Diversification of Pseudomonas aeruginosa Biofilm Populations under Repeated Phage Exposures Decreases the Efficacy of the Treatment
Abstract
:1. Introduction
2. Material and Methods
2.1. Bacterial Strains and Phages
2.2. Sequencing of Phages
2.3. Propagation and Titration of Phages
2.4. Biofilm Formation
2.5. Phage Treatment
2.6. Crystal Violet Staining
2.7. Viable Bacteria Count
2.8. Twitching Motility
2.9. Growth Experiments
2.10. Phage Susceptibility Assay
2.11. Sequencing of the Bacteria
2.12. Illumina Sequencing, Assembly, and Annotation
2.13. Sequence Analysis and Variant Calling
2.14. Statistical Analysis
3. Results
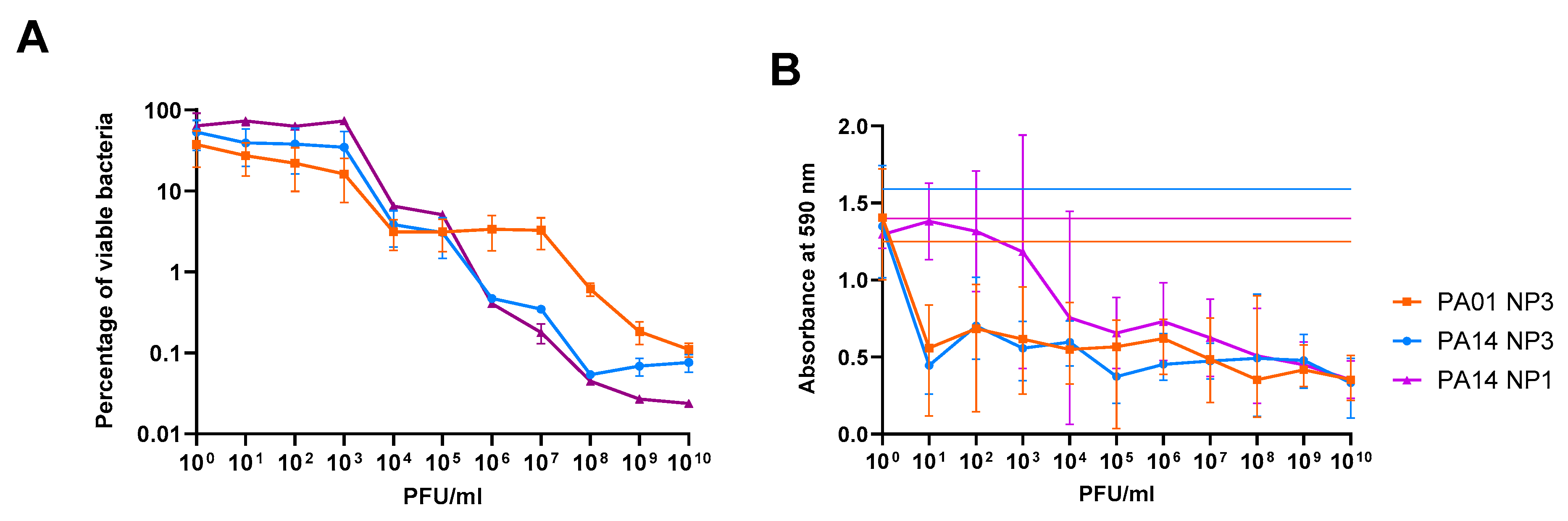
3.1. The Anti-Biofilm Effect Is Phage-Concentration-Dependent
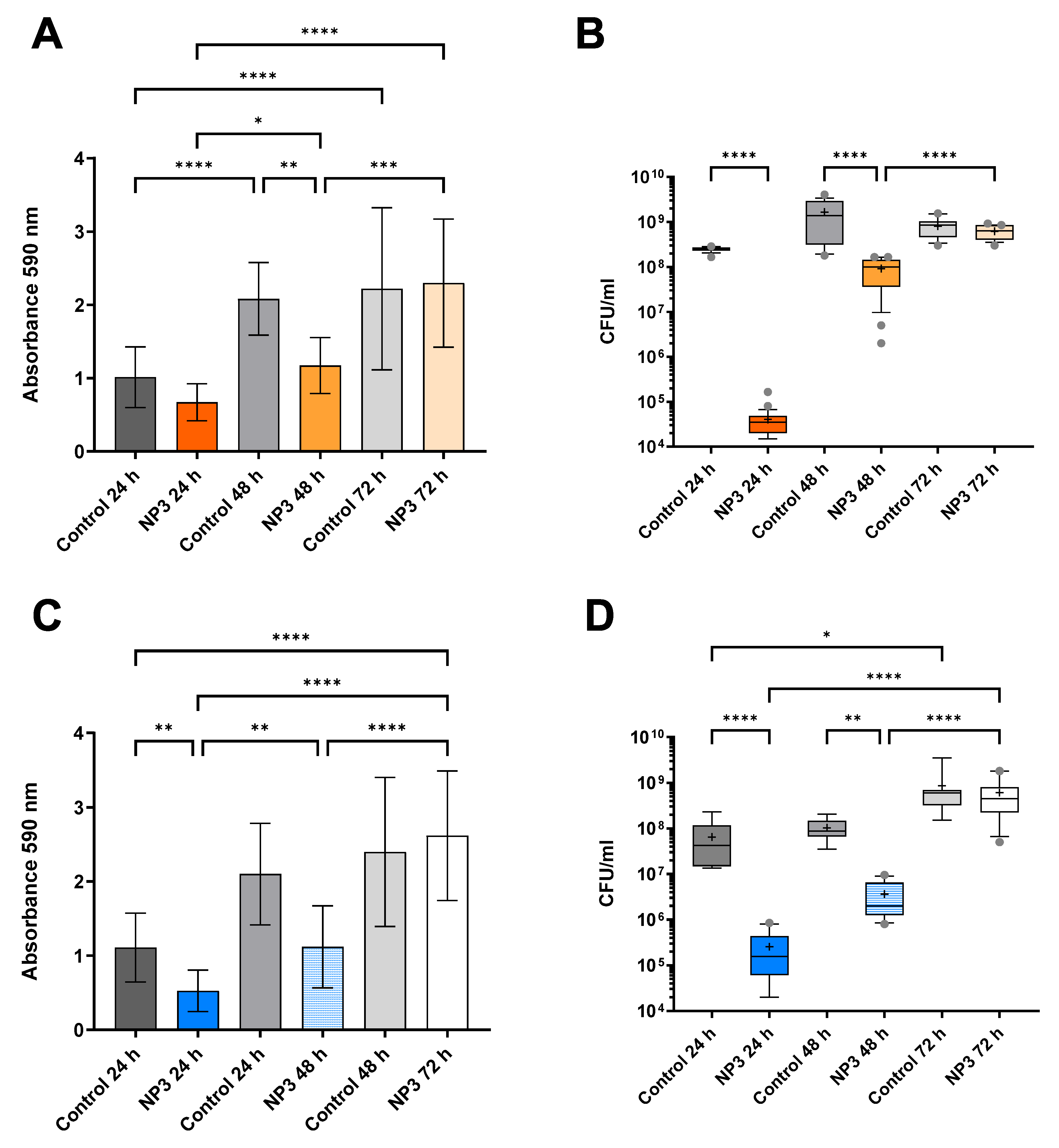
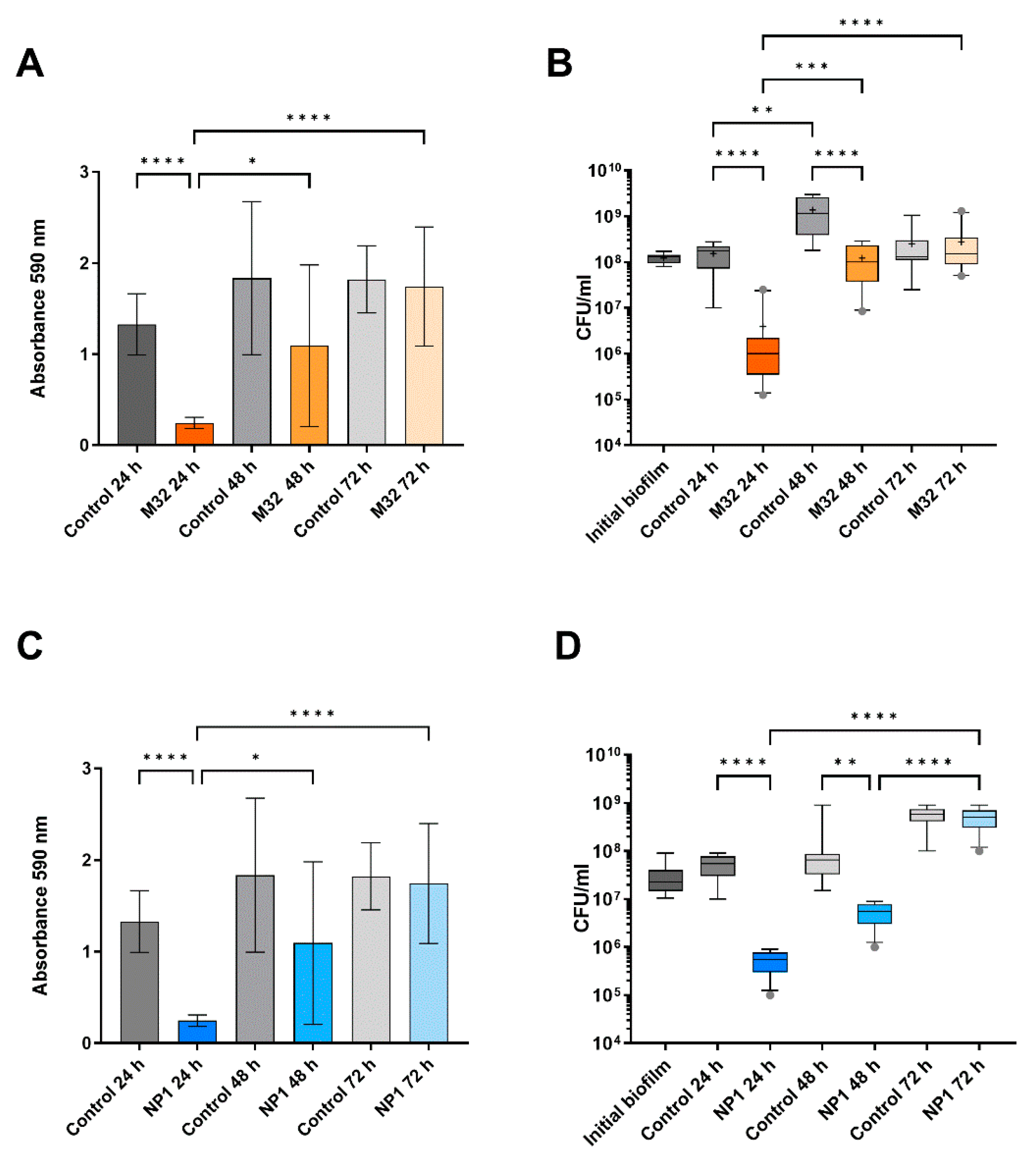
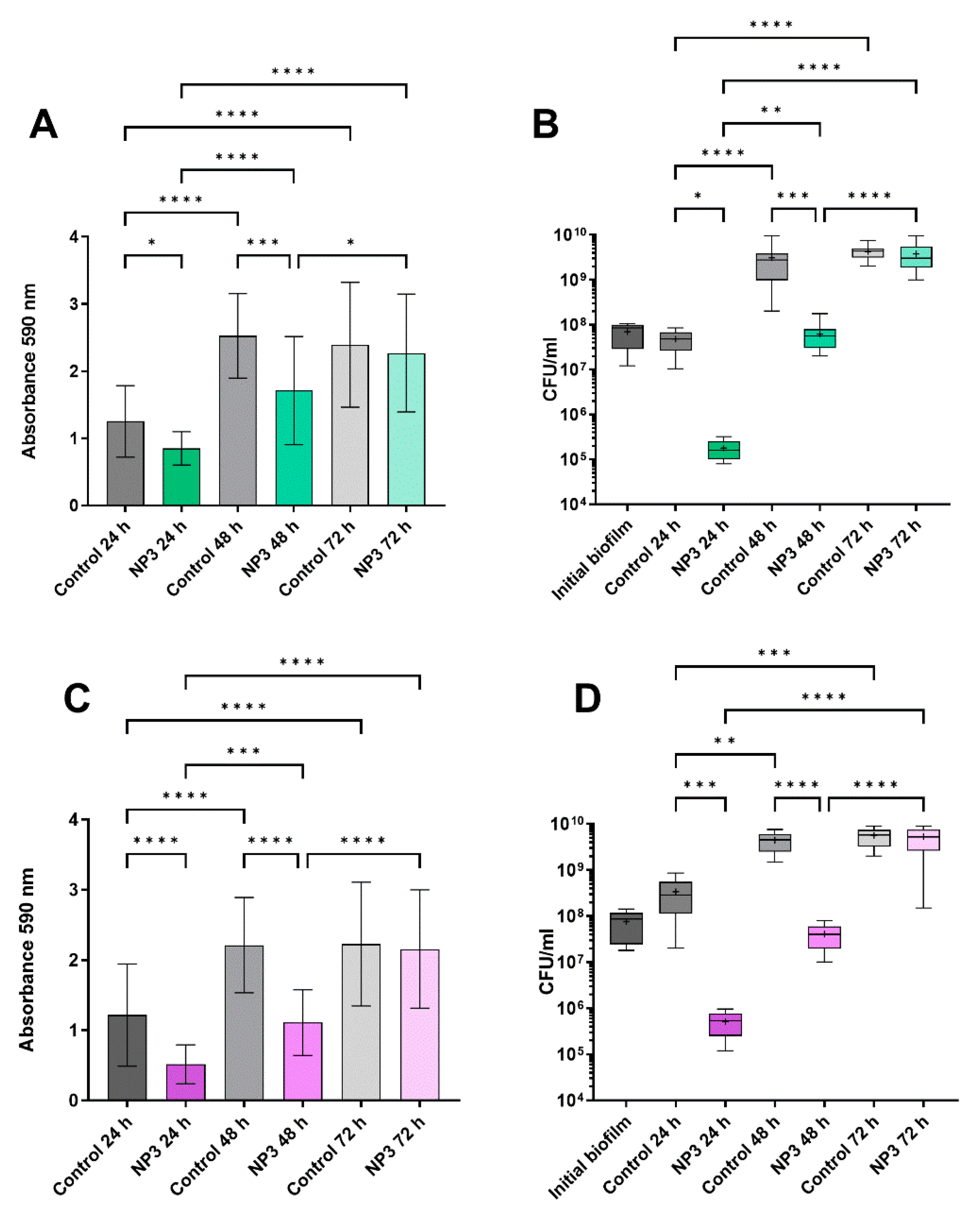
3.2. The Anti-Biofilm Effect Decreases with Repeated Phage Treatments
3.3. Sensitivity to Phages of Clones from Phage-Treated Biofilms
3.4. Twitching Motility of the Clones after Treatment with Phages
3.5. Effect of Phage Treatment on Growth Rate
3.6. Effect of Phage Treatment on Colony Morphotypes
3.7. Spontaneous Genetic Alterations in Clones Isolated from Biofilms
3.8. Genetic Alterations Related to Exposure by M32 and NP3 Phages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ciofu, O.; Moser, C.; Jensen, P.; Høiby, N. Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 2022, 20, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Hoiby, N.; Bjarnsholt, T.; Moser, C.; Bassi, G.L.; Coenye, T.; Donelli, G.; Hall-Stoodley, L.; Hola, V.; Imbert, C.; Kirketerp-Moller, K.; et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 2015, 21 (Suppl. S1), S1–S25. [Google Scholar] [CrossRef]
- Pires, D.P.; Melo, L.; Vilas, B.D.; Sillankorva, S.; Azeredo, J. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr. Opin. Microbiol. 2017, 39, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Ng, R.N.; Tai, A.S.; Chang, B.J.; Stick, S.M.; Kicic, A. Overcoming Challenges to Make Bacteriophage Therapy Standard Clinical Treatment Practice for Cystic Fibrosis. Front. Microbiol. 2020, 11, 593988. [Google Scholar] [CrossRef]
