Efficacy of Indigenous Bacteria in the Biodegradation of Hydrocarbons Isolated from Agricultural Soils in Huamachuco, Peru
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Microbial Isolation
2.3. Selection of Hydrocarbon Degrading Bacteria
2.4. Analytical Method
2.5. Identification of Selected Bacteria
2.6. Statistical Analysis
3. Results
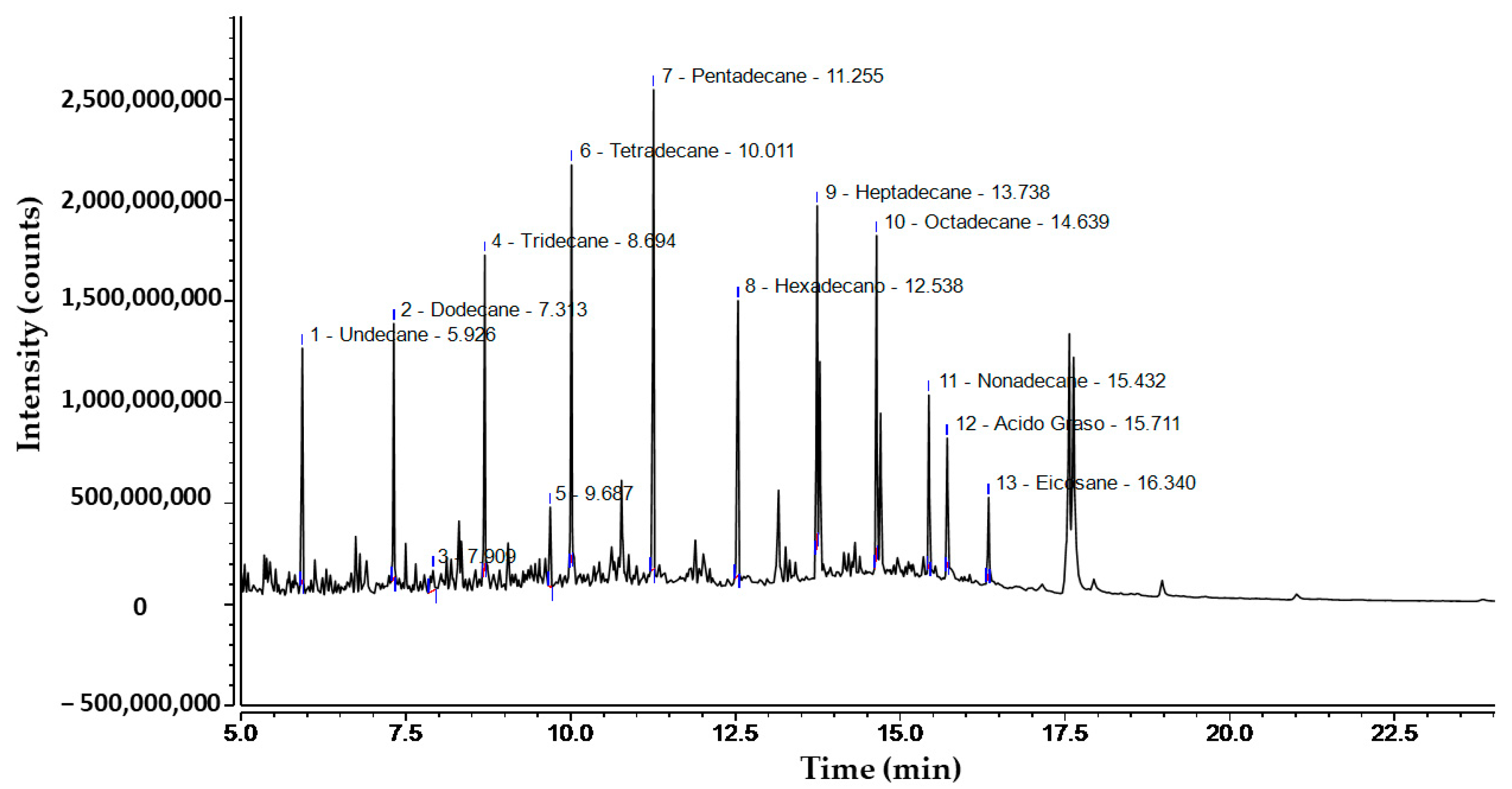
3.1. Chromatographic Characterization of Diesel
3.2. Characterization of the Isolated and Selected Bacteria
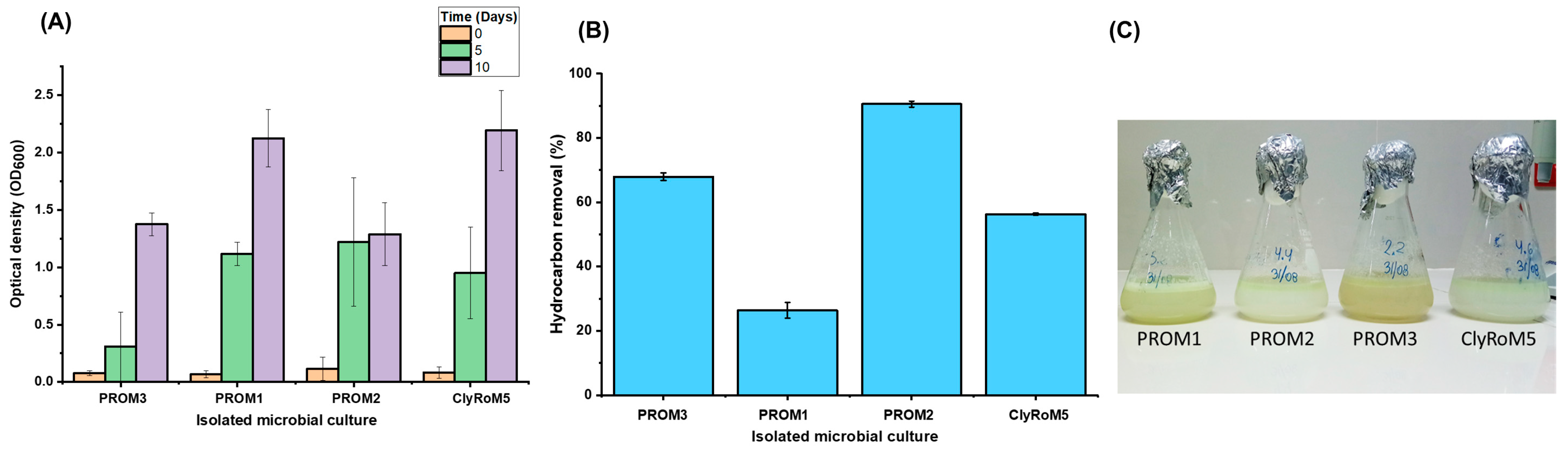
3.3. Diesel Biodegradation Tests
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sarıkoç, S. Fuels of the diesel-gasoline engines and their properties. In Diesel and Gasoline Engines, 1st ed.; Viskup, R., Ed.; IntechOpen: London, UK, 2020; Volume 1, pp. 1–16. [Google Scholar] [CrossRef]
- Olivera, C.; Laura-Tondo, M.; Girardi, V.; Sol-Herrero, M.; Lucía-Balaban, C.; Matías-Salvatierra, L. High-performance diesel biodegradation using biogas digestate as microbial inoculum in lab-scale solid supported bioreactors. Chemosphere 2024, 352, 141384. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Li, Q.; Ding, X.; Sun, J.; Li, D.; Fu, H.; Teich, M.; Ye, X.; Chen, J. Primary particulate matter emitted from heavy fuel and diesel oil combustion in a typical container ship: Characteristics and Toxicity. Environ. Sci. Technol. 2018, 52, 12943–12951. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.T.; Chun-Te, J.; Chen, S.H.; Verpoort, F.; Hong, K.L.; Surampalli, R.Y.; Kao, C.M. Remediation of diesel-oil contaminated soils using an innovative nanobubble and electrolyzed catalytic system. J. Clean. Prod. 2023, 432, 139776. [Google Scholar] [CrossRef]
- Nzila, A. Current status of the degradation of aliphatic and aromatic petroleum hydrocarbons by thermophilic microbes and future perspectives. Int. J. Environ. Res. Public Health 2018, 15, 2782. [Google Scholar] [CrossRef]
- Chaudhary, D.K.; Kim, J. New insights into bioremediation strategies for oil-contaminated soil in cold environments. Int. Biodeterior. Biodegrad. 2019, 142, 58–72. [Google Scholar] [CrossRef]
- Gao, J.; Ming, J.; Xu, M.; Fu, X.; Duan, L.F.; Xu, C.C.; Gao, Y.; Xue, J.L.; Xiao, X.F. Isolation and characterization of a high-efficiency marine diesel oil-degrading bacterium. Pet. Sci. 2021, 18, 641–653. [Google Scholar] [CrossRef]
- Imron, M.F.; Kurniawan, S.B.; Titah, H.S. Potential of bacteria isolated from diesel-contaminated seawater in diesel biodegradation. Environ. Technol. Innov. 2019, 14, 100368. [Google Scholar] [CrossRef]
