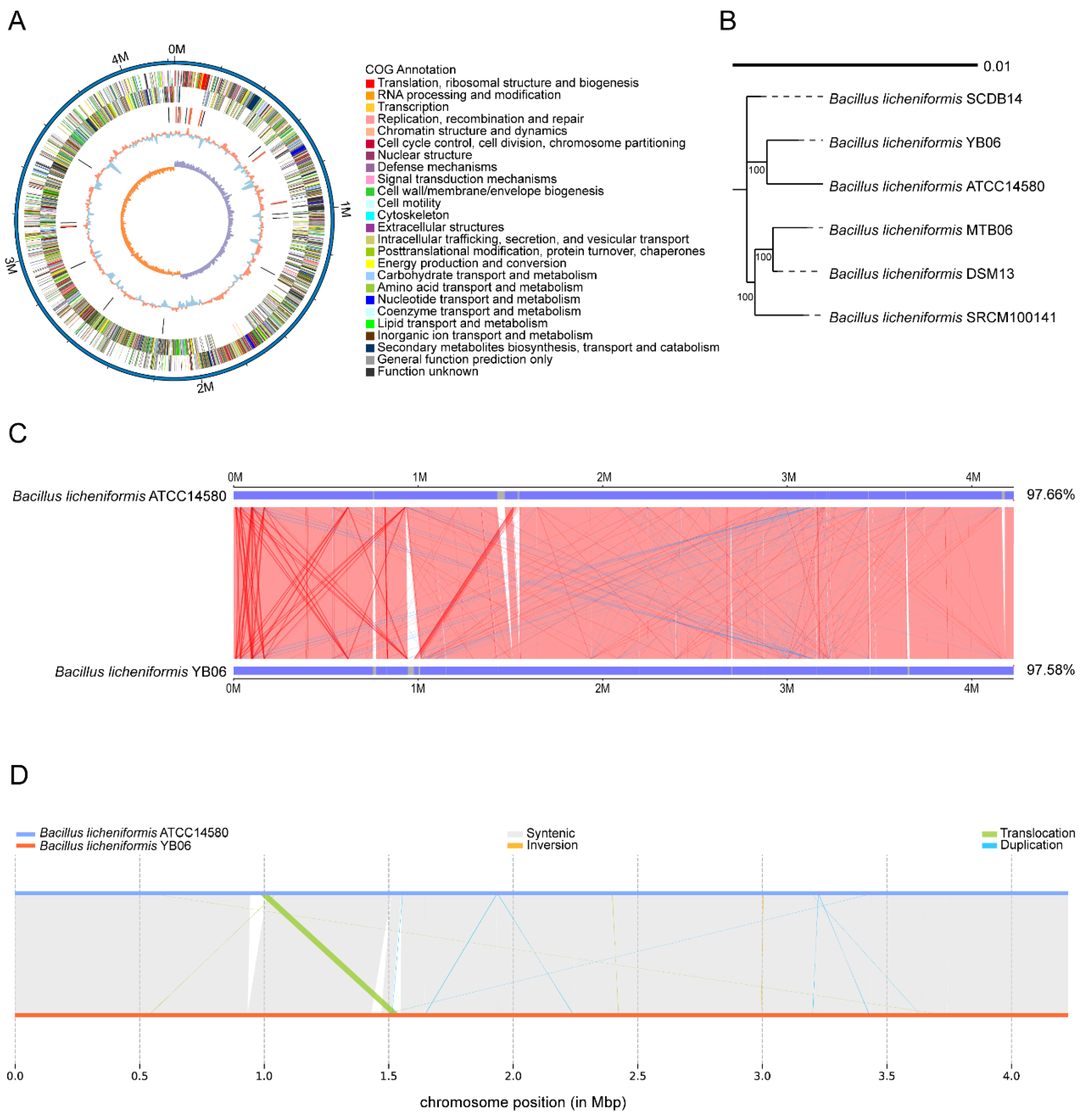
3.2.1. Genomic Features and Comparative Genomic Analysis of B. licheniformis YB06
3.2.2. Functional Genomic Annotation of B. licheniformis YB06 Genome
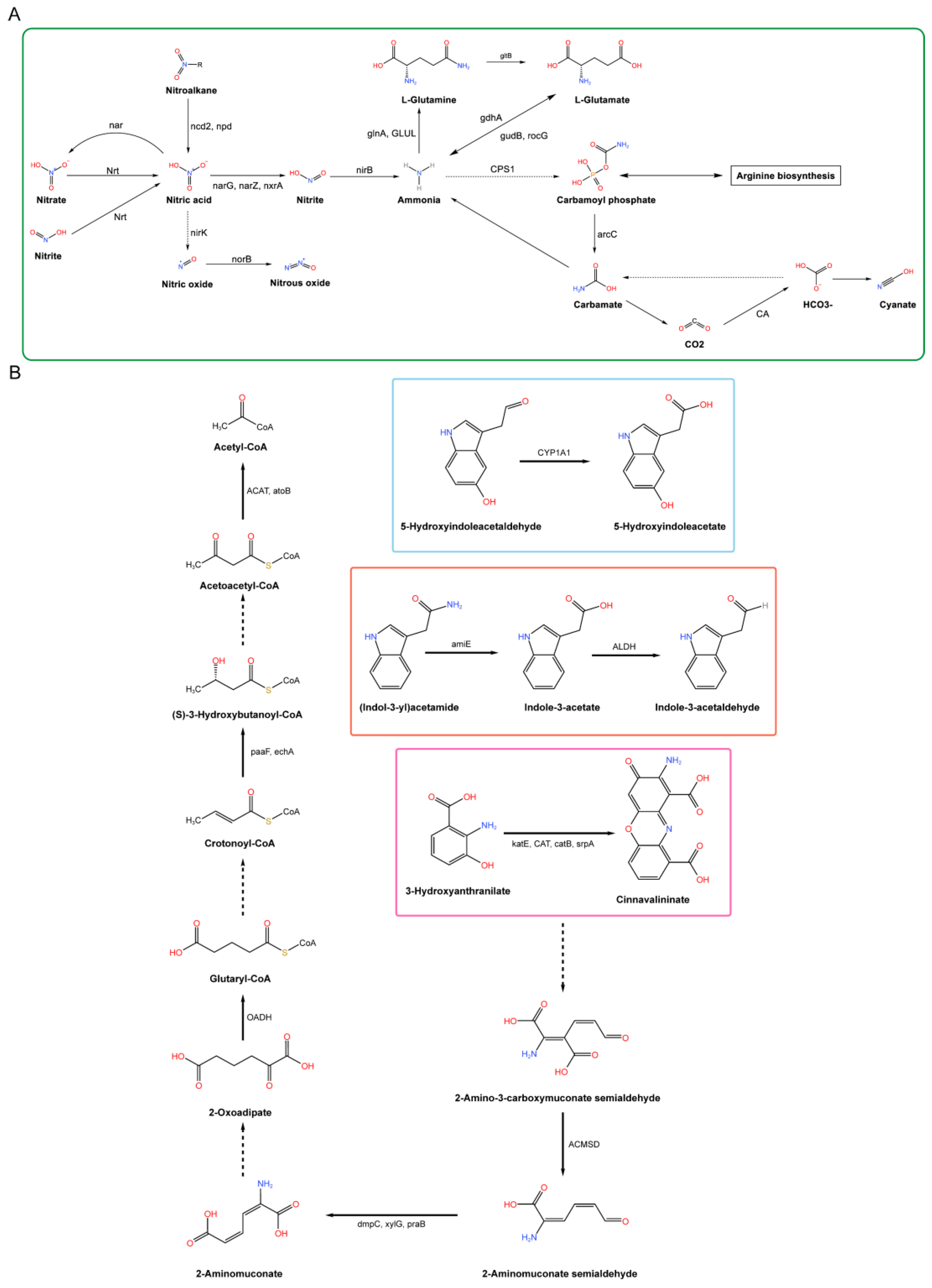
3.2.3. Metabolic Pathway Analysis of Plant Growth Promotion in B. licheniformis YB06
3.3. Physicochemical Characteristics of Soil under Various Fertilization Regimes
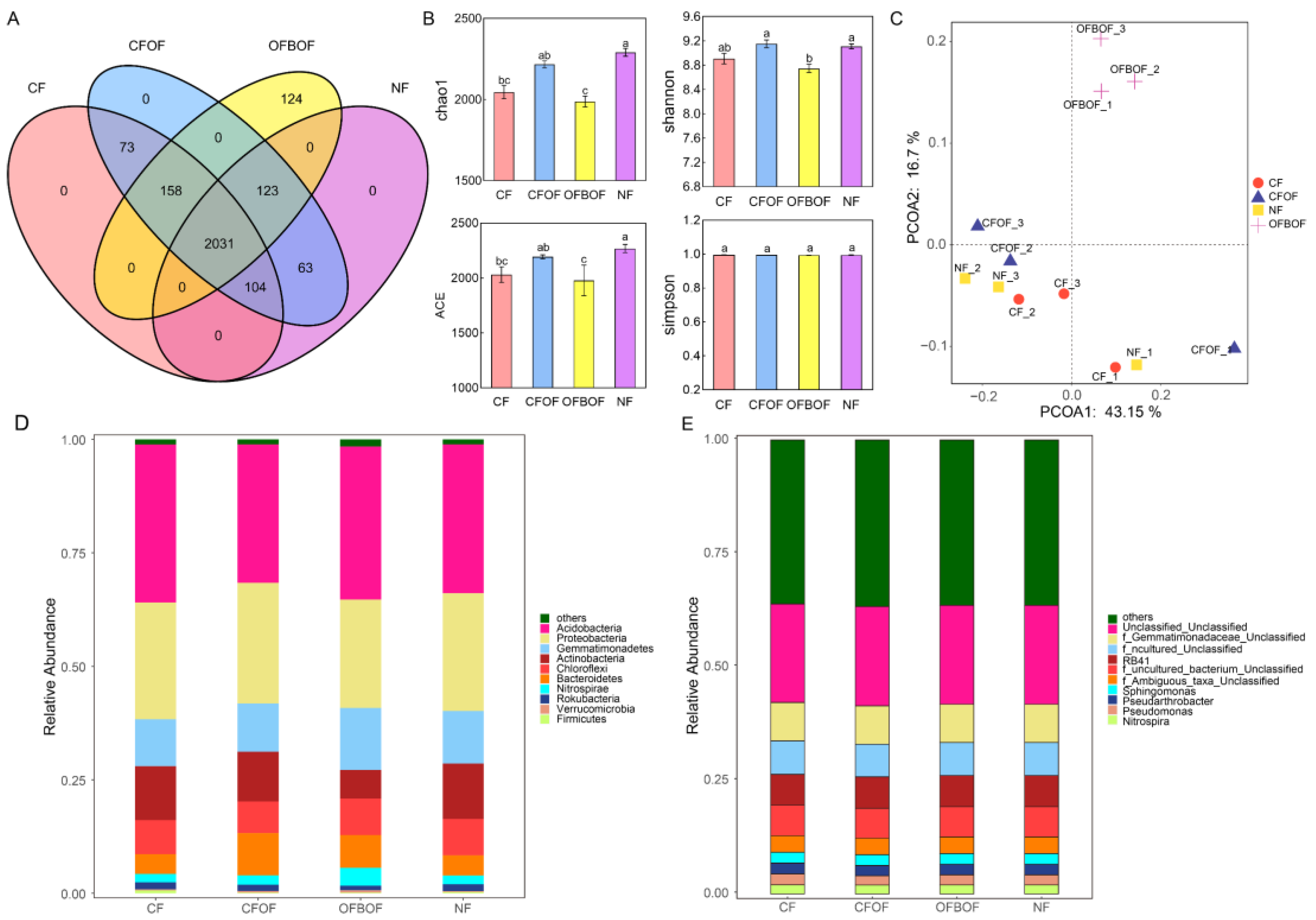
3.4. Effects of Different Fertilization Treatments on Rhizosphere Bacterial Diversity of C. pilosula
3.4.1. Analysis of Soil Bacterial Diversity under Different Fertilization Conditions
3.4.2. Soil Bacterial Community Structure under Various Fertilization Regimes
3.4.3. Correlation between Soil Microbial Community Composition and Environmental Factors
4. Discussion
4.1. Taxonomic Classification and Biological Characteristics of B. licheniformis YB06
4.2. Identification of Growth-Promoting Genes Associated with YB06 via Genome-Wide Analysis
4.3. Effects of Different Fertilization Conditions on Soil Physicochemical Properties and Bacterial Microbial Diversity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luan, F.; Ji, Y.; Peng, L.; Liu, Q.; Cao, H.; Yang, Y.; He, X.; Zeng, N. Extraction, purification, structural characteristics and biological properties of the polysaccharides from Codonopsis pilosula: A review. Carbohydr. Polym. 2021, 261, 117863. [Google Scholar] [CrossRef]
- Guo, H.; Lou, Y.; Hou, X.; Han, Q.; Guo, Y.; Li, Z.; Guan, X.; Liu, H.; Zhang, C. A systematic review of the mechanism of action and potential medicinal value of Codonopsis pilosula in diseases. Front. Pharmacol. 2024, 15, 1415147. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.-F.; Zhang, Y.-Y.; Paulsen, B.S.; Fu, Y.-P.; Huang, C.; Feng, B.; Li, L.-X.; Chen, X.-F.; Jia, R.-Y.; Song, X. Prospects of Codonopsis pilosula polysaccharides: Structural features and bioactivities diversity. Trends Food Sci. Technol. 2020, 103, 937581. [Google Scholar] [CrossRef]
- Lv, B.; Yang, X.; Xue, H.; Nan, M.; Zhang, Y.; Liu, Z.; Bi, Y.; Shang, S. Isolation of main pathogens causing postharvest disease in fresh Codonopsis pilosula during different storage stages and ozone control against disease and mycotoxin accumulation. J. Fungi 2023, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Vasco, C.; Torres, B.; Jácome, E.; Torres, A.; Eche, D.; Velasco, C. Use of chemical fertilizers and pesticides in frontier areas: A case study in the Northern Ecuadorian Amazon. Land Use Policy 2021, 107, 105490. [Google Scholar] [CrossRef]
- Cetin, M.; Aljama, A.M.O.; Alrabiti, O.B.M.; Adiguzel, F.; Sevik, H.; Zeren Cetin, I. Using topsoil analysis to determine and map changes in Ni Co pollution. Water Air Soil Pollut. 2022, 233, 293. [Google Scholar] [CrossRef]
- Ma, J.; Chen, Y.; Wang, K.; Huang, Y.; Wang, H. Re-utilization of Chinese medicinal herbal residues improved soil fertility and maintained maize yield under chemical fertilizer reduction. Chemosphere 2021, 283, 131262. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wu, Z.; Liu, C.; Zhang, Z.; Liu, X. Biochar combined with Bacillus subtilis SL-44 as an eco-friendly strategy to improve soil fertility, reduce Fusarium wilt, and promote radish growth. Ecotoxicol. Environ. Saf. 2023, 251, 114509. [Google Scholar] [CrossRef]
- Ha-Tran, D.M.; Nguyen, T.T.M.; Hung, S.-H.; Huang, E.; Huang, C.-C. Roles of plant growth-promoting rhizobacteria (PGPR) in stimulating salinity stress defense in plants: A review. Int. J. Mol. Sci. 2021, 22, 3154. [Google Scholar] [CrossRef]
- Wang, J.; He, Y.; Li, T.; Li, C.; Xu, X.; Xiang, H.; Wang, X.; Wu, Z. Complex biochemical synergistic interactions between two rhizobacteria grown in consortium, Bacillus subtilis SL-44 and Enterobacter hormaechei Wu-15. Rhizosphere 2022, 24, 100587. [Google Scholar] [CrossRef]
- Vocciante, M.; Grifoni, M.; Fusini, D.; Petruzzelli, G.; Franchi, E. The role of plant growth-promoting rhizobacteria (PGPR) in mitigating plant’s environmental stresses. Appl. Sci. 2022, 12, 1231. [Google Scholar] [CrossRef]
