Unravelling the Roles of Bacterial Nanomachines Bistability in Pathogens’ Life Cycle
Abstract
1. Introduction
2. Molecular Pathways Underlying Phenotypic Heterogeneity: Stochasticity versus Determinism
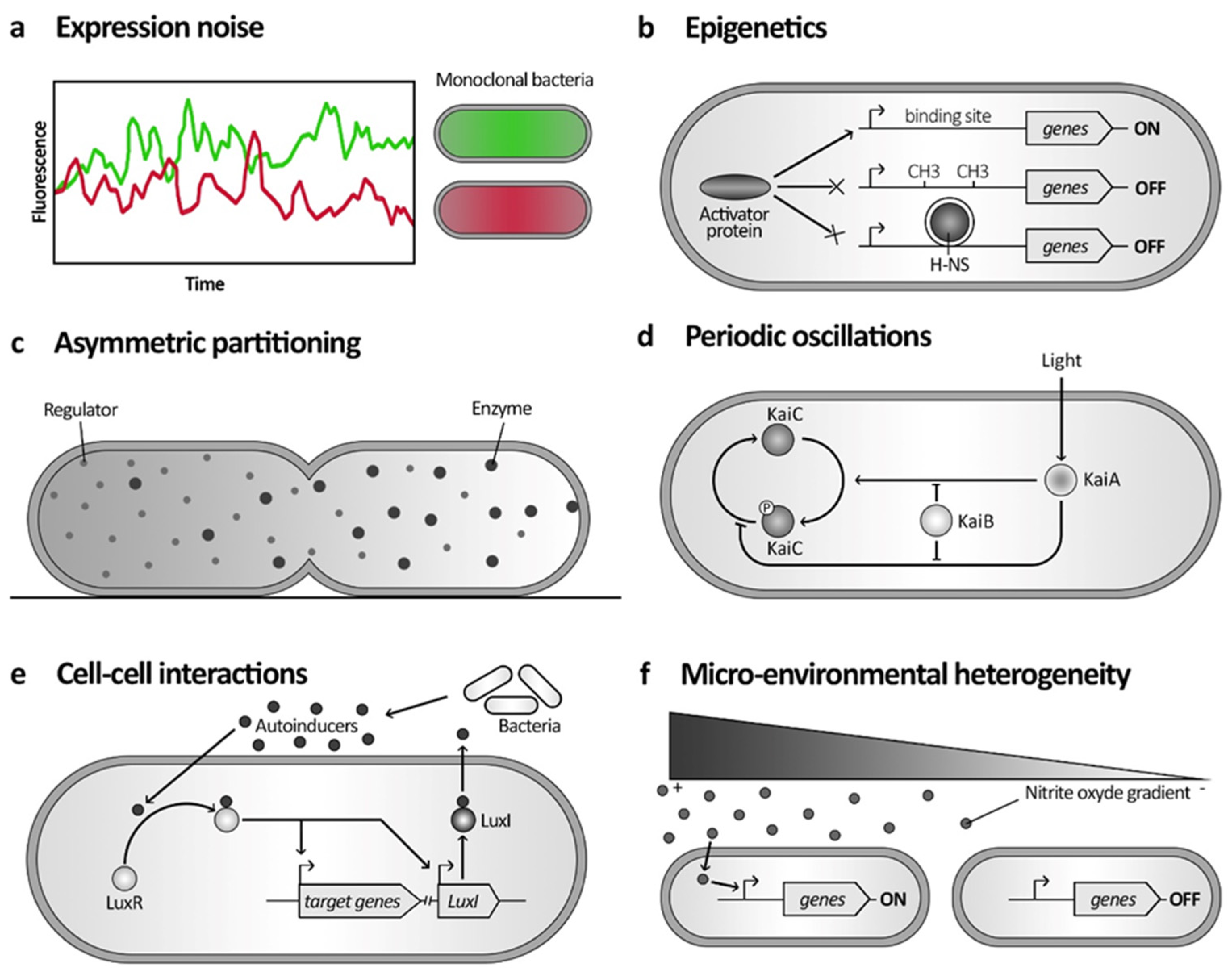
2.1. Stochasticity in Gene Expression
2.1.1. Expression Noise
2.1.2. Epigenetics
2.1.3. Asymmetric Partitioning
2.2. Determinism in Phenotypic Variation
2.2.1. Intrinsic Cellular Factors
2.2.2. Cell–Cell Interactions
2.2.3. Heterogeneous Microenvironment
2.3. Approaches and Challenges in Studying Phenotypic Heterogeneity
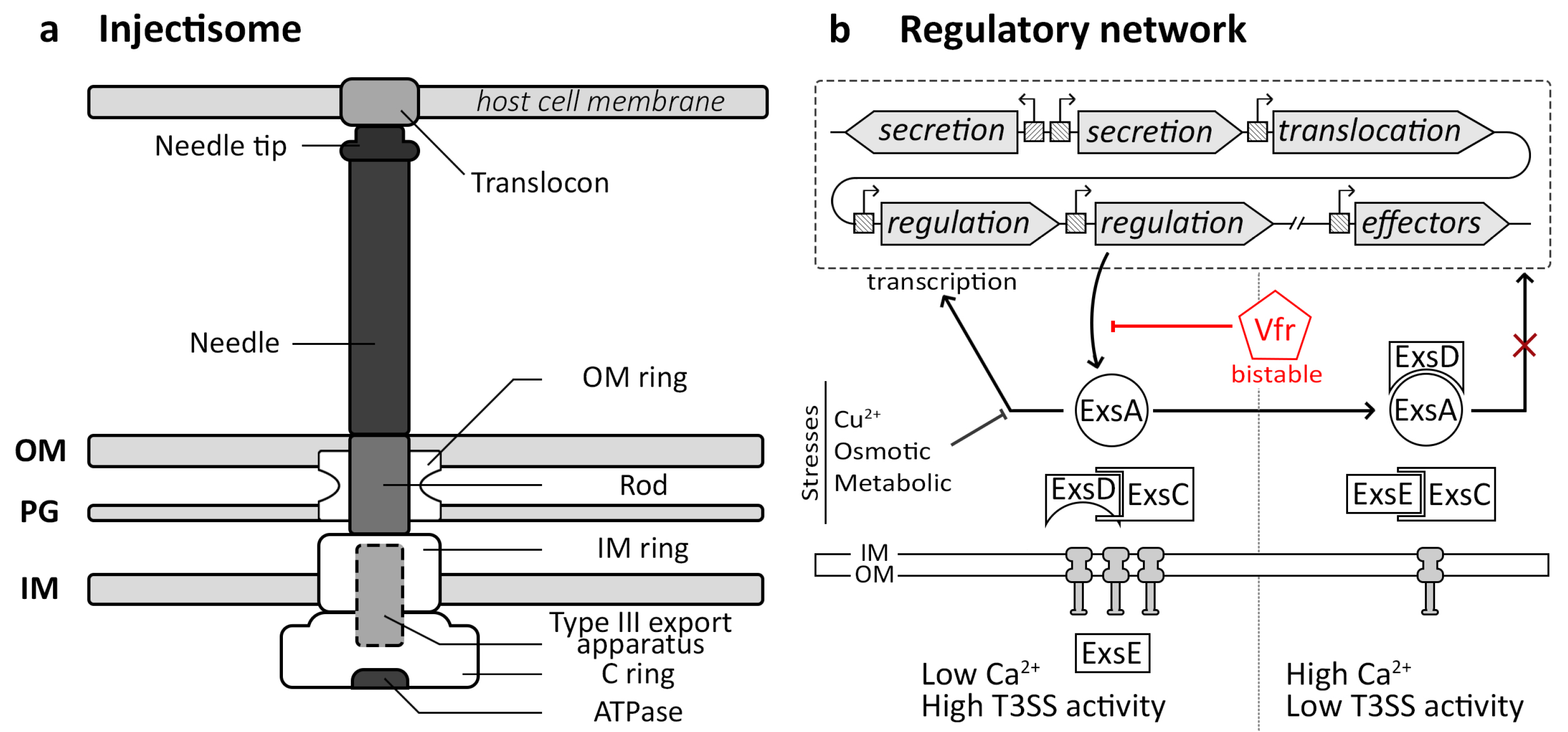
3. The Injectisome Nanomachine
3.1. Injectisome Structure and Functions
3.2. Injectisome Regulatory Pathway
3.3. Molecular Bases of the Bacterial Injectisome Bistability
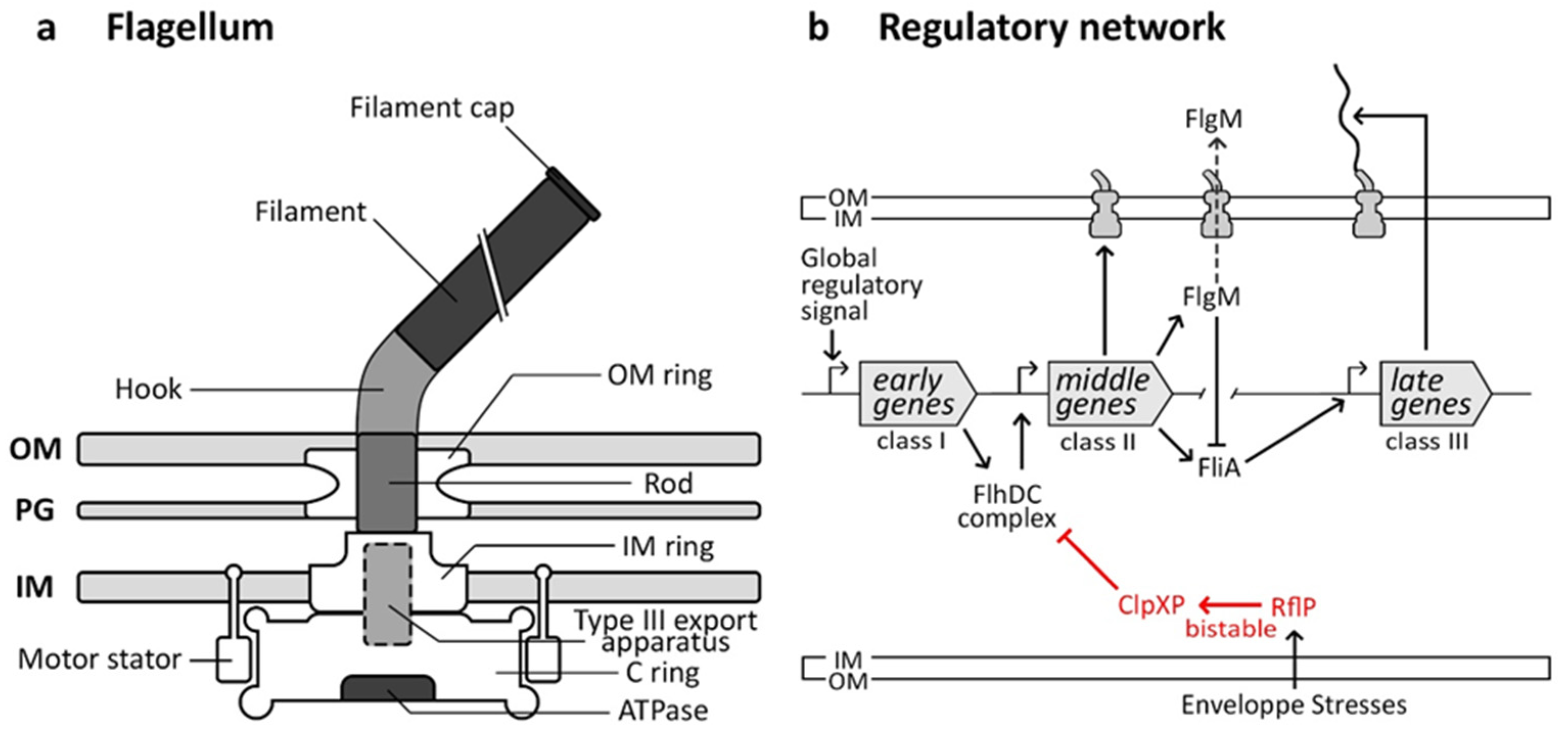
4. Flagellum Nanomachine
4.1. Flagellum Structure and Functions
4.2. Flagellar Regulatory Pathway
4.3. Molecular Bases of the Flagellum Bistability
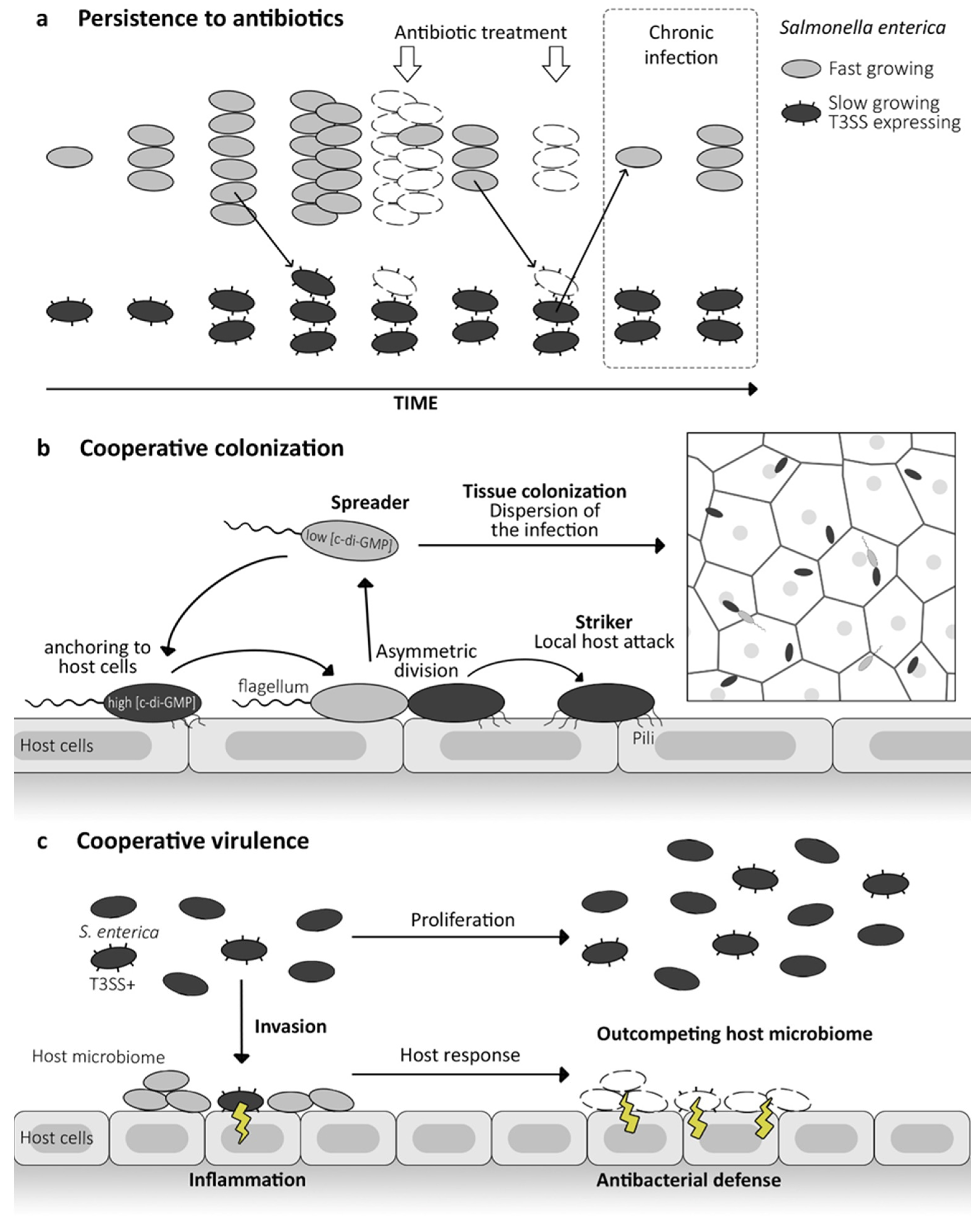
5. Nanomachines Bistability and Pathogen Community Behavior
5.1. Persistence against Antibiotics
5.2. Cooperative Colonization
5.3. Cooperative Virulence
5.4. Evolution
6. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- van Vliet, S.; Ackermann, M. Bacterial Ventures into Multicellularity: Collectivism through Individuality. PLoS Biol. 2015, 13, e1002162. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ackermann, M. A Functional Perspective on Phenotypic Heterogeneity in Microorganisms. Nat. Rev. Microbiol. 2015, 13, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Schröter, L.; Dersch, P. Phenotypic Diversification of Microbial Pathogens—Cooperating and Preparing for the Future. J. Mol. Biol. 2019, 431, 4645–4655. [Google Scholar] [CrossRef] [PubMed]
- Hare, P.J.; LaGree, T.J.; Byrd, B.A.; DeMarco, A.M.; Mok, W.W.K. Single-Cell Technologies to Study Phenotypic Heterogeneity and Bacterial Persisters. Microorganisms 2021, 9, 2277. [Google Scholar] [CrossRef]
- Davis, K.M.; Isberg, R.R. Defining Heterogeneity within Bacterial Populations via Single Cell Approaches. BioEssays 2016, 38, 782–790. [Google Scholar] [CrossRef]
- Kreibich, S.; Hardt, W.-D. Experimental Approaches to Phenotypic Diversity in Infection. Curr. Opin. Microbiol. 2015, 27, 25–36. [Google Scholar] [CrossRef]
- Dubnau, D.; Losick, R. Bistability in Bacteria. Mol. Microbiol. 2006, 61, 564–572. [Google Scholar] [CrossRef]
- Veening, J.-W.; Smits, W.K.; Kuipers, O.P. Bistability, Epigenetics, and Bet-Hedging in Bacteria. Annu. Rev. Microbiol. 2008, 62, 193–210. [Google Scholar] [CrossRef]
- Evans, T.D.; Zhang, F. Bacterial Metabolic Heterogeneity: Origins and Applications in Engineering and Infectious Disease. Curr. Opin. Biotechnol. 2020, 64, 183–189. [Google Scholar] [CrossRef]
- Spacapan, M.; Bez, C.; Venturi, V. Quorum Sensing Going Wild. iScience 2023, 26, 108000. [Google Scholar] [CrossRef]
- Sánchez-Romero, M.A.; Casadesús, J. Single Cell Analysis of Bistable Expression of Pathogenicity Island 1 and the Flagellar Regulon in Salmonella enterica. Microorganisms 2021, 9, 210. [Google Scholar] [CrossRef] [PubMed]
- Goldlust, K.; Ducret, A.; Halte, M.; Dedieu-Berne, A.; Erhardt, M.; Lesterlin, C. The F Pilus Serves as a Conduit for the DNA during Conjugation between Physically Distant Bacteria. Proc. Natl. Acad. Sci. USA 2023, 120, e2310842120. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.D.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion Systems in Gram-Negative Bacteria: Structural and Mechanistic Insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-Q.; Dixon, N.E. Bacterial Replisomes. Curr. Opin. Struct. Biol. 2018, 53, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Worrall, L.J.; Majewski, D.D.; Strynadka, N.C.J. Structural Insights into Type III Secretion Systems of the Bacterial Flagellum and Injectisome. Annu. Rev. Microbiol. 2023, 77, 669–698. [Google Scholar] [CrossRef]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.J.; Finlay, B.B. Assembly, Structure, Function and Regulation of Type III Secretion Systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef]