- Yap, H.S.; Khalid, F.E.; Wong, R.R.; Convey, P.; Sabri, S.; Khalil, K.A.; Zulkharnain, A.; Merican, F.; Shaari, H.; Ahmad, S.A. Diesel−biodegradation and biosurfactant—Production by Janthinobacterium lividum AQ5-29 and Pseudomonas fildesensis AQ5-41 isolated from Antarctic soil. Int. Biodeterior. Biodegrad. 2024, 188, 105731. [Google Scholar] [CrossRef]
- Sawadogo, A.; Cissé, H.; Otoidobiga, H.C.; Odetokun, I.A.; Zongo, C.; Dianou, D.; Savadogo, A. Characterization of two bacterial strains isolated from wastewater and exhibiting in-vitro degradation of diesel and used oils. Sci. Afr. 2024, 25, e02289. [Google Scholar] [CrossRef]
- Patowary, K.; Patowary, R.; Kalita, M.C.; Deka, S. Development of an Efficient Bacterial Consortium for the Potential Remediation of Hydrocarbons from Contaminated Sites. Front. Microbiol. 2016, 7, 1092. [Google Scholar] [CrossRef]
- Alsberi, H.; Hamad, A.A.; Hassan, M.M. Biodegradation of petroleum hydrocarbons using indigenous bacterial and actinomycetes cultures. Pak. J. Biol. Sci. 2020, 23, 726–734. [Google Scholar] [CrossRef]
- Bharali, P.; Bashir, Y.; Ray, A.; Dutta, N.; Mudoi, P.; Alemtoshi; Sorhie, V.; Vishwakarma, V.; Debnath, P.; Konwar, B.K. Bioprospecting of indigenous biosurfactant-producing oleophilic bacteria for green remediation: An eco-sustainable approach for the management of petroleum contaminated soil. 3 Biotech 2022, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- Oba, A.; John, B.; Garba, J.; Oba, A.J.; John, K.V.; Balami, S.B.; Uchechukwu, O.; Musa, J.A.; Ofili, A. Bioprospecting of hydrocarbonoclastic representative bacteria. J. Environ. Prot. 2022, 13, 449–458. [Google Scholar] [CrossRef]
- Wang, M.; Ding, M.; Yuan, Y. Bioengineering for the microbial degradation of petroleum hydrocarbon contaminants. Bioengineering 2023, 10, 347. [Google Scholar] [CrossRef]
- Gao, Y.; Du, J.; Bahar, M.M.; Wang, H.; Subashchandrabose, S.; Duan, L.; Yang, X.; Megharaj, M.; Zhao, Q.; Zhang, W.; et al. Metagenomics analysis identifies nitrogen metabolic pathway in bioremediation of diesel contaminated soil. Chemosphere 2021, 271, 129566. [Google Scholar] [CrossRef]
- Lostaunau-Silvera, C.A.; Puris-Naupay, J.E.; Zaldivar-Alvarez, W.F.; King-Santos, M.E.; Anahua-Balcon, E.A.; Reátegui-Romero, W. Removal of organic contaminants from the water used to wash the tanks of trucks transporting diesel: Electrocoagulation in batch mode. Desalination Water Treat. 2023, 315, 241–250. [Google Scholar] [CrossRef]