- Ali, S.; Hameed, S.; Shahid, M.; Iqbal, M.; Lazarovits, G.; Imran, A. Functional characterization of potential PGPR exhibiting broad-spectrum antifungal activity. Microbiol. Res. 2020, 232, 126389. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, W.; Zhang, Z.; Kou, Z.; Wang, X.; Wang, Y.; Su, X.; Zhang, J.; Liu, L.; Yan, F. Biocontrol effects of three antagonistic bacteria strains against Codonopsis pilosula wilt disease caused by Fusarium oxysporum. Biol. Control. 2024, 190, 105446. [Google Scholar] [CrossRef]
- Feng, M.-Q.; Feng, Y.-C.; Gou, W. Identification of the pathogen of Codonopsis pilosula stem base rot and screening of its control agents. J. Fungi 2023, 9, 146. [Google Scholar]
- Verma, J.; Kumar, C.; Sharma, M.; Shukla, A.C.; Saxena, S. Exploitation of microbial consortia for formulating biofungicides, biopesticides, and biofertilizers for plant growth promotion. In Entrepreneurship with Microorganisms; Elsevier: Amsterdam, The Netherlands, 2024; pp. 227–257. [Google Scholar]
- Samada, L.H.; Tambunan, U.S.F. Biopesticides as promising alternatives to chemical pesticides: A review of their current and future status. Online J. Biol. Sci. 2020, 20, 66–76. [Google Scholar] [CrossRef]
- Muras, A.; Romero, M.; Mayer, C.; Otero, A. Biotechnological applications of Bacillus licheniformis. Crit. Rev. Biotechnol. 2021, 41, 609–627. [Google Scholar] [CrossRef]
- Arnaouteli, S.; Bamford, N.C.; Stanley-Wall, N.R.; Kovács, Á.T. Bacillus subtilis biofilm formation and social interactions. Nat. Rev. Microbiol. 2021, 19, 600–614. [Google Scholar] [CrossRef]
- Miljaković, D.; Marinković, J.; Balešević-Tubić, S. The significance of Bacillus spp. in disease suppression and growth promotion of field and vegetable crops. Microorganisms 2020, 8, 1037. [Google Scholar] [CrossRef]
- Wang, Q.; Ou, E.-L.; Wang, P.-C.; Chen, Y.; Wang, Z.-Y.; Wang, Z.-W.; Fang, X.-W.; Zhang, J.-L. Bacillus amyloliquefaciens GB03 augmented tall fescue growth by regulating phytohormone and nutrient homeostasis under nitrogen deficiency. Front. Plant Sci. 2022, 13, 979883. [Google Scholar] [CrossRef]
- Han, X.; Shen, D.; Xiong, Q.; Bao, B.; Zhang, W.; Dai, T.; Zhao, Y.; Borriss, R.; Fan, B. The plant-beneficial rhizobacterium Bacillus velezensis FZB42 controls the soybean pathogen Phytophthora sojae due to bacilysin production. Appl. Environ. Microbiol. 2021, 87, e01601–e01621. [Google Scholar] [CrossRef]
- Lane, D. 16S/23S rRNA Sequencing. Nucleic Acid Techniques in Bacterial Systematics; Wiley: Hoboken, NJ, USA, 1991. [Google Scholar]
- Shahab, S.; Ahmed, N.; Khan, N.S. Indole acetic acid production and enhanced plant growth promotion by indigenous PSBs. Afr. J. Agric. Res. 2009, 4, 1312–1316. [Google Scholar]
- Payne, S.M. [25] Detection, isolation, and characterization of siderophores. Methods Enzymol. 1994, 235, 329–344. [Google Scholar] [PubMed]
- Li, Z.; Chang, S.; Lin, L.; Li, Y.; An, Q. A colorimetric assay of 1-aminocyclopropane-1-carboxylate (ACC) based on ninhydrin reaction for rapid screening of bacteria containing ACC deaminase. Lett. Appl. Microbiol. 2011, 53, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Meddeb-Mouelhi, F.; Moisan, J.K.; Beauregard, M. A comparison of plate assay methods for detecting extracellular cellulase and xylanase activity. Enzym. Microb. Technol. 2014, 66, 16–19. [Google Scholar] [CrossRef] [PubMed]
- de Veras, B.O.; dos Santos, Y.Q.; Diniz, K.M.; Carelli, G.S.C.; dos Santos, E.A. Screening of protease, cellulase, amylase and xylanase from the salt-tolerant and thermostable marine Bacillus subtilis strain SR60. F1000Research 2018, 7, 1704. [Google Scholar] [CrossRef]
- Fasiku, S.A.; Ogunsola, O.F.; Fakunle, A.; Olanbiwoninu, A.A. Isolation of bacteria with potential of producing extracellular enzymes (Amylase, Cellulase and Protease) from soil samples. J. Adv. Microbiol. 2020, 20, 21–26. [Google Scholar] [CrossRef]