- Hansen, M.F.; Svenningsen, S.L.; Roder, H.L.; Middelboe, M.; Burmolle, M. Big Impact of the Tiny: Bacteriophage-Bacteria Interactions in Biofilms. Trends Microbiol. 2019, 27, 739–752. [Google Scholar] [CrossRef]
- Pires, D.P.; Meneses, L.; Brandão, A.C.; Azeredo, J. An overview of the current state of phage therapy for the treatment of biofilm-related infections. Curr. Opin. Virol. 2022, 53, 101209. [Google Scholar] [CrossRef]
- Pires, D.P.; Melo, L.D.R.; Azeredo, J. Understanding the Complex Phage-Host Interactions in Biofilm Communities. Annu. Rev. Virol. 2021, 8, 73–94. [Google Scholar] [CrossRef]
- Chen, Q.; Dharmaraj, T.; Cai, P.C.; Burgener, E.B.; Haddock, N.L.; Spakowitz, A.J.; Bollyky, P.L. Bacteriophage and Bacterial Susceptibility, Resistance, and Tolerance to Antibiotics. Pharmaceutics 2022, 14, 1425. [Google Scholar] [CrossRef]
- Henriksen, K.; Rorbo, N.; Rybtke, M.L.; Martinet, M.G.; Tolker-Nielsen, T.; Hoiby, N.; Middelboe, M.; Ciofu, O. P. aeruginosa flow-cell biofilms are enhanced by repeated phage treatments but can be eradicated by phage-ciprofloxacin combination. Pathog. Dis. 2019, 77, ftz011. [Google Scholar] [CrossRef]
- Hosseinidoust, Z.; Tufenkji, N.; van de Ven, T.G. Formation of biofilms under phage predation: Considerations concerning a biofilm increase. Biofouling 2013, 29, 457–468. [Google Scholar] [CrossRef]
- Chaudhry, W.N.; Concepcion-Acevedo, J.; Park, T.; Andleeb, S.; Bull, J.J.; Levin, B.R. Synergy and Order Effects of Antibiotics and Phages in Killing Pseudomonas aeruginosa Biofilms. PLoS ONE 2017, 12, e0168615. [Google Scholar] [CrossRef] [PubMed]
- Grace, A.; Sahu, R.; Owen, D.R.; Dennis, V.A. Pseudomonas aeruginosa reference strains PAO1 and PA14: A genomic, phenotypic, and therapeutic review. Front. Microbiol. 2022, 13, 1023523. [Google Scholar] [CrossRef]