- Cruz, J.; Quiñones, C.; Saavedra, J.B.; Urquizo, D.; Esparza, M. Biodegradation of phenol by Pseudomonas aeruginosa isolated from oil contaminated environments in Peru. Biosci. Res. 2021, 18, 1294–1300. [Google Scholar] [CrossRef]
- Landrigan, P.J.; Stegeman, J.J.; Fleming, L.E.; Allemand, D.; Anderson, D.M.; Backer, L.C.; Brucker-Davis, F.; Chevalier, N.; Corra, L.; Czerucka, D.; et al. Human health and ocean pollution. Ann. Glob. Health 2020, 86, 151. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.D.; Oliveira, A.F.; Golin, R.; Lopes, V.C.P.; Caixeta, D.S.; Lima, Z.M.; Morais, E.B. Isolation and characterization of hydrocarbon-degrading bacteria from gas station leaking-contaminated groundwater in the Southern Amazon, Brazil. Braz. J. Biol. 2020, 80, 354–361. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, M.; Xue, J.; Shi, K.; Gu, M. Characterization and Enhanced Degradation Potentials of Biosurfactant-Producing Bacteria Isolated from a Marine Environment. ACS Omega 2019, 4, 1645–1651. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, Y.; Li, C.; Fang, Y.; Li, X. Physicochemical characteristics of particulate matter emitted from the oxygenated fuel/diesel blend engine. Aerosol. Air Qual. Res. 2021, 21, 210175. [Google Scholar] [CrossRef]
- Akbari, A.; David, C.; Rahim, A.A.; Ghoshal, S. Salt selected for hydrocarbon-degrading bacteria and enhanced hydrocarbon biodegradation in slurry bioreactors. Water Res. 2021, 202, 117424. [Google Scholar] [CrossRef] [PubMed]
- Xin, Q.; Saborimanesh, N.; Greer, C.W.; Farooqi, H.; Dettman, H.D. The effect of temperature on hydrocarbon profiles and the microbial community composition in North Saskatchewan River water during mesoscale tank tests of diluted bitumen spills. Sci. Total Environ. 2023, 859, 160161. [Google Scholar] [CrossRef]
- Donner, J.; Reck, M.; Bunk, B.; Jarek, M.; App, C.; Meier-Kolthoff, J.; Overmann, J.; Müller, R.; Kirschning, A.; Wagner-Döbler, I. The Biofilm Inhibitor Carolacton Enters Gram-Negative Cells: Studies Using a TolC-Deficient Strain of Escherichia coli Jannik. mSphere 2017, 2, e00375-17. [Google Scholar] [CrossRef] [PubMed]
- Tejada, K.C.; Quiñones, C.E.; Salirrosas, D.; Huanes, J.E.; Valdivieso, S.C.; Cruz, J.A.; Haro, D.; Rodriguez, J. Production of Prodigiosin using Serratia marcescens from tilapia scale hydrolysates: Influence of stirring speed and NaCl concentration. Chem. Eng. Trans. 2024, 108, 37–42. [Google Scholar] [CrossRef]