- Schollenberger, C. Determination of soil organic matter. Soil Sci. 1945, 59, 53–56. [Google Scholar] [CrossRef]
- Kirk, P.L. Kjeldahl method for total nitrogen. Anal. Chem. 1950, 22, 354–358. [Google Scholar] [CrossRef]
- Rowland, A.; Haygarth, P. Determination of Total Dissolved Phosphorus in Soil Solutions; Wiley Online Library: Hoboken, NJ, USA, 1997. [Google Scholar]
- Broderick, E.; Zack, P. Flame Spectrophotometry for Determination of Sodium, Potassium, and Lithium in Glass. Anal. Chem. 1951, 23, 1455–1458. [Google Scholar] [CrossRef]
- Chen, S.; Lin, B.; Li, Y.; Zhou, S. Spatial and temporal changes of soil properties and soil fertility evaluation in a large grain-production area of subtropical plain, China. Geoderma 2020, 357, 113937. [Google Scholar] [CrossRef]
- Koetsier, G.; Cantor, E. A practical guide to analyzing nucleic acid concentration and purity with microvolume spectrophotometers. N. Engl. Biolabs Inc 2019, 12, 1–8. [Google Scholar]
- Saxena, A.K.; Kumar, M.; Chakdar, H.; Anuroopa, N.; Bagyaraj, D. Bacillus species in soil as a natural resource for plant health and nutrition. J. Appl. Microbiol. 2020, 128, 1583–1594. [Google Scholar] [CrossRef]
- de, O.; Nunes, P.S.; De Medeiros, F.H.; De Oliveira, T.S.; de Almeida Zago, J.R.; Bettiol, W. Bacillus subtilis and Bacillus licheniformispromote tomato growth. Braz. J. Microbiol. 2023, 54, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Bhutani, N.; Maheshwari, R.; Sharma, N.; Kumar, P.; Dang, A.S.; Suneja, P. Characterization of halo-tolerant plant growth promoting endophytic Bacillus licheniformisMHN 12. J. Genet. Eng. Biotechnol. 2022, 20, 113. [Google Scholar] [CrossRef]
- Devi, S.; Sharma, S.; Tiwari, A.; Bhatt, A.K.; Singh, N.K.; Singh, M.; Kaushalendra; Kumar, A. Screening for multifarious plant growth promoting and biocontrol attributes in bacillus strains isolated from indo gangetic soil for enhancing growth of rice crops. Microorganisms 2023, 11, 1085. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, J.; Zhu, M.; Wan, H.; Chen, Z.; Yang, N.; Duan, J.; Wei, Z.; Hu, T.; Liu, F. Effects of plant growth promoting rhizobacteria (PGPR) strain Bacillus licheniformis with biochar amendment on potato growth and water use efficiency under reduced irrigation regime. Agronomy 2022, 12, 1031. [Google Scholar] [CrossRef]
- Wan, G.-Z.; Wang, L.; Jin, L.; Chen, J. Evaluation of environmental factors affecting the quality of Codonopsis pilosula based on chromatographic fingerprint and MaxEnt model. Ind. Crops Prod. 2021, 170, 113783. [Google Scholar] [CrossRef]
- Yan, H.; He, J.; Xu, X.; Yao, X.; Wang, G.; Tang, L.; Feng, L.; Zou, L.; Gu, X.; Qu, Y. Prediction of potentially suitable distributions of Codonopsis pilosula in China based on an optimized MaxEnt model. Front. Ecol. Evol. 2021, 9, 773396. [Google Scholar] [CrossRef]
- Ahmad, E.; Sharma, P.K.; Khan, M.S. IAA biosynthesis in bacteria and its role in plant-microbe interaction for drought stress management. In Plant Stress Mitigators: Action and Application; Springer: Berlin/Heidelberg, Germany, 2022; pp. 235–258. [Google Scholar]
- Wagi, S.; Ahmed, A. Bacillus spp.: Potent microfactories of bacterial IAA. PeerJ 2019, 7, e7258. [Google Scholar] [CrossRef]
- Sansinenea, E. Bacillus spp.: As plant growth-promoting bacteria. In Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms: Discovery and Applications; Springer: Berlin/Heidelberg, Germany, 2019; pp. 225–237. [Google Scholar]
- Shah, R.; Amaresan, N.; Patel, P.; Jinal, H.N.; Krishnamurthy, R. Isolation and characterization of Bacillus spp. endowed with multifarious plant growth-promoting traits and their potential effect on tomato (Lycopersicon esculentum) seedlings. Arab. J. Sci. Eng. 2020, 45, 4579–4587. [Google Scholar] [CrossRef]
- Choo, Q.-C.; Samian, M.-R.; Najimudin, N. Phylogeny and characterization of three nifH-homologous genes from Paenibacillus azotofixans. Appl. Environ. Microbiol. 2003, 69, 3658–3662. [Google Scholar] [CrossRef] [PubMed]