- Hajra, D.; Nair, A.V.; Chakravortty, D. An Elegant Nano-Injection Machinery for Sabotaging the Host: Role of Type III Secretion System in Virulence of Different Human and Animal Pathogenic Bacteria. Phys. Life Rev. 2021, 38, 25–54. [Google Scholar] [CrossRef]
- Colin, R.; Ni, B.; Laganenka, L.; Sourjik, V. Multiple Functions of Flagellar Motility and Chemotaxis in Bacterial Physiology. FEMS Microbiol. Rev. 2021, 45, fuab038. [Google Scholar] [CrossRef]
- Hajam, I.A.; Dar, P.A.; Shahnawaz, I.; Jaume, J.C.; Lee, J.H. Bacterial Flagellin—A Potent Immunomodulatory Agent. Exp. Mol. Med. 2017, 49, e373. [Google Scholar] [CrossRef]
- Jouault, A.; Saliba, A.M.; Touqui, L. Modulation of the Immune Response by the Pseudomonas aeruginosa Type-III Secretion System. Front. Cell. Infect. Microbiol. 2022, 12, 1064010. [Google Scholar] [CrossRef]
- Cui, Z.; Yuan, X.; Yang, C.-H.; Huntley, R.B.; Sun, W.; Wang, J.; Sundin, G.W.; Zeng, Q. Development of a Method to Monitor Gene Expression in Single Bacterial Cells during the Interaction with Plants and Use to Study the Expression of the Type III Secretion System in Single Cells of Dickeya Dadantii in Potato. Front. Microbiol. 2018, 9, 1429. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Bossi, N.; Sánchez-Romero, M.A.; Kerboriou, P.; Naquin, D.; Mendes, C.; Bouloc, P.; Casadesús, J.; Bossi, L. Pervasive Transcription Enhances the Accessibility of H-NS–Silenced Promoters and Generates Bistability in Salmonella Virulence Gene Expression. Proc. Natl. Acad. Sci. USA 2022, 119, e2203011119. [Google Scholar] [CrossRef] [PubMed]
- Marsden, A.E.; Intile, P.J.; Schulmeyer, K.H.; Simmons-Patterson, E.R.; Urbanowski, M.L.; Wolfgang, M.C.; Yahr, T.L. Vfr Directly Activates exsA Transcription to Regulate Expression of the Pseudomonas aeruginosa Type III Secretion System. J. Bacteriol. 2016, 198, 1442–1450. [Google Scholar] [CrossRef] [PubMed]
- Ronin, I.; Katsowich, N.; Rosenshine, I.; Balaban, N.Q. A Long-Term Epigenetic Memory Switch Controls Bacterial Virulence Bimodality. eLife 2017, 6, e19599. [Google Scholar] [CrossRef]
- Rufián, J.S.; Sánchez-Romero, M.-A.; López-Márquez, D.; Macho, A.P.; Mansfield, J.W.; Arnold, D.L.; Ruiz-Albert, J.; Casadesús, J.; Beuzón, C.R. Pseudomonas syringae Differentiates into Phenotypically Distinct Subpopulations During Colonization of a Plant Host. Environ. Microbiol. 2016, 18, 3593–3605. [Google Scholar] [CrossRef]
- Kearns, D.B.; Losick, R. Cell Population Heterogeneity during Growth of Bacillus subtilis. Genes Dev. 2005, 19, 3083–3094. [Google Scholar] [CrossRef]
- Kim, J.M.; Garcia-Alcala, M.; Balleza, E.; Cluzel, P. Stochastic Transcriptional Pulses Orchestrate Flagellar Biosynthesis in Escherichia coli. Sci. Adv. 2020, 6, eaax0947. [Google Scholar] [CrossRef]
- Kreiling, V.; Thormann, K.M. Polarity of C-Di-GMP Synthesis and Degradation. microLife 2023, 4, uqad014. [Google Scholar] [CrossRef]
- Spöring, I.; Felgner, S.; Preuße, M.; Eckweiler, D.; Rohde, M.; Häussler, S.; Weiss, S.; Erhardt, M. Regulation of Flagellum Biosynthesis in Response to Cell Envelope Stress in Salmonella enterica Serovar Typhimurium. mBio 2018, 9, e00736-17. [Google Scholar] [CrossRef]