- Simmons, E.L.; Bond, M.C.; Koskella, B.; Drescher, K.; Bucci, V.; Nadell, C.D. Biofilm Structure Promotes Coexistence of Phage-Resistant and Phage-Susceptible Bacteria. mSystems 2020, 5, e00877-19. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef]
- Freschi, L.; Vincent, A.T.; Jeukens, J.; Emond-Rheault, J.G.; Kukavica-Ibrulj, I.; Dupont, M.J.; Charette, S.J.; Boyle, B.; Levesque, R.C. The Pseudomonas aeruginosa Pan-Genome Provides New Insights on Its Population Structure, Horizontal Gene Transfer, and Pathogenicity. Genome Biol. Evol. 2019, 11, 109–120. [Google Scholar] [CrossRef]
- Jørgensen, K.M.; Wassermann, T.; Johansen, H.K.; Christiansen, L.E.; Molin, S.; Hoiby, N.; Ciofu, O. Diversity of metabolic profiles of cystic fibrosis Pseudomonas aeruginosa during the early stages of lung infection. Microbiology 2015, 161, 1447–1462. [Google Scholar] [CrossRef]
- Karumidze, N.; Thomas, J.A.; Kvatadze, N.; Goderdzishvili, M.; Hakala, K.W.; Weintraub, S.T.; Alavidze, Z.; Hardies, S.C. Characterization of lytic Pseudomonas aeruginosa bacteriophages via biological properties and genomic sequences. Appl. Microbiol. Biotechnol. 2012, 94, 1609–1617. [Google Scholar] [CrossRef]
- Hengzhuang, W.; Hoiby, N.; Ciofu, O. Pharmacokinetics and Pharmacodynamics of Antibiotics in Biofilm Infections of Pseudomonas aeruginosa In Vitro and In Vivo. Methods Mol. Biol. 2014, 1147, 239–254. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinformatics 2020, 70, e102. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 685. [Google Scholar] [CrossRef]
- Marquet, M.; Hölzer, M.; Pletz, M.W.; Viehweger, A.; Makarewicz, O.; Ehricht, R.; Brandt, C. What the Phage: A scalable workflow for the identification and analysis of phage sequences. GigaScience 2022, 11, giac110. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zeng, Y.; Wang, M.; Bao, R.; Chen, Y.; Li, X.; Pan, J.; Zhu, T.; Hu, B.; Tan, D. Characterization of Phage Resistance and Their Impacts on Bacterial Fitness in Pseudomonas aeruginosa. Microbiol. Spectr. 2022, 10, e0207222. [Google Scholar] [CrossRef]