- Cueva-almendras, L.C.; Alva, J.; Fuentes-Olivera, A.; Llontop-Bernabé, K.; Quiñones, C.; Rodriguez-Soto, J.; Cruz-Monzon, J.; Quezada-Alvarez, M. Production of polyhydroxyalkanoate by Bacillus thuringiensis isolated from agricultural soils of Cascas-Peru. Braz. Arch. Biol. Technol. 2022, 65, e22220107. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, H.; Li, J.; Wang, B.; Qi, C.; Hu, X. Nutrient-enhanced n-alkanes biodegradation and succession of bacterial communities. J. Oceanol. Limnol. 2018, 36, 1294–1303. [Google Scholar] [CrossRef]
- Yang, Z.; Shah, K.; Pilon-McCullough, C.; Faragher, R.; Azmi, P.; Hollebone, B.; Fieldhouse, B.; Yang, C.; Dey, D.; Lambert, P.; et al. Characterization of renewable diesel, petroleum diesel and renewable diesel/biodiesel/petroleum diesel blends. Renew. Energy 2024, 224, 120151. [Google Scholar] [CrossRef]
- Bekele, G.K.; Gebrie, S.A.; Mekonen, E.; Fida, T.T.; Woldesemayat, A.A.; Abda, E.M.; Tafesse, M.; Assefa, F. Isolation and Characterization of Diesel-Degrading Bacteria from Hydrocarbon-Contaminated Sites, Flower Farms, and Soda Lakes. Int. J. Microbiol. 2022, 2022, 5655767. [Google Scholar] [CrossRef]
- Pagès, S.; Ogier, J.C.; Gaudriault, S. A novel semi-selective medium for Pseudomonas protegens isolation from soil samples. J. Microbiol. Methods 2020, 172, 105911. [Google Scholar] [CrossRef]
- Ravishankar, P.; Srinivas, R.M.; Bharathi, K.; Subramanian, S.K.; Asiedu, S.K.; Selvaraj, D. Unlocking nature’s toolbox: Kinetin-producing Priestia flexa VL1 paves the way for efficient bioremediation of chromium-contaminated environments. J. Environ. Chem. Eng. 2024, 12, 112065. [Google Scholar] [CrossRef]
- Gao, H.; Feng, G.; Feng, Z.; Yao, Q.; Li, J.; Deng, X.; Li, X.; Zhu, H. Pseudomonas citri sp. nov., a potential novel plant growth promoting bacterium isolated from rhizosphere soil of citrus. Antonie Van Leeuwenhoek 2023, 116, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Faiza, B.; Mohamed, B.B.H.; Sidi-Mohammed, E.A.A. Crude oil degradation potential of indigenous hydrocarbonoclastic bacterial strain Acinetobacter johnsonii firstly isolated from marine sediments of Oran Port, Algeria. J. Environ. Sci. Eng. A 2019, 8, 131–140. [Google Scholar] [CrossRef]
- Dohare, S.; Kumar, H.; Bhargava, Y.; Kango, N. Characterization of Diesel Degrading Indigenous Bacterial Strains, Acinetobacter pittii and Pseudomonas aeruginosa, Isolated from Oil Contaminated Soils. Indian J. Microbiol. 2024, 64, 749–757. [Google Scholar] [CrossRef]
- Palanisamy, N.; Ramya, J.; Kumar, S.; Vasanthi, N.S.; Chandran, P.; Khan, S. Diesel biodegradation capacities of indigenous bacterial species isolated from diesel contaminated soil. Environ. Health Sci. Eng. 2014, 12, 142. [Google Scholar] [CrossRef] [PubMed]