- Striednig, B.; Lanner, U.; Niggli, S.; Katic, A.; Vormittag, S.; Brülisauer, S.; Hochstrasser, R.; Kaech, A.; Welin, A.; Flieger, A.; et al. Quorum Sensing Governs a Transmissive Legionella Subpopulation at the Pathogen Vacuole Periphery. EMBO Rep. 2021, 22, e52972. [Google Scholar] [CrossRef]
- Warren Norris, M.A.H.; Plaskon, D.M.; Tamayo, R. Phase Variation of Flagella and Toxins in Clostridioides difficile Is Mediated by Selective Rho-Dependent Termination. J. Mol. Biol. 2024, 436, 168456. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yuan, Z.; Liu, P.; Zhou, T. Effects of Promoter Leakage on Dynamics of Gene Expression. BMC Syst. Biol. 2015, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Kiviet, D.J.; Nghe, P.; Walker, N.; Boulineau, S.; Sunderlikova, V.; Tans, S.J. Stochasticity of Metabolism and Growth at the Single-Cell Level. Nature 2014, 514, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Melderen, L.V.; Bast, M.S.D. Bacterial Toxin–Antitoxin Systems: More Than Selfish Entities? PLoS Genet. 2009, 5, e1000437. [Google Scholar] [CrossRef] [PubMed]
- Quigley, J.; Lewis, K. Noise in a Metabolic Pathway Leads to Persister Formation in Mycobacterium Tuberculosis. Microbiol. Spectr. 2022, 10, e02948-22. [Google Scholar] [CrossRef]
- Elowitz, M.B.; Levine, A.J.; Siggia, E.D.; Swain, P.S. Stochastic Gene Expression in a Single Cell. Science 2002, 297, 1183–1186. [Google Scholar] [CrossRef]
- Sanchez, A.; Golding, I. Genetic Determinants and Cellular Constraints in Noisy Gene Expression. Science 2013, 342, 1188–1193. [Google Scholar] [CrossRef]
- Chowdhury, D.; Wang, C.; Lu, A.; Zhu, H. Cis-Regulatory Logic Produces Gene-Expression Noise Describing Phenotypic Heterogeneity in Bacteria. Front. Genet. 2021, 12, 698910. [Google Scholar] [CrossRef]
- Raj, A.; van Oudenaarden, A. Nature, Nurture, or Chance: Stochastic Gene Expression and Its Consequences. Cell 2008, 135, 216–226. [Google Scholar] [CrossRef]
- Jones, D.L.; Brewster, R.C.; Phillips, R. Promoter Architecture Dictates Cell-to-Cell Variability in Gene Expression. Science 2014, 346, 1533–1536. [Google Scholar] [CrossRef]
- Ozbudak, E.M.; Thattai, M.; Kurtser, I.; Grossman, A.D.; van Oudenaarden, A. Regulation of Noise in the Expression of a Single Gene. Nat. Genet. 2002, 31, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, J.; Xu, H.; Golding, I. Measuring Transcription at a Single Gene Copy Reveals Hidden Drivers of Bacterial Individuality. Nat. Microbiol. 2019, 4, 2118–2127. [Google Scholar] [CrossRef] [PubMed]
- Smits, W.K.; Kuipers, O.P.; Veening, J.-W. Phenotypic Variation in Bacteria: The Role of Feedback Regulation. Nat. Rev. Microbiol. 2006, 4, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Casadesús, J.; Low, D. Epigenetic Gene Regulation in the Bacterial World. Microbiol. Mol. Biol. Rev. 2006, 70, 830–856. [Google Scholar] [CrossRef]
- Casadesús, J.; Low, D.A. Programmed Heterogeneity: Epigenetic Mechanisms in Bacteria. J. Biol. Chem. 2013, 288, 13929–13935. [Google Scholar] [CrossRef]
- Riber, L.; Hansen, L.H. Epigenetic Memories: The Hidden Drivers of Bacterial Persistence? Trends Microbiol. 2021, 29, 190–194. [Google Scholar] [CrossRef]
- Huh, D.; Paulsson, J. Non-Genetic Heterogeneity from Stochastic Partitioning at Cell Division. Nat. Genet. 2011, 43, 95–100. [Google Scholar] [CrossRef]
- Knorre, D.A.; Azbarova, A.V.; Galkina, K.V.; Feniouk, B.A.; Severin, F.F. Replicative Aging as a Source of Cell Heterogeneity in Budding Yeast. Mech. Ageing Dev. 2018, 176, 24–31. [Google Scholar] [CrossRef]
- Levy, S.F.; Ziv, N.; Siegal, M.L. Bet Hedging in Yeast by Heterogeneous, Age-Correlated Expression of a Stress Protectant. PLoS Biol. 2012, 10, e1001325. [Google Scholar] [CrossRef]