- Chen, G.; Gan, J.; Yang, C.; Zuo, Y.; Peng, J.; Li, M.; Huo, W.; Xie, Y.; Zhang, Y.; Wang, T.; et al. The SiaA/B/C/D signaling network regulates biofilm formation in Pseudomonas aeruginosa. EMBO J. 2020, 39, e103412. [Google Scholar] [CrossRef]
- Leighton, T.L.; Buensuceso, R.N.; Howell, P.L.; Burrows, L.L. Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ. Microbiol. 2015, 17, 4148–4163. [Google Scholar] [CrossRef]
- Kuchma, S.L.; O’Toole, G.A. Surface-Induced cAMP Signaling Requires Multiple Features of the Pseudomonas aeruginosa Type IV Pili. J. Bacteriol. 2022, 204, e0018622. [Google Scholar] [CrossRef]
- Melamed, J.; Kocev, A.; Torgov, V.; Veselovsky, V.; Brockhausen, I. Biosynthesis of the Pseudomonas aeruginosa common polysaccharide antigen by D-Rhamnosyltransferases WbpX and WbpY. Glycoconj. J. 2022, 39, 393–411. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.T.; Lam, J.S. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can. J. Microbiol. 2014, 60, 697–716. [Google Scholar] [CrossRef]
- Nordstrom, H.R.; Evans, D.R.; Finney, A.G.; Westbrook, K.J.; Zamora, P.F.; Hofstaedter, C.E.; Yassin, M.H.; Pradhan, A.; Iovleva, A.; Ernst, R.K.; et al. Genomic characterization of lytic bacteriophages targeting genetically diverse Pseudomonas aeruginosa clinical isolates. iScience 2022, 25, 104372. [Google Scholar] [CrossRef]
- Khandekar, S.; Liebens, V.; Fauvart, M.; Tulkens, P.M.; Michiels, J.; Van Bambeke, F. The Putative De-N-acetylase DnpA Contributes to Intracellular and Biofilm-Associated Persistence of Pseudomonas aeruginosa Exposed to Fluoroquinolones. Front. Microbiol. 2018, 9, 1455. [Google Scholar] [CrossRef]
- Pulcrano, G.; Iula, D.V.; Raia, V.; Rossano, F.; Catania, M.R. Different mutations in mucA gene of Pseudomonas aeruginosa mucoid strains in cystic fibrosis patients and their effect on algU gene expression. New Microbiol. 2012, 35, 295–305. [Google Scholar]
- Ciofu, O.; Mandsberg, L.F.; Bjarnsholt, T.; Wassermann, T.; Hoiby, N. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: Strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology 2010, 156, 1108–1119. [Google Scholar] [CrossRef]