- Ashikodi, A.O.; Abu, G.O. Hydrocarbon degradation potential of some hydrocarbon—Utilizing bacterial species associated with Kenaf (Hibiscus cannabinus L.) plant. Int. Res. J. Biol. Sci. 2019, 8, 10–19. [Google Scholar]
- Talaiekhozani, A.; Jafarzadeh, N.; Fulazzaky, M.A.; Talaie, M.R. Kinetics of substrate utilization and bacterial growth of crude oil degraded by Pseudomonas aeruginosa. J. Environ. Health Sci. Eng. 2015, 13, 64. [Google Scholar] [CrossRef]
- Ho, M.T.; Li, M.S.; Mcdowell, T.; Macdonald, J.; Yuan, Z. Characterization and genomic analysis of a diesel-degrading bacterium, Acinetobacter calcoaceticus CA16, isolated from Canadian soil. BMC Biotechnol. 2020, 20, 39. [Google Scholar] [CrossRef]
- Stancu, M.M. Characterization of new diesel-degrading bacteria isolated from freshwater sediments. Int. Microbiol. 2023, 26, 109–122. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Xu, H.; Zhang, S.; Kim, H.; Chiang, P. Comparison of Petroleum Hydrocarbons Degradation by Klebsiella pneumoniae and Pseudomonas aeruginosa. Appl. Sci. 2018, 8, 2551. [Google Scholar] [CrossRef]
- Chaudhary, D.K.; Bajagain, R.; Jeong, S.; Kim, J. Biodegradation of diesel oil and n-alkanes (C18, C20 and C22) by a novel strain Acinetobacter sp. K-6 in unsaturated soil. Environ. Eng. Res. 2020, 25, 290–298. [Google Scholar] [CrossRef]
- Panda, S.K.; Kar, R.N.; Panda, C.R. Isolation and identification of petroleum hydrocarbon degrading microorganisms from oil contaminated environment. Int. J. Environ. Sci. 2013, 3, 1314–1321. [Google Scholar] [CrossRef]
- Otiniano, N.M.; Rojas-Villacorta, W.; De La Cruz-Noriega, M.; Lora-Cahuas, C.; Mendoza-Villanueva, K.; Benites, S.M.; Gallozzo-Cardenas, M.; Rojas-Flores, S. Effect of Inoculum concentration on the degradation of diesel 2 by a microbial consortium. Sustain. 2022, 14, 16750. [Google Scholar] [CrossRef]
- Bensidhoum, L.; Nabti, E.; Tabli, N.; Kupferschmied, P.; Weiss, A.; Rothballer, M.; Schmid, M.; Keel, C.; Hartmann, A. Heavy metal tolerant Pseudomonas protegens isolates from agricultural well water in northeastern Algeria with plant growth promoting, insecticidal and antifungal activities. Eur. J. Soil Biol. 2016, 75, 38–46. [Google Scholar] [CrossRef]
- Liu, H.; Gao, H.; Wu, M.; Ma, C.; Wu, J.; Ye, X. Distribution Characteristics of Bacterial Communities and Hydrocarbon Degradation Dynamics During the Remediation of Petroleum-Contaminated Soil by Enhancing Moisture Content. Microb. Ecol. 2020, 80, 202–211. [Google Scholar] [CrossRef]