- Aldridge, B.B.; Fernandez-Suarez, M.; Heller, D.; Ambravaneswaran, V.; Irimia, D.; Toner, M.; Fortune, S.M. Asymmetry and Aging of Mycobacterial Cells Lead to Variable Growth and Antibiotic Susceptibility. Science 2012, 335, 100–104. [Google Scholar] [CrossRef]
- Vaubourgeix, J.; Lin, G.; Dhar, N.; Chenouard, N.; Jiang, X.; Botella, H.; Lupoli, T.; Mariani, O.; Yang, G.; Ouerfelli, O.; et al. Stressed Mycobacteria Use the Chaperone ClpB to Sequester Irreversibly Oxidized Proteins Asymmetrically within and between Cells. Cell Host Microbe 2015, 17, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, L.; McAdams, H.H.; Losick, R. Generating and Exploiting Polarity in Bacteria. Science 2002, 298, 1942–1946. [Google Scholar] [CrossRef]
- Jenal, U.; Stephens, C. The Caulobacter Cell Cycle: Timing, Spatial Organization and Checkpoints. Curr. Opin. Microbiol. 2002, 5, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Collier, J. Cell Division Control in Caulobacter crescentus. Biochim. Biophys. Acta—Gene Regul. Mech. 2019, 1862, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.; Kaur, M.; Jang, H.-I.; Kim, Y.-I. The Circadian Clock—A Molecular Tool for Survival in Cyanobacteria. Life 2020, 10, 365. [Google Scholar] [CrossRef] [PubMed]
- Seef, S.; Herrou, J.; de Boissier, P.; My, L.; Brasseur, G.; Robert, D.; Jain, R.; Mercier, R.; Cascales, E.; Habermann, B.H.; et al. A Tad-like Apparatus Is Required for Contact-Dependent Prey Killing in Predatory Social Bacteria. eLife 2021, 10, e72409. [Google Scholar] [CrossRef]
- Flores, E.; Herrero, A. Compartmentalized Function through Cell Differentiation in Filamentous Cyanobacteria. Nat. Rev. Microbiol. 2010, 8, 39–50. [Google Scholar] [CrossRef]
- Grote, J.; Krysciak, D.; Streit, W.R. Phenotypic Heterogeneity, a Phenomenon That May Explain Why Quorum Sensing Does Not Always Result in Truly Homogenous Cell Behavior. Appl. Environ. Microbiol. 2015, 81, 5280–5289. [Google Scholar] [CrossRef]
- Bettenworth, V.; Steinfeld, B.; Duin, H.; Petersen, K.; Streit, W.R.; Bischofs, I.; Becker, A. Phenotypic Heterogeneity in Bacterial Quorum Sensing Systems. J. Mol. Biol. 2019, 431, 4530–4546. [Google Scholar] [CrossRef]
- Striednig, B.; Hilbi, H. Bacterial Quorum Sensing and Phenotypic Heterogeneity: How the Collective Shapes the Individual. Trends Microbiol. 2022, 30, 379–389. [Google Scholar] [CrossRef]
- Choudhary, D.; Lagage, V.; Foster, K.R.; Uphoff, S. Phenotypic Heterogeneity in the Bacterial Oxidative Stress Response Is Driven by Cell-Cell Interactions. Cell Rep. 2023, 42, 112168. [Google Scholar] [CrossRef] [PubMed]
- Gallegos-Monterrosa, R.; Kovács, Á.T. Phenotypic Plasticity: The Role of a Phosphatase Family Rap in the Genetic Regulation of Bacilli. Mol. Microbiol. 2023, 120, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.M.; Mohammadi, S.; Isberg, R.R. Community Behavior and Spatial Regulation within a Bacterial Microcolony in Deep Tissue Sites Serves to Protect against Host Attack. Cell Host Microbe 2015, 17, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Schwartzman, J.A.; Ebrahimi, A.; Chadwick, G.; Sato, Y.; Roller, B.R.K.; Orphan, V.J.; Cordero, O.X. Bacterial Growth in Multicellular Aggregates Leads to the Emergence of Complex Life Cycles. Curr. Biol. 2022, 32, 3059–3069.e7. [Google Scholar] [CrossRef]
- Stewart, P.S.; Franklin, M.J. Physiological Heterogeneity in Biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.; Price-Whelan, A.; Dietrich, L.E.P. Gradients and Consequences of Heterogeneity in Biofilms. Nat. Rev. Microbiol. 2022, 20, 593–607. [Google Scholar] [CrossRef]