- Masood, S.; Zhao, X.Q.; Shen, R.F. Bacillus pumilus promotes the growth and nitrogen uptake of tomato plants under nitrogen fertilization. Sci. Hortic. 2020, 272, 109581. [Google Scholar] [CrossRef]
- Groß, C.; Hossen, S.; Hartmann, H.; Noll, M.; Borken, W. Biological nitrogen fixation and nifH gene abundance in deadwood of 13 different tree species. Biogeochemistry 2022, 161, 353–371. [Google Scholar] [CrossRef]
- Krewulak, K.D.; Vogel, H.J. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta (BBA)-Biomembr. 2008, 1778, 1781–1804. [Google Scholar] [CrossRef]
- Parmar, H.Y.; Chakraborty, H. Effect of siderophore on plant growth promotion. Int. J. Appl. Pure Sci. Agric. 2016, 2, 60–68. [Google Scholar]
- Naing, A.H.; Maung, T.T.; Kim, C.K. The ACC deaminase-producing plant growth-promoting bacteria: Influences of bacterial strains and ACC deaminase activities in plant tolerance to abiotic stress. Physiol. Plant. 2021, 173, 1992–2012. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Chauhan, P.S. ACC deaminase-producing rhizosphere competent Bacillus spp. mitigate salt stress and promote Zea mays growth by modulating ethylene metabolism. 3 Biotech. 2020, 10, 119. [Google Scholar] [CrossRef]
- Shahid, M.; Singh, U.B.; Khan, M.S.; Singh, P.; Kumar, R.; Singh, R.N.; Kumar, A.; Singh, H.V. Bacterial ACC deaminase: Insights into enzymology, biochemistry, genetics, and potential role in amelioration of environmental stress in crop plants. Front. Microbiol. 2023, 14, 1132770. [Google Scholar] [CrossRef]
- Gowtham, H.; Singh, B.; Murali, M.; Shilpa, N.; Prasad, M.; Aiyaz, M.; Amruthesh, K.; Niranjana, S. Induction of drought tolerance in tomato upon the application of ACC deaminase producing plant growth promoting rhizobacterium Bacillus subtilis Rhizo SF 48. Microbiol. Res. 2020, 234, 126422. [Google Scholar] [CrossRef]
- Mukhtar, T.; Rehman, S.U.; Smith, D.; Sultan, T.; Seleiman, M.F.; Alsadon, A.A.; Amna; Ali, S.; Chaudhary, H.J.; Solieman, T.H. Mitigation of heat stress in Solanum lycopersicum L. by ACC-deaminase and exopolysaccharide producing Bacillus cereus: Effects on biochemical profiling. Sustainability 2020, 12, 2159. [Google Scholar] [CrossRef]
- Kulkova, I.; Dobrzyński, J.; Kowalczyk, P.; Bełżecki, G.; Kramkowski, K. Plant growth promotion using Bacillus cereus. Int. J. Mol. Sci. 2023, 24, 9759. [Google Scholar] [CrossRef] [PubMed]
- Medison, R.G.; Jiang, J.; Medison, M.B.; Tan, L.-T.; Kayange, C.D.; Sun, Z.; Zhou, Y. Evaluating the potential of Bacillus licheniformisYZCUO202005 isolated from lichens in maize growth promotion and biocontrol. Heliyon 2023, 9, e20204. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Xie, J.; Li, Y.; Gao, T.; Xu, C.; Wang, Q. Comparative genomic and functional analyses of four sequenced Bacillus cereus genomes reveal conservation of genes relevant to plant-growth-promoting traits. Sci. Rep. 2018, 8, 17009. [Google Scholar] [CrossRef]
- Yang, F.; Liu, Y.; Chen, L.; Li, J.; Wang, L.; Du, G. Genome sequencing and flavor compound biosynthesis pathway analyses of Bacillus licheniformisisolated from Chinese Maotai-flavor liquor-brewing microbiome. Food Biotechnol. 2020, 34, 193–211. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, M.; Kang, J.; Wang, S.; Zha, Z.; Zhan, Y.; Wang, Z.; Li, J.; Cai, D.; Chen, S. Engineering Bacillus licheniformis as industrial chassis for efficient bioproduction from starch. Bioresour. Technol. 2024, 131061. [Google Scholar] [CrossRef]
- He, H.; Yu, Q.; Ding, Z.; Zhang, L.; Shi, G.; Li, Y. Biotechnological and food synthetic biology potential of platform strain: Bacillus licheniformis. Synth. Syst. Biotechnol. 2023, 8, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Cao, K.; Deng, C.; Li, Y.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wu, J.; Guan, L. An integrated peach genome structural variation map uncovers genes associated with fruit traits. Genome Biol. 2020, 21, 258. [Google Scholar] [CrossRef]