- Nnabuife, O.O.; Ogbonna, J.C.; Anyanwu, C.; Ike, A.C.; Eze, C.N.; Enemuor, S.C. Mixed bacterial consortium can hamper the efficient degradation of crude oil hydrocarbons. Arch. Microbiol. 2022, 204, 306. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Pandey, L.M. Integration of biosorption and biodegradation in a fed- batch mode for the enhanced crude oil remediation. Lett. Appl. Microbiol. 2021, 73, 471–476. [Google Scholar] [CrossRef]
- Azzahra, F.; Rinanti, A.; Hadisoebroto, R.; Minarti, A.; Aphirta, S. Removal of Total Petroleum Hydrocarbon (TPH) crude oil by consortium bacteria Acetobacter tropicalis and Lactobacillus casei. E3S Web Conf. 2023, 420, 09009. [Google Scholar] [CrossRef]
- Zannotti, M.; Vassallo, A.; Ramasamy, P.; Loggi, V. Hydrocarbon degradation strategy and pyoverdine production using the salt tolerant Antarctic. RSC Adv. 2023, 13, 19276–19285. [Google Scholar] [CrossRef]
- Safitri, R.A.; Mangunwardoyo, W.; Ambarsari, H. Biodegradation of diesel oil hydrocarbons using Bacillus subtilis InaCC B289 and Pseudomonas aeruginosa InaCC B290 in single and mixed cultures. AIP Conf. Proc. 2018, 2021, 030013. [Google Scholar] [CrossRef]
- Su, Q.; Yu, J.; Fang, K.; Dong, P.; Li, Z.; Zhang, W.; Liu, M.; Xiang, L.; Cai, J. Microbial removal of petroleum hydrocarbons from contaminated soil under arsenic stress. Toxics 2023, 11, 143. [Google Scholar] [CrossRef] [PubMed]
- Acer, Ö.; Johnston, G.P.; Lineman, D.; Johnston, C.G. Evaluating degradation of polycyclic aromatic hydrocarbon (PAH) potential by indigenous bacteria isolated from highly contaminated riverbank sediments. Environ. Earth Sci. 2021, 80, 773. [Google Scholar] [CrossRef]




| Microbial Isolation | Natural Source of Insulation | Diesel Biodegradation Yield | Reference |
|---|---|---|---|
| Bacillus megaterium MJ4 | Seawater from Jiaozhou Bay, China. | Degradation rate at 26.54% in 5 days, using 10 g/L of diesel. | [7] |
| Vibrio alginolyticus MFI5 | Diesel-contaminated seawater in Tanjung Jati, Madura Island, Indonesia. | Diesel degradation capacity up to 26.78% during 14 days of incubation at 10% initial diesel (v/v). | [8] |
| Janthinobacterium lividum AQ5-29 and Pseudomonas fildesensis AQ5-41 | Antarctic soil of Signy Island. | Strains AQ5-29 and AQ5-41 removed 2.9 and 4.2 mg/mL of diesel, respectively, with biodegradation of C10 to C30 hydrocarbons ranging from 40 to 100% at 10 °C in less than 8 days. | [9] |