- Olvera, C.; Goldberg, J.B.; Sánchez, R.; Soberón-Chávez, G. The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol. Lett. 1999, 179, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Gallique, M.; Bouteiller, M.; Merieau, A. The Type VI Secretion System: A Dynamic System for Bacterial Communication? Front. Microbiol. 2017, 8, 1454. [Google Scholar] [CrossRef]
- Tagel, M.; Ilves, H.; Leppik, M.; Jurgenstein, K.; Remme, J.; Kivisaar, M. Pseudouridines of tRNA Anticodon Stem-Loop Have Unexpected Role in Mutagenesis in Pseudomonas sp. Microorganisms 2020, 9, 25. [Google Scholar] [CrossRef]
- Wang, X.; Xie, Z.; Zhao, J.; Zhu, Z.; Yang, C.; Liu, Y. Prospects of Inhaled Phage Therapy for Combatting Pulmonary Infections. Front. Cell. Infect. Microbiol. 2021, 11, 758392. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, N.M.; Devequi Gomes Nunes, D.; Shiach, J.; Valeria Saraiva Hodel, K.; Dantas Viana Barbosa, J.; Alencar Pereira Rodrigues, L.; Coler, B.S.; Botelho Pereira Soares, M.; Badaró, R. Current Clinical Landscape and Global Potential of Bacteriophage Therapy. Viruses 2023, 15, 1020. [Google Scholar] [CrossRef]
- Chegini, Z.; Khoshbayan, A.; Taati Moghadam, M.; Farahani, I.; Jazireian, P.; Shariati, A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: A review. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 45. [Google Scholar] [CrossRef]
- Markwitz, P.; Olszak, T.; Gula, G.; Kowalska, M.; Arabski, M.; Drulis-Kawa, Z. Emerging Phage Resistance in Pseudomonas aeruginosa PAO1 Is Accompanied by an Enhanced Heterogeneity and Reduced Virulence. Viruses 2021, 13, 1332. [Google Scholar] [CrossRef]
- Visnapuu, A.; Van der Gucht, M.; Wagemans, J.; Lavigne, R. Deconstructing the Phage-Bacterial Biofilm Interaction as a Basis to Establish New Antibiofilm Strategies. Viruses 2022, 14, 1057. [Google Scholar] [CrossRef]
- Kilmury, S.L.N.; Burrows, L.L. The Pseudomonas aeruginosa PilSR Two-Component System Regulates Both Twitching and Swimming Motilities. mBio 2018, 9, e01310-18. [Google Scholar] [CrossRef]