- Veith, B.; Herzberg, C.; Steckel, S.; Feesche, J.; Maurer, K.H.; Ehrenreich, P.; Bäumer, S.; Henne, A.; Liesegang, H.; Merkl, R. The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J. Mol. Microbiol. Biotechnol. 2004, 7, 204–211. [Google Scholar] [CrossRef]
- Harwood, C.R.; Mouillon, J.-M.; Pohl, S.; Arnau, J. Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol. Rev. 2018, 42, 721–738. [Google Scholar] [CrossRef]
- Thilagar, G.; Bagyaraj, D.; Podile, A.R.; Vaikuntapu, P.R. Bacillus sonorensis, a novel plant growth promoting rhizobacterium in improving growth, nutrition and yield of chilly (Capsicum annuum L.). Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018, 88, 813–818. [Google Scholar] [CrossRef]
- Deng, J.; Kong, S.; Wang, F.; Liu, Y.; Jiao, J.; Lu, Y.; Zhang, F.; Wu, J.; Wang, L.; Li, X. Identification of a new Bacillus sonorensis strain KLBC GS-3 as a biocontrol agent for postharvest green mould in grapefruit. Biol. Control 2020, 151, 104393. [Google Scholar] [CrossRef]
- Harinathan, B.; Sankaralingam, S.; Palpperumal, S.; Balachandran, C.; Hashem, A.; Alqarawi, A.A.; Abd_Allah, E.F.; Arokiyaraj, S.; Baskar, K. Impact of rhizobacterium Bacillus sonorensis on propagation of Abelmoschus esculentus and its antimicrobial activity. J. King Saud Univ. -Sci. 2021, 33, 101496. [Google Scholar] [CrossRef]
- Baslam, M.; Mitsui, T.; Sueyoshi, K.; Ohyama, T. Recent advances in carbon and nitrogen metabolism in C3 plants. Int. J. Mol. Sci. 2020, 22, 318. [Google Scholar] [CrossRef] [PubMed]
- Nuti, M.; Lepidi, A.; Prakash, R.; Schilperoort, R.; Cannon, F. Evidence for nitrogen fixation (nif) genes on indigenous Rhizobium plasmids. Nature 1979, 282, 533–535. [Google Scholar] [CrossRef]
- Ghosh, S.; Ghosh, P.; Maiti, T. Production and metabolism of indole acetic acid (IAA) by root nodule bacteria (Rhizobium): A review. J. Pure Appl. Microbiol. 2011, 5, 523–540. [Google Scholar]
- Sun, S.-L.; Yang, W.-L.; Fang, W.-W.; Zhao, Y.-X.; Guo, L.; Dai, Y.-J. The plant growth-promoting rhizobacterium Variovorax boronicumulans CGMCC 4969 regulates the level of indole-3-acetic acid synthesized from indole-3-acetonitrile. Appl. Environ. Microbiol. 2018, 84, e00298-18. [Google Scholar] [CrossRef]
- Chaudhary, T.; Gera, R.; Shukla, P. Deciphering the potential of Rhizobium pusense MB-17a, a plant growth-promoting root endophyte, and functional annotation of the genes involved in the metabolic pathway. Front. Bioeng. Biotechnol. 2021, 8, 617034. [Google Scholar] [CrossRef]
- Cai, D.; Zhu, J.; Li, Y.; Li, L.; Zhang, M.; Wang, Z.; Yang, H.; Li, J.; Yang, Z.; Chen, S. Systematic engineering of branch chain amino acid supply modules for the enhanced production of bacitracin from Bacillus licheniformis. Metab. Eng. Commun. 2020, 11, e00136. [Google Scholar] [CrossRef]
- Zhou, W.; Zeng, S.; Yu, J.; Xiang, J.; Zhang, F.; Takriff, M.S.; Ding, G.; Ma, Z.; Zhou, X. Complete genome sequence of Bacillus Licheniformis NWMCC0046, a candidate for the laundry industry. J. Basic Microbiol. 2023, 63, 223–234. [Google Scholar] [CrossRef]
- Paria, P.; Chakraborty, H.J.; Behera, B.K. Identification of novel salt tolerance-associated proteins from the secretome of Enterococcus faecalis. World J. Microbiol. Biotechnol. 2022, 38, 177. [Google Scholar] [CrossRef]
- Rakesh, B.; Sudheer, W.; Nagella, P. Role of polyamines in plant tissue culture: An overview. Plant Cell Tissue Organ. Cult. 2021, 145, 487–506. [Google Scholar] [CrossRef]
- Zhang, B.-X.; Li, P.-S.; Wang, Y.-Y.; Wang, J.-J.; Liu, X.-L.; Wang, X.-Y.; Hu, X.-M. Characterization and synthesis of indole-3-acetic acid in plant growth promoting Enterobacter sp. RSC Adv. 2021, 11, 31601–31607. [Google Scholar] [CrossRef] [PubMed]
- Khazaal, M.T.; Faraag, A.H.; Hamada, M.A.; El-Hendawy, H.H. Characterization and Statistical Optimization of Enterobatin Synthesized by Escherichia coli OQ866153. Biochem. Genet. 2024, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Nigris, S.; Baldan, E.; Tondello, A.; Zanella, F.; Vitulo, N.; Favaro, G.; Guidolin, V.; Bordin, N.; Telatin, A.; Barizza, E. Biocontrol traits of Bacillus licheniformis GL174, a culturable endophyte of Vitis vinifera cv. Glera. BMC Microbiol. 2018, 18, 133. [Google Scholar] [CrossRef] [PubMed]