| Acinetobacter calcoaceticus and Pseudomonas aeruginosa | Wastewater samples with effluents from mechanical workshops, Burkina Faso (Africa). | Highest biodegradation rate at 65.53%, using both S2+S7 in diesel. | [10] |
| Isolated Microbial Culture | Name | Similarity Strain | Pairwise Similarity (%) | Access Number | Link (accessed on 11 June 2024) |
|---|---|---|---|---|---|
| PROM1 | Priestia flexa | NBRC 15715 | 99.59 | PP886133 | https://www.ncbi.nlm.nih.gov/nuccore/PP886133 |
| PROM2 | Pseudomonas protegens | CHA0 | 100 | PP886146 | https://www.ncbi.nlm.nih.gov/nuccore/PP886146 |
| PROM3 | Pseudomonas citri | OPS13-3 | 99.18 | PP886148 | https://www.ncbi.nlm.nih.gov/nuccore/PP886148 |
| ClyRoM5 | Acinetobacter guillouiae | CIP 63.46 | 100 | PP892527 | https://www.ncbi.nlm.nih.gov/nuccore/PP892527 |
| Isolated Culture | MeanDiff | q Value | Prob |
|---|---|---|---|
| PROM1 and PROM3 | −41.51 | 64.58 | 0.0 (p < 0.05) |
| PROM2 and PROM3 | 23.87 | 37.13 | 0.0 (p < 0.05) |
| PROM2 and PROM1 | 65.38 | 101.71 | 0.0 (p < 0.05) |
| ClyRoM5 and PROM3 | −10.20 | 15.88 | 4.20 × 10−1 |
| ClyRoM5 and PROM1 | 31.30 | 48.70 | 0 (p < 0.05) |
| ClyRoM5 and PROM2 | −34.07 | 53.01 | 0 (p < 0.05) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quiñones-Cerna, C.; Castañeda-Aspajo, A.; Tirado-Gutierrez, M.; Salirrosas-Fernández, D.; Rodríguez-Soto, J.C.; Cruz-Monzón, J.A.; Hurtado-Butrón, F.; Ugarte-López, W.; Gutiérrez-Araujo, M.; Quezada-Alvarez, M.A.; et al. Efficacy of Indigenous Bacteria in the Biodegradation of Hydrocarbons Isolated from Agricultural Soils in Huamachuco, Peru. Microorganisms 2024, 12, 1896. https://doi.org/10.3390/microorganisms12091896
Quiñones-Cerna C, Castañeda-Aspajo A, Tirado-Gutierrez M, Salirrosas-Fernández D, Rodríguez-Soto JC, Cruz-Monzón JA, Hurtado-Butrón F, Ugarte-López W, Gutiérrez-Araujo M, Quezada-Alvarez MA, et al. Efficacy of Indigenous Bacteria in the Biodegradation of Hydrocarbons Isolated from Agricultural Soils in Huamachuco, Peru. Microorganisms. 2024; 12(9):1896. https://doi.org/10.3390/microorganisms12091896
Chicago/Turabian StyleQuiñones-Cerna, Claudio, Alina Castañeda-Aspajo, Marycielo Tirado-Gutierrez, David Salirrosas-Fernández, Juan Carlos Rodríguez-Soto, José Alfredo Cruz-Monzón, Fernando Hurtado-Butrón, Wilmer Ugarte-López, Mayra Gutiérrez-Araujo, Medardo Alberto Quezada-Alvarez, and et al. 2024. "Efficacy of Indigenous Bacteria in the Biodegradation of Hydrocarbons Isolated from Agricultural Soils in Huamachuco, Peru" Microorganisms 12, no. 9: 1896. https://doi.org/10.3390/microorganisms12091896