- Weaver, S.J.; Ortega, D.R.; Sazinsky, M.H.; Dalia, T.N.; Dalia, A.B.; Jensen, G.J. CryoEM structure of the type IVa pilus secretin required for natural competence in Vibrio cholerae. Nat. Commun. 2020, 11, 5080. [Google Scholar] [CrossRef] [PubMed]
- Koo, J.; Tammam, S.; Ku, S.Y.; Sampaleanu, L.M.; Burrows, L.L.; Howell, P.L. PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa Type IV pilus secretin. J. Bacteriol. 2008, 190, 6961–6969. [Google Scholar] [CrossRef] [PubMed]
- Webb, J.S.; Lau, M.; Kjelleberg, S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 2004, 186, 8066–8073. [Google Scholar] [CrossRef] [PubMed]
- Hesse, S.; Rajaure, M.; Wall, E.; Johnson, J.; Bliskovsky, V.; Gottesman, S.; Adhya, S. Phage Resistance in Multidrug-Resistant Klebsiella pneumoniae ST258 Evolves via Diverse Mutations That Culminate in Impaired Adsorption. mBio 2020, 11, e02530-19. [Google Scholar] [CrossRef]
- Priebe, G.P.; Dean, C.R.; Zaidi, T.; Meluleni, G.J.; Coleman, F.T.; Coutinho, Y.S.; Noto, M.J.; Urban, T.A.; Pier, G.B.; Goldberg, J.B. The galU Gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect. Immun. 2004, 72, 4224–4232. [Google Scholar] [CrossRef]
- Vaitekenas, A.; Tai, A.S.; Ramsay, J.P.; Stick, S.M.; Kicic, A. Pseudomonas aeruginosa Resistance to Bacteriophages and Its Prevention by Strategic Therapeutic Cocktail Formulation. Antibiotics 2021, 10, 145. [Google Scholar] [CrossRef]
- Viars, S.; Valentine, J.; Hernick, M. Structure and function of the LmbE-like superfamily. Biomolecules 2014, 4, 527–545. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.R. Mucoid variation in Pseudomonas aeruginosa induced by the action of phage. J. Med. Microbiol. 1973, 6, 111–118. [Google Scholar] [CrossRef]
- Katharios-Lanwermeyer, S.; Whitfield, G.B.; Howell, P.L.; O’Toole, G.A. Pseudomonas aeruginosa Uses c-di-GMP Phosphodiesterases RmcA and MorA To Regulate Biofilm Maintenance. mBio 2021, 12, e03384-20. [Google Scholar] [CrossRef]