- Gollan, B.; Grabe, G.; Michaux, C.; Helaine, S. Bacterial Persisters and Infection: Past, Present, and Progressing. Annu. Rev. Microbiol. 2019, 73, 359–385. [Google Scholar] [CrossRef]
- Claudi, B.; Spröte, P.; Chirkova, A.; Personnic, N.; Zankl, J.; Schürmann, N.; Schmidt, A.; Bumann, D. Phenotypic Variation of Salmonella in Host Tissues Delays Eradication by Antimicrobial Chemotherapy. Cell 2014, 158, 722–733. [Google Scholar] [CrossRef]
- Galán, J.E.; Lara-Tejero, M.; Marlovits, T.C.; Wagner, S. Bacterial Type III Secretion Systems: Specialized Nanomachines for Protein Delivery into Target Cells. Annu. Rev. Microbiol. 2014, 68, 415–438. [Google Scholar] [CrossRef]
- Cornelis, G.R. The Type III Secretion Injectisome. Nat. Rev. Microbiol. 2006, 4, 811–825. [Google Scholar] [CrossRef]
- Erhardt, M.; Namba, K.; Hughes, K.T. Bacterial Nanomachines: The Flagellum and Type III Injectisome. Cold Spring Harb. Perspect. Biol. 2010, 2, a000299. [Google Scholar] [CrossRef] [PubMed]
- Lou, L.; Zhang, P.; Piao, R.; Wang, Y. Salmonella Pathogenicity Island 1 (SPI-1) and Its Complex Regulatory Network. Front. Cell. Infect. Microbiol. 2019, 9, 270. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, J.; Finlay, B.B. Type III Effector-Mediated Processes in Salmonella Infection. Future Microbiol. 2012, 7, 685–703. [Google Scholar] [CrossRef] [PubMed]
- Figueira, R.; Holden, D.W. Functions of the Salmonella Pathogenicity Island 2 (SPI-2) Type III Secretion System Effectors. Microbiology 2012, 158, 1147–1161. [Google Scholar] [CrossRef]
- Raymond, B.; Young, J.C.; Pallett, M.; Endres, R.G.; Clements, A.; Frankel, G. Subversion of Trafficking, Apoptosis, and Innate Immunity by Type III Secretion System Effectors. Trends Microbiol. 2013, 21, 430–441. [Google Scholar] [CrossRef]
- Ratu, S.T.N.; Amelia, L.; Okazaki, S. Type III Effector Provides a Novel Symbiotic Pathway in Legume–Rhizobia Symbiosis. Biosci. Biotechnol. Biochem. 2023, 87, 28–37. [Google Scholar] [CrossRef]
- Brown, N.F.; Finlay, B. Potential Origins and Horizontal Transfer of Type III Secretion Systems and Effectors. Mob. Genet. Elem. 2011, 1, 118–121. [Google Scholar] [CrossRef][Green Version]
- Fàbrega, A.; Vila, J. Salmonella enterica Serovar Typhimurium Skills To Succeed in the Host: Virulence and Regulation. Clin. Microbiol. Rev. 2013, 26, 308–341. [Google Scholar] [CrossRef]
- Schroeder, G.N.; Hilbi, H. Molecular Pathogenesis of Shigella Spp.: Controlling Host Cell Signaling, Invasion, and Death by Type III Secretion. Clin. Microbiol. Rev. 2008, 21, 134–156. [Google Scholar] [CrossRef]
- Bajunaid, W.; Haidar-Ahmad, N.; Kottarampatel, A.H.; Ourida Manigat, F.; Silué, N.; Tchagang, C.F.; Tomaro, K.; Campbell-Valois, F.-X. The T3SS of Shigella: Expression, Structure, Function, and Role in Vacuole Escape. Microorganisms 2020, 8, 1933. [Google Scholar] [CrossRef]
- Yahr, T.L.; Wolfgang, M.C. Transcriptional Regulation of the Pseudomonas aeruginosa Type III Secretion System. Mol. Microbiol. 2006, 62, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Horna, G.; Ruiz, J. Type 3 Secretion System of Pseudomonas aeruginosa. Microbiol. Res. 2021, 246, 126719. [Google Scholar] [CrossRef] [PubMed]
- Hamed, S.; Wang, X.; Shawky, R.M.; Emara, M.; Aldridge, P.D.; Rao, C.V. Synergistic Action of SPI-1 Gene Expression in Salmonella enterica Serovar Typhimurium through Transcriptional Crosstalk with the Flagellar System. BMC Microbiol. 2019, 19, 211. [Google Scholar] [CrossRef] [PubMed]
- Koirala, S.; Mears, P.; Sim, M.; Golding, I.; Chemla, Y.R.; Aldridge, P.D.; Rao, C.V. A Nutrient-Tunable Bistable Switch Controls Motility in Salmonella enterica Serovar Typhimurium. mBio 2014, 5, e01611-14. [Google Scholar] [CrossRef]