- Tans-Kersten, J.; Brown, D.; Allen, C. Swimming motility, a virulence trait of Ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Mol. Plant-Microbe Interact. 2004, 17, 686–695. [Google Scholar] [CrossRef]
- Bowen, G.; Rovira, A. Microbial colonization of plant roots. Annu. Rev. Phytopathol. 1976, 14, 121–144. [Google Scholar] [CrossRef]
- Kobayashi, K.; Kanesaki, Y.; Yoshikawa, H. Genetic analysis of collective motility of Paenibacillus sp. NAIST15-1. PLoS Genet. 2016, 12, e1006387. [Google Scholar] [CrossRef]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O. Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef]
- Chen, J.; Yang, W.; Li, J.; Anwar, S.; Wang, K.; Yang, Z.; Gao, Z. Effects of herbicides on the microbial community and urease activity in the rhizosphere soil of maize at maturity stage. J. Sens. 2021, 2021, 6649498. [Google Scholar] [CrossRef]
- Machado, R.M.A.; Serralheiro, R.P. Soil salinity: Effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, H.; Leng, J.; Niu, H.; Chen, X.; Liu, D.; Chen, Y.; Gao, N.; Ying, H. Isolation and characterization of plant growth-promoting rhizobacteria and their effects on the growth of Medicago sativa L. under salinity conditions. Antonie Van Leeuwenhoek 2020, 113, 1263–1278. [Google Scholar] [CrossRef]
- Zhou, H.; Chen, C.; Wang, D.; Arthur, E.; Zhang, Z.; Guo, Z.; Peng, X.; Mooney, S.J. Effect of long-term organic amendments on the full-range soil water retention characteristics of a Vertisol. Soil Tillage Res. 2020, 202, 104663. [Google Scholar] [CrossRef]
- Lu, H.; Zhang, Y.; Tian, T.; Li, X.; Wu, J.; Yang, H.; Huang, H. Preparation and properties of Sanxan gel based fertilizer for water retention and slow-release. Int. J. Biol. Macromol. 2023, 238, 124104. [Google Scholar] [CrossRef]
- Mekhzoum, M.E.M.; Aasfar, A.; Mzibra, A.; El Mernissi, N.; Farrie, Y.; Khouloud, M.; Boulif, R.; Qaiss, A.e.k.; Kadmiri, I.M.; Bouhfid, R. Phosphorus fertilizer coated with polysaccharide-enriched extracts from the red seaweed Schizymenia dubyi for slow release and water retention. J. Appl. Phycol. 2023, 35, 935–948. [Google Scholar] [CrossRef]
- Soliman, E.; Mansour, M.M. Enhancing soil organic carbon content and water retention using polyvinyl alcohol cross-linked with chitosan and pectin. J. Soil Sci. Plant Nutr. 2024, 24, 791–803. [Google Scholar] [CrossRef]
- Wei, X.; Xie, B.; Wan, C.; Song, R.; Zhong, W.; Xin, S.; Song, K. Enhancing soil health and plant growth through microbial fertilizers: Mechanisms, benefits, and sustainable agricultural practices. Agronomy 2024, 14, 609. [Google Scholar] [CrossRef]
- Shao, F.; Tao, W.; Yan, H.; Wang, Q. Effects of Microbial Organic Fertilizer (MOF) Application on Desert Soil Enzyme Activity and Jujube Yield and Quality. Agronomy 2023, 13, 2427. [Google Scholar] [CrossRef]
- Tian, J.; Ge, F.; Zhang, D.; Deng, S.; Liu, X. Roles of phosphate solubilizing microorganisms from managing soil phosphorus deficiency to mediating biogeochemical P cycle. Biology 2021, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-P.; Han, Q.-Q.; Liu, Q.-M.; Gan, Y.-N.; Rensing, C.; Rivera, W.L.; Zhao, Q.; Zhang, J.-L. Roles of phosphate-solubilizing bacteria in mediating soil legacy phosphorus availability. Microbiol. Res. 2023, 272, 127375. [Google Scholar] [CrossRef]
- Cheng, Y.; Narayanan, M.; Shi, X.; Chen, X.; Li, Z.; Ma, Y. Phosphate-solubilizing bacteria: Their agroecological function and optimistic application for enhancing agro-productivity. Sci. Total Environ. 2023, 166468. [Google Scholar] [CrossRef]