- Schumann, A.R.; Sue, A.D.; Roach, D.R. Hypoxia Increases the Tempo of Phage Resistance and Mutational Bottlenecking of Pseudomonas aeruginosa. Front. Microbiol. 2022, 13, 905343. [Google Scholar] [CrossRef]
- Boles, B.R.; Singh, P.K. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl. Acad. Sci. USA 2008, 105, 12503–12508. [Google Scholar] [CrossRef]
- Da Cruz Nizer, W.S.; Inkovskiy, V.; Versey, Z.; Strempel, N.; Cassol, E.; Overhage, J. Oxidative Stress Response in Pseudomonas aeruginosa. Pathogens 2021, 10, 1187. [Google Scholar] [CrossRef] [PubMed]
- Menon, N.D.; Penziner, S.; Montaño, E.T.; Zurich, R.; Pride, D.T.; Nair, B.G.; Kumar, G.B.; Nizet, V. Increased Innate Immune Susceptibility in Hyperpigmented Bacteriophage-Resistant Mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2022, 66, e0023922. [Google Scholar] [CrossRef]
- Moreau, P.; Diggle, S.P.; Friman, V.P. Bacterial cell-to-cell signaling promotes the evolution of resistance to parasitic bacteriophages. Ecol. Evol. 2017, 7, 1936–1941. [Google Scholar] [CrossRef]
- Mauritzen, J.J.; Søndberg, E.; Kalatzis, P.G.; Roager, L.; Gram, L.; Svenningsen, S.L.; Middelboe, M. Strain-specific quorum-sensing responses determine virulence properties in Vibrio anguillarum. Environ. Microbiol. 2023, 25, 1344–1362. [Google Scholar] [CrossRef] [PubMed]
- Kaern, M.; Elston, T.C.; Blake, W.J.; Collins, J.J. Stochasticity in gene expression: From theories to phenotypes. Nat. Rev. Genet. 2005, 6, 451–464. [Google Scholar] [CrossRef]
- Blasco, L.; López-Hernández, I.; Rodríguez-Fernández, M.; Pérez-Florido, J.; Casimiro-Soriguer, C.S.; Djebara, S.; Merabishvili, M.; Pirnay, J.P.; Rodríguez-Baño, J.; Tomás, M.; et al. Case report: Analysis of phage therapy failure in a patient with a Pseudomonas aeruginosa prosthetic vascular graft infection. Front. Med. 2023, 10, 1199657. [Google Scholar] [CrossRef]

| Parameter | PAO1 + M32 | PAO1 + NP3 | PA14 + NP3 | CF341_06 + NP3 | CF341_08 + NP3 | |
|---|---|---|---|---|---|---|
| Number of clones | S | 8 | 0 | 10 | 14 | 16 |
| R | 40 | 48 | 38 | 34 | 32 | |