- Fuchs, E.L.; Brutinel, E.D.; Jones, A.K.; Fulcher, N.B.; Urbanowski, M.L.; Yahr, T.L.; Wolfgang, M.C. The Pseudomonas aeruginosa Vfr Regulator Controls Global Virulence Factor Expression through Cyclic AMP-Dependent and -Independent Mechanisms. J. Bacteriol. 2010, 192, 3553–3564. [Google Scholar] [CrossRef]
- Lin, C.K.; Lee, D.S.W.; McKeithen-Mead, S.; Emonet, T.; Kazmierczak, B. A Primed Subpopulation of Bacteria Enables Rapid Expression of the Type 3 Secretion System in Pseudomonas aeruginosa. mBio 2021, 12, e0083121. [Google Scholar] [CrossRef]
- O’Malley, M.R.; Anderson, J.C. Regulation of the Pseudomonas syringae Type III Secretion System by Host Environment Signals. Microorganisms 2021, 9, 1227. [Google Scholar] [CrossRef]
- Gophna, U.; Ron, E.Z.; Graur, D. Bacterial Type III Secretion Systems Are Ancient and Evolved by Multiple Horizontal-Transfer Events. Gene 2003, 312, 151–163. [Google Scholar] [CrossRef]
- Macnab, R.M. Genetics and Biogenesis of Bacterial Flagella. Annu. Rev. Genet. 1992, 26, 131–158. [Google Scholar] [CrossRef]
- Wadhwa, N.; Berg, H.C. Bacterial Motility: Machinery and Mechanisms. Nat. Rev. Microbiol. 2022, 20, 161–173. [Google Scholar] [CrossRef]
- Chilcott, G.S.; Hughes, K.T. Coupling of Flagellar Gene Expression to Flagellar Assembly in Salmonella enterica Serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 2000, 64, 694–708. [Google Scholar] [CrossRef]
- Bouteiller, M.; Dupont, C.; Bourigault, Y.; Latour, X.; Barbey, C.; Konto-Ghiorghi, Y.; Merieau, A. Pseudomonas Flagella: Generalities and Specificities. Int. J. Mol. Sci. 2021, 22, 3337. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, R. State of the Art of Bacterial Chemotaxis. J. Basic Microbiol. 2021, 61, 366–379. [Google Scholar] [CrossRef] [PubMed]
- Baraquet, C.; Harwood, C.S. Cyclic Diguanosine Monophosphate Represses Bacterial Flagella Synthesis by Interacting with the Walker A Motif of the Enhancer-Binding Protein FleQ. Proc. Natl. Acad. Sci. USA 2013, 110, 18478–18483. [Google Scholar] [CrossRef]
- Hochstrasser, R.; Hilbi, H. Intra-Species and Inter-Kingdom Signaling of Legionella pneumophila. Front. Microbiol. 2017, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Takaya, A.; Erhardt, M.; Karata, K.; Winterberg, K.; Yamamoto, T.; Hughes, K.T. YdiV: A Dual Function Protein That Targets FlhDC for ClpXP-Dependent Degradation by Promoting Release of DNA-Bound FlhDC Complex. Mol. Microbiol. 2012, 83, 1268–1284. [Google Scholar] [CrossRef]
- Rodesney, C.A.; Roman, B.; Dhamani, N.; Cooley, B.J.; Katira, P.; Touhami, A.; Gordon, V.D. Mechanosensing of Shear by Pseudomonas aeruginosa Leads to Increased Levels of the Cyclic-Di-GMP Signal Initiating Biofilm Development. Proc. Natl. Acad. Sci. USA 2017, 114, 5906–5911. [Google Scholar] [CrossRef]
- Laventie, B.-J.; Jenal, U. Surface Sensing and Adaptation in Bacteria. Annu. Rev. Microbiol. 2020, 74, 735–760. [Google Scholar] [CrossRef]
- Hengge, R. Principles of C-Di-GMP Signalling in Bacteria. Nat. Rev. Microbiol. 2009, 7, 263–273. [Google Scholar] [CrossRef]
- Laventie, B.-J.; Sangermani, M.; Estermann, F.; Manfredi, P.; Planes, R.; Hug, I.; Jaeger, T.; Meunier, E.; Broz, P.; Jenal, U. A Surface-Induced Asymmetric Program Promotes Tissue Colonization by Pseudomonas aeruginosa. Cell Host Microbe 2019, 25, 140–152.e6. [Google Scholar] [CrossRef]
- Anjuwon-Foster, B.R.; Tamayo, R. A Genetic Switch Controls the Production of Flagella and Toxins in Clostridium Difficile. PLoS Genet. 2017, 13, e1006701. [Google Scholar] [CrossRef] [PubMed]