- Saeid, A.; Prochownik, E.; Dobrowolska-Iwanek, J. Phosphorus solubilization by Bacillus species. Molecules 2018, 23, 2897. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Ahmad, M.; Hussain, A.; Jamil, M. Integrated use of phosphate-solubilizing Bacillus subtilis strain IA6 and zinc-solubilizing Bacillus sp. strain IA16: A promising approach for improving cotton growth. Folia Microbiol. 2021, 66, 115–125. [Google Scholar] [CrossRef] [PubMed]
- James, N.; Umesh, M.; Sarojini, S.; Shanmugam, S.; Nasif, O.; Alharbi, S.A.; Lan Chi, N.T.; Brindhadevi, K. Unravelling the potential plant growth activity of halotolerant Bacillus licheniformis NJ04 isolated from soil and its possible use as a green bioinoculant on Solanum lycopersicum L. Environ. Res. 2023, 216, 114620. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Ramirez, L.F.; Uribe-Velez, D. Phosphorus solubilizing and mineralizing Bacillus spp. contribute to rice growth promotion using soil amended with rice straw. Curr. Microbiol. 2021, 78, 932–943. [Google Scholar] [CrossRef] [PubMed]
- Lili, W.; Shijun, Z.; Rui, D.; Jinbo, Z.; Shuquan, J. Effects of bio-organic fertilizer and microbial agent on the growth, yield and quality of tomato. Soils Crops 2022, 11, 88–95. [Google Scholar]
- Liu, Y.; Shi, J.; Feng, Y.; Yang, X.; Li, X.; Shen, Q. Tobacco bacterial wilt can be biologically controlled by the application of antagonistic strains in combination with organic fertilizer. Biol. Fertil. Soils 2013, 49, 447–464. [Google Scholar] [CrossRef]
- Tripathi, A.; Meena, B.; Pandey, K.; Singh, J. Microbial bioagents in agriculture: Current status and prospects. New Front. Stress Manag. Durable Agric. 2020, 331–368. [Google Scholar] [CrossRef]
- Lv, L.; Huang, H.; Lv, J.; Xu, X.; Cao, D.; Rao, Z.; Geng, F.; Kang, Y. Unique dissolved organic matter molecules and microbial communities in rhizosphere of three typical crop soils and their significant associations based on FT-ICR-MS and high-throughput sequencing analysis. Sci. Total Environ. 2024, 919, 170904. [Google Scholar] [CrossRef]
- del Barrio-Duque, A.; Samad, A.; Nybroe, O.; Antonielli, L.; Sessitsch, A.; Compant, S. Interaction between endophytic Proteobacteria strains and Serendipita indica enhances biocontrol activity against fungal pathogens. Plant Soil 2020, 451, 277–305. [Google Scholar] [CrossRef]
- Bovio-Winkler, P.; Guerrero, L.D.; Erijman, L.; Oyarzúa, P.; Suárez-Ojeda, M.E.; Cabezas, A.; Etchebehere, C. Genome-centric metagenomic insights into the role of Chloroflexi in anammox, activated sludge and methanogenic reactors. BMC Microbiol. 2023, 23, 45. [Google Scholar] [CrossRef]
- Yang, W.; Gong, T.; Wang, J.; Li, G.; Liu, Y.; Zhen, J.; Ning, M.; Yue, D.; Du, Z.; Chen, G. Effects of compound microbial fertilizer on soil characteristics and yield of wheat (Triticum aestivum L.). J. Soil Sci. Plant Nutr. 2020, 20, 2740–2748. [Google Scholar] [CrossRef]
- Javed, Z.; Tripathi, G.D.; Mishra, M.; Dashora, K. Actinomycetes–the microbial machinery for the organic-cycling, plant growth, and sustainable soil health. Biocatal. Agric. Biotechnol. 2021, 31, 101893. [Google Scholar] [CrossRef]
- Chen, Y.; Li, X.; Li, S.; Xu, Y. Novel-integrated process for production of bio-organic fertilizer via swine manure composting. Environ. Eng. Res. 2021, 26. [Google Scholar] [CrossRef]


| Pathways | Function | Genes | Details of Genes |
|---|---|---|---|
| Carbon metabolism | The glycolysis cycle | pyk | pyruvate kinase [EC:2.7.1.40] |
| Organic acid synthesis | ackA | acetate kinase [EC:2.7.2.1] | |
| fumC | fumarate hydratase, class II [EC:4.2.1.2] | ||
| gltA | citrate synthase [EC:2.3.3.1] | ||
| aceA | isocitrate lyase [EC:4.1.3.1] | ||
| acnA | aconitate hydratase [EC:4.2.1.3] | ||
| mdh | malate dehydrogenase [EC:1.1.1.37] | ||
| sdhB | succinate dehydrogenase [EC:1.3.5.1 1.3.5.4] | ||
| Nitrogen metabolism | Nitrogen metabolism | arcC | carbamate kinase [EC:2.7.2.2] |
| gudB | glutamate dehydrogenase [EC:1.4.1.2] | ||
| gltB | glutamate synthase (NADPH) large chain [EC:1.4.1.13] | ||
| glnA | glutamine synthetase [EC:6.3.1.2] | ||
| nirB | nitrite reductase (NADH) large subunit [EC:1.7.1.15] | ||