| Ʃ | 48 | 48 | 48 | 48 | 48 | |
| Phage susceptibility | S [%] | 16.7 | 0 | 20.8 | 29.2 | 33.3 |
| R [%] | 83.3 | 100 | 79.2 | 70.8 | 66.7 | |
| (Ø ± SD) for S [PFU/mL] | (1.43 ± 3.78) × 109 | / | (7.93 ± 14.7) × 1011 | (2.94 ± 3.32) × 1011 | (2.77 ± 2.95) × 1011 | |
| Twitching motility (Ø ± SD) | dS [mm] | 13.25 ± 18.83 | / | 21.80 ± 15.60 | 17.79 ± 12.21 | 9.64 ± 7.13 |
| dR [mm] | 13.33 ± 16.99 | 7.89 ± 13.47 | 6.34 ± 10.30 | 13.59 ± 10.04 | 14.31 ± 6.59 | |
| p-value (S vs. R) | 0.9901 | / | 0.0005 | 0.2228 | 0.0388 | |
| dC [mm] | 34 ± 25.5 | 41.5 ± 26.7 | 34.6 ± 5.7 | 40. 3 ± 10.1 | 17.9 ± 10.2 | |
| p-value (C vs. S) | 0.051 | / | <0.014 | <0.0001 | <0.0240 | |
| p-value (C vs. R) | 0.019 | 0.0006 | <0.0001 | <0.0001 | 0.2814 | |
| Growth rates (Ø ± SD) | Ø µR [1/h] | 0.26 ±0.03 | 0.20 ± 0.04 | 0.23 ± 0.03 | 0.14 ± 0.04 | 0.10 ± 0.04 |
| Ø µS [1/h] | 0.23 ± 0.03 | / | 0.18 ± 0.02 | 0.16 ± 0.08 | 0.15 ± 0.04 | |
| p-value | 0.0945 | / | 0.0196 | 0.6503 | 0.0972 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinet, M.G.; Lohde, M.; Higazy, D.; Brandt, C.; Pletz, M.W.; Middelboe, M.; Makarewicz, O.; Ciofu, O. Diversification of Pseudomonas aeruginosa Biofilm Populations under Repeated Phage Exposures Decreases the Efficacy of the Treatment. Microorganisms 2024, 12, 1880. https://doi.org/10.3390/microorganisms12091880
Martinet MG, Lohde M, Higazy D, Brandt C, Pletz MW, Middelboe M, Makarewicz O, Ciofu O. Diversification of Pseudomonas aeruginosa Biofilm Populations under Repeated Phage Exposures Decreases the Efficacy of the Treatment. Microorganisms. 2024; 12(9):1880. https://doi.org/10.3390/microorganisms12091880
Chicago/Turabian StyleMartinet, Mark Grevsen, Mara Lohde, Doaa Higazy, Christian Brandt, Mathias W. Pletz, Mathias Middelboe, Oliwia Makarewicz, and Oana Ciofu. 2024. "Diversification of Pseudomonas aeruginosa Biofilm Populations under Repeated Phage Exposures Decreases the Efficacy of the Treatment" Microorganisms 12, no. 9: 1880. https://doi.org/10.3390/microorganisms12091880