- Syvertsson, S.; Wang, B.; Staal, J.; Gao, Y.; Kort, R.; Hamoen, L.W. Different Resource Allocation in a Bacillus subtilis Population Displaying Bimodal Motility. J. Bacteriol. 2021, 203, e0003721. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Kearns, D.B. The Structure and Regulation of Flagella in Bacillus subtilis. Annu. Rev. Genet. 2014, 48, 319–340. [Google Scholar] [CrossRef] [PubMed]
- Grimbergen, A.J.; Siebring, J.; Solopova, A.; Kuipers, O.P. Microbial Bet-Hedging: The Power of Being Different. Curr. Opin. Microbiol. 2015, 25, 67–72. [Google Scholar] [CrossRef]
- de Jong, I.G.; Haccou, P.; Kuipers, O.P. Bet Hedging or Not? A Guide to Proper Classification of Microbial Survival Strategies. BioEssays 2011, 33, 215–223. [Google Scholar] [CrossRef]
- Miao, C.; Cui, Y.; Yan, Z.; Jiang, Y. Pilus of Streptococcus pneumoniae: Structure, Function and Vaccine Potential. Front. Cell. Infect. Microbiol. 2023, 13, 1270848. [Google Scholar] [CrossRef]
- Sturm, A.; Heinemann, M.; Arnoldini, M.; Benecke, A.; Ackermann, M.; Benz, M.; Dormann, J.; Hardt, W.-D. The Cost of Virulence: Retarded Growth of Salmonella Typhimurium Cells Expressing Type III Secretion System 1. PLoS Pathog. 2011, 7, e1002143. [Google Scholar] [CrossRef]
- Arnoldini, M.; Vizcarra, I.A.; Peña-Miller, R.; Stocker, N.; Diard, M.; Vogel, V.; Beardmore, R.E.; Hardt, W.-D.; Ackermann, M. Bistable Expression of Virulence Genes in Salmonella Leads to the Formation of an Antibiotic-Tolerant Subpopulation. PLoS Biol. 2014, 12, e1001928. [Google Scholar] [CrossRef]
- Helaine, S.; Kugelberg, E. Bacterial Persisters: Formation, Eradication, and Experimental Systems. Trends Microbiol. 2014, 22, 417–424. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent Bacterial Infections and Persister Cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef]
- Lyu, Z.; Yang, A.; Villanueva, P.; Singh, A.; Ling, J. Heterogeneous Flagellar Expression in Single Salmonella Cells Promotes Diversity in Antibiotic Tolerance. mBio 2021, 12, e0237421. [Google Scholar] [CrossRef] [PubMed]
- West, S.A.; Cooper, G.A. Division of Labour in Microorganisms: An Evolutionary Perspective. Nat. Rev. Microbiol. 2016, 14, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Diard, M.; Garcia, V.; Maier, L.; Remus-Emsermann, M.N.P.; Regoes, R.R.; Ackermann, M.; Hardt, W.-D. Stabilization of Cooperative Virulence by the Expression of an Avirulent Phenotype. Nature 2013, 494, 353–356. [Google Scholar] [CrossRef]
- Rocha, C.L.; Coburn, J.; Rucks, E.A.; Olson, J.C. Characterization of Pseudomonas aeruginosa Exoenzyme S as a Bifunctional Enzyme in J774A.1 Macrophages. Infect. Immun. 2003, 71, 5296–5305. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kroken, A.R.; Chen, C.K.; Evans, D.J.; Yahr, T.L.; Fleiszig, S.M.J. The Impact of ExoS on Pseudomonas aeruginosa Internalization by Epithelial Cells Is Independent of fleQ and Correlates with Bistability of Type Three Secretion System Gene Expression. mBio 2018, 9, e00668-18. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.G.; Nieto, V.; Kroken, A.R.; Jedel, E.; Grosser, M.R.; Hallsten, M.E.; Mettrucio, M.M.E.; Yahr, T.L.; Evans, D.J.; Fleiszig, S.M.J. Pseudomonas aeruginosa Can Diversify after Host Cell Invasion to Establish Multiple Intracellular Niches. mBio 2022, 13, e02742-22. [Google Scholar] [CrossRef]
- Ackermann, M.; Stecher, B.; Freed, N.E.; Songhet, P.; Hardt, W.-D.; Doebeli, M. Self-Destructive Cooperation Mediated by Phenotypic Noise. Nature 2008, 454, 987–990. [Google Scholar] [CrossRef]
- Stewart, M.K.; Cummings, L.A.; Johnson, M.L.; Berezow, A.B.; Cookson, B.T. Regulation of Phenotypic Heterogeneity Permits Salmonella Evasion of the Host Caspase-1 Inflammatory Response. Proc. Natl. Acad. Sci. USA 2011, 108, 20742–20747. [Google Scholar] [CrossRef]