| narG narZ nxrA | nitrate reductase nitrite oxidoreductase, alpha subunit [EC:1.7.5.1 1.7.99.-] | ||
| norB | nitric oxide reductase subunit B [EC:1.7.2.5] | ||
| ncd2 npd | nitronate monooxygenase [EC:1.13.12.16] | ||
| nirD | nitrite reductase (NADH) small subunit [EC:1.7.1.15] | ||
| Tryptophan metabolism | Tryptophan metabolism | ALDH | aldehyde dehydrogenase (NAD+) [EC:1.2.1.3] |
| amiE | amidase [EC:3.5.1.4] | ||
| katE | catalase [EC:1.11.1.6] | ||
| atoB | acetyl-CoA C-acetyltransferase [EC:2.3.1.9] | ||
| Biosynthesis of siderophore group nonribosomal peptides | Biosynthesis of siderophore group nonribosomal peptides | entA | 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase [EC:1.3.1.28] |
| entB | bifunctional isochorismate lyase/ aryl carrier protein [EC:3.3.2.1 6.3.2.14] | ||
| entC | isochorismate synthase [EC:5.4.4.2] | ||
| mbtH | nocI; MbtH protein | ||
| Flagellar assembly | Flagellar assembly | flhA | flagellar biosynthesis protein FlhA |
| fliD | flagellar hook-associated protein 2 | ||
| fliI | flagellum-specific ATP synthase [EC:7.4.2.8] | ||
| fliF | flagellar M-ring protein FliF | ||
| fliD | flagellar hook-associated protein 2 | ||
| Flagellar motor proteins | motA | chemotaxis protein MotA | |
| motB | chemotaxis protein MotB | ||
| Bacterial chemotaxis | Bacterial chemotaxis | mcp | methyl-accepting chemotaxis protein |
| cheR | chemotaxis protein methyltransferase CheR [EC:2.1.1.80] | ||
| cheA | chemotaxis family, sensor kinase CheA [EC:2.7.13.3] |
| Group | Hydration (%) | pH | EC (μs/cm) | Salinity (mol/L) | TN (mg/kg) | AN (mg/kg) | TP (mg/kg) | AP (mg/kg) | TK (mg/kg) | AK (mg/kg) | SOM (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CF | 9.31 ± 0.54 c | 7.02 ± 0.09 c | 377.33 ± 9.45 a | 0.051 ± 0.04 a | 103.51 ± 6.25 c | 86 ± 0.19 b | 280.83 ± 7.04 a | 0.18 ± 0.07 a | 23.02 ± 2.3 a | 0.16 ± 0.05 a | 2.90 ± 0.27 b |
| OFCF | 10.47 ± 0.26 b | 7.23 ± 0.08 b | 329.33 ± 5.03 b | 0.037 ± 0.02 a | 139.70 ± 5.73 b | 89.33 ± 7.57 b | 235.00 ± 25.92 b | 0.17 ± 0.04 a | 25.94 ± 1.13 a | 0.12 ± 0.001 a | 5.09 ± 0.78 a |
| NF | 9.81 ± 0.13 bc | 7.47 ± 0.04 a | 295 ± 3 c | 0.025 ± 0.0 a | 191.01 ± 12.41 a | 119.67 ± 6.03 a | 91.46 ± 2.87 d | 0.24 ± 0.12 a | 25.69 ± 0.46 a | 0.104 ± 0.058 a | 3.31 ± 0.19 b |
| OFBOF | 11.67 ± 0.6 a | 7.41 ± 0.02 ab | 241 ± 7 d | 0.029 ± 0.01 a | 121.13 ± 0.04 bc | 79.67 ± 8.08 b | 133.68 ± 3.75 c | 0.28 ± 0.03 a | 26.34 ± 6.32 a | 0.1 ± 0.035 a | 5.58 ± 0.1 a |
| Sample | Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|---|
| CF | 28 | 84 | 167 | 232 | 342 | 268 |
| CFOF | 28 | 77 | 154 | 219 | 321 | 252 |
| OFBOF | 28 | 85 | 171 | 235 | 348 | 275 |
| NF | 29 | 84 | 162 | 220 | 321 | 253 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, S.; Wu, Y.; Zhu, N.; Leng, F.; Wang, Y. Bacillus licheniformisYB06: A Rhizosphere–Genome-Wide Analysis and Plant Growth-Promoting Analysis of a Plant Growth-Promoting Rhizobacterium Isolated from Codonopsis pilosula. Microorganisms 2024, 12, 1861. https://doi.org/10.3390/microorganisms12091861
Ni S, Wu Y, Zhu N, Leng F, Wang Y. Bacillus licheniformisYB06: A Rhizosphere–Genome-Wide Analysis and Plant Growth-Promoting Analysis of a Plant Growth-Promoting Rhizobacterium Isolated from Codonopsis pilosula. Microorganisms. 2024; 12(9):1861. https://doi.org/10.3390/microorganisms12091861
Chicago/Turabian StyleNi, Shuo, Yamiao Wu, Ning Zhu, Feifan Leng, and Yonggang Wang. 2024. "Bacillus licheniformisYB06: A Rhizosphere–Genome-Wide Analysis and Plant Growth-Promoting Analysis of a Plant Growth-Promoting Rhizobacterium Isolated from Codonopsis pilosula" Microorganisms 12, no. 9: 1861. https://doi.org/10.3390/microorganisms12091861
APA StyleNi, S., Wu, Y., Zhu, N., Leng, F., & Wang, Y. (2024). Bacillus licheniformisYB06: A Rhizosphere–Genome-Wide Analysis and Plant Growth-Promoting Analysis of a Plant Growth-Promoting Rhizobacterium Isolated from Codonopsis pilosula. Microorganisms, 12(9), 1861. https://doi.org/10.3390/microorganisms12091861