- Miao, E.A.; Alpuche-Aranda, C.M.; Dors, M.; Clark, A.E.; Bader, M.W.; Miller, S.I.; Aderem, A. Cytoplasmic Flagellin Activates Caspase-1 and Secretion of Interleukin 1β via Ipaf. Nat. Immunol. 2006, 7, 569–575. [Google Scholar] [CrossRef]
- Personnic, N.; Striednig, B.; Lezan, E.; Manske, C.; Welin, A.; Schmidt, A.; Hilbi, H. Quorum Sensing Modulates the Formation of Virulent Legionella Persisters within Infected Cells. Nat. Commun. 2019, 10, 5216. [Google Scholar] [CrossRef]
- Jiang, X.; Hall, A.B.; Arthur, T.D.; Plichta, D.R.; Covington, C.T.; Poyet, M.; Crothers, J.; Moses, P.L.; Tolonen, A.C.; Vlamakis, H.; et al. Invertible Promoters Mediate Bacterial Phase Variation, Antibiotic Resistance, and Host Adaptation in the Gut. Science 2019, 363, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Behrens, A.-J.; Kaever, V.; Kazmierczak, B.I. Type IV Pilus Assembly in Pseudomonas aeruginosa over a Broad Range of Cyclic Di-GMP Concentrations. J. Bacteriol. 2012, 194, 4285–4294. [Google Scholar] [CrossRef] [PubMed]
- Moscoso, J.A.; Mikkelsen, H.; Heeb, S.; Williams, P.; Filloux, A. The Pseudomonas aeruginosa Sensor RetS Switches Type III and Type VI Secretion via C-Di-GMP Signalling. Environ. Microbiol. 2011, 13, 3128–3138. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.T.; Dolganov, N.A.; Rasmussen, T.; Otto, G.; Miller, M.C.; Felt, S.A.; Torreilles, S.; Schoolnik, G.K. A Bistable Switch and Anatomical Site Control Vibrio Cholerae Virulence Gene Expression in the Intestine. PLoS Pathog. 2010, 6, e1001102. [Google Scholar] [CrossRef]
- Basset, A.; Turner, K.H.; Boush, E.; Sayeed, S.; Dove, S.L.; Malley, R. An Epigenetic Switch Mediates Bistable Expression of the Type 1 Pilus Genes in Streptococcus pneumoniae. J. Bacteriol. 2012, 194, 1088–1091. [Google Scholar] [CrossRef]
- Weber, B.S.; Ly, P.M.; Irwin, J.N.; Pukatzki, S.; Feldman, M.F. A Multidrug Resistance Plasmid Contains the Molecular Switch for Type VI Secretion in Acinetobacter Baumannii. Proc. Natl. Acad. Sci. USA 2015, 112, 9442–9447. [Google Scholar] [CrossRef]
- Bumann, D. Heterogeneous Host-Pathogen Encounters: Act Locally, Think Globally. Cell Host Microbe 2015, 17, 13–19. [Google Scholar] [CrossRef]
- Dadole, I.; Blaha, D.; Personnic, N. The Macrophage–Bacterium Mismatch in Persister Formation. Trends Microbiol. 2024. Online ahead of print. [Google Scholar] [CrossRef]
- Ruiz, L.M.R.; Williams, C.L.; Tamayo, R. Enhancing Bacterial Survival through Phenotypic Heterogeneity. PLoS Pathog. 2020, 16, e1008439. [Google Scholar] [CrossRef]
- Tsai, C.N.; Coombes, B.K. The Role of the Host in Driving Phenotypic Heterogeneity in Salmonella. Trends Microbiol. 2019, 27, 508–523. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gory, R.; Personnic, N.; Blaha, D. Unravelling the Roles of Bacterial Nanomachines Bistability in Pathogens’ Life Cycle. Microorganisms 2024, 12, 1930. https://doi.org/10.3390/microorganisms12091930
Gory R, Personnic N, Blaha D. Unravelling the Roles of Bacterial Nanomachines Bistability in Pathogens’ Life Cycle. Microorganisms. 2024; 12(9):1930. https://doi.org/10.3390/microorganisms12091930
Chicago/Turabian StyleGory, Romain, Nicolas Personnic, and Didier Blaha. 2024. "Unravelling the Roles of Bacterial Nanomachines Bistability in Pathogens’ Life Cycle" Microorganisms 12, no. 9: 1930. https://doi.org/10.3390/microorganisms12091930
APA StyleGory, R., Personnic, N., & Blaha, D. (2024). Unravelling the Roles of Bacterial Nanomachines Bistability in Pathogens’ Life Cycle. Microorganisms, 12(9), 1930. https://doi.org/10.3390/microorganisms12091930





