Association Analysis of the Genomic and Functional Characteristics of Halotolerant Glutamicibacter endophyticus J2-5-19 from the Rhizosphere of Suaeda salsa
Abstract
:1. Introduction
| Bacterial Species | Source Halophyte | Applied Crops | References |
|---|---|---|---|
| Agrobacierium tumefaciens Klebsiella sp. Ochrobactrum anthropi Pseudomonas stutzeri Pseudomonas sp. | Arthrocnemum indicum | peanut | [10] |
| Arthrobacter woluwensis AK1 Microbacterium oxydans AK2 Arthrobacter aurescens AK3 Bacillus megaterium AK4 Bacillus aryabhattai AK5 | Artemisia princeps Chenopodium ficifolium Oenothera biennis Echinochloa crus-galli | soybean | [11] |
| Bacillus sp. Arthrobacter pascens | Atriplex leucoclada Suaeda fruticosa | maize | [12] |
| Bacillus pumilus HR Zhihengliuella halotolerans SB | Seidlitzia rosmarinus Halostachys belangeriana | wheat | [13] |
| Bacillus pumilus STR2 Halomonas desiderata STR8 Exiguobacterium oxidotolerans STR36 | Poaceae | Mentha arvensis | [14] |
| Bacillus inaquosorum Bacillus thuringiensis Bacillus proteolyticus | Limoniastrum monopetalum Arthrocnemum indicum Holocnemum strobilaceum | quinoa (Chenopodium quinoa) | [15] |
| Enterobacter asburiae A103 | Salix linearistipularis | Medicago sativa | [16] |
| Glutamicibacter halophytocola KLBMP 5180 | Limonium sinense | tomato | [17] |
| Kocuria turfanensis 2M4 | Suaeda fruticosa | peanut | [18] |
| Pseudarthrobacter oxydans B9 Staphylococcus pasteuri B10 | sorghum | tomato | [19] |
| Paraburkholderia sp. GD17 | Glycine soja | rice | [20] |
| Pseudomonas putida CO1 Bacillus paramycoides CO8 | Cocos nucifera | French bean (Phaseolus vulgaris) | [21] |
| Pseudomonas sp. TE7 Providencia rettgeri SE5 Pantoea agglomerans SE19 | Tamarix gallica Suaeda fruticosa | barley tomato | [22] |
| Pseudomonas chloritidismutans 6L11 | Salicornia | wheat | [23] |
| Streptomyces sp. KLBMP5084 | Limonium sinense | tomato | [24] |
| Serratia marcescens Bacillus velezensis Kocuria rhizophila Kosakonia radicincitans | Sesuvium portulacastrum | Vigna mungo | [25] |
| Serratia sp. NTN6 | Puccinellia tenuiflora | maize | [26] |
| Stenotrophomonas maltophilia BJ01 | Cyperus laevigatus | peanut | [27] |
| Variovorax sp. P1R9 | Distichlis spicata | wheat | [28] |
2. Materials and Methods
2.1. Soil Sample Collection and Strain Isolation
2.2. Strain Identification
2.2.1. Morphological Identification
2.2.2. Biochemical Identification
2.2.3. 16S rRNA Gene Identification
2.3. Functional Characterization and Plant Growth Promotion Experiments
2.3.1. Salt Tolerance Evaluation of Strains
2.3.2. Evaluation of Plant Growth-Promoting Abilities
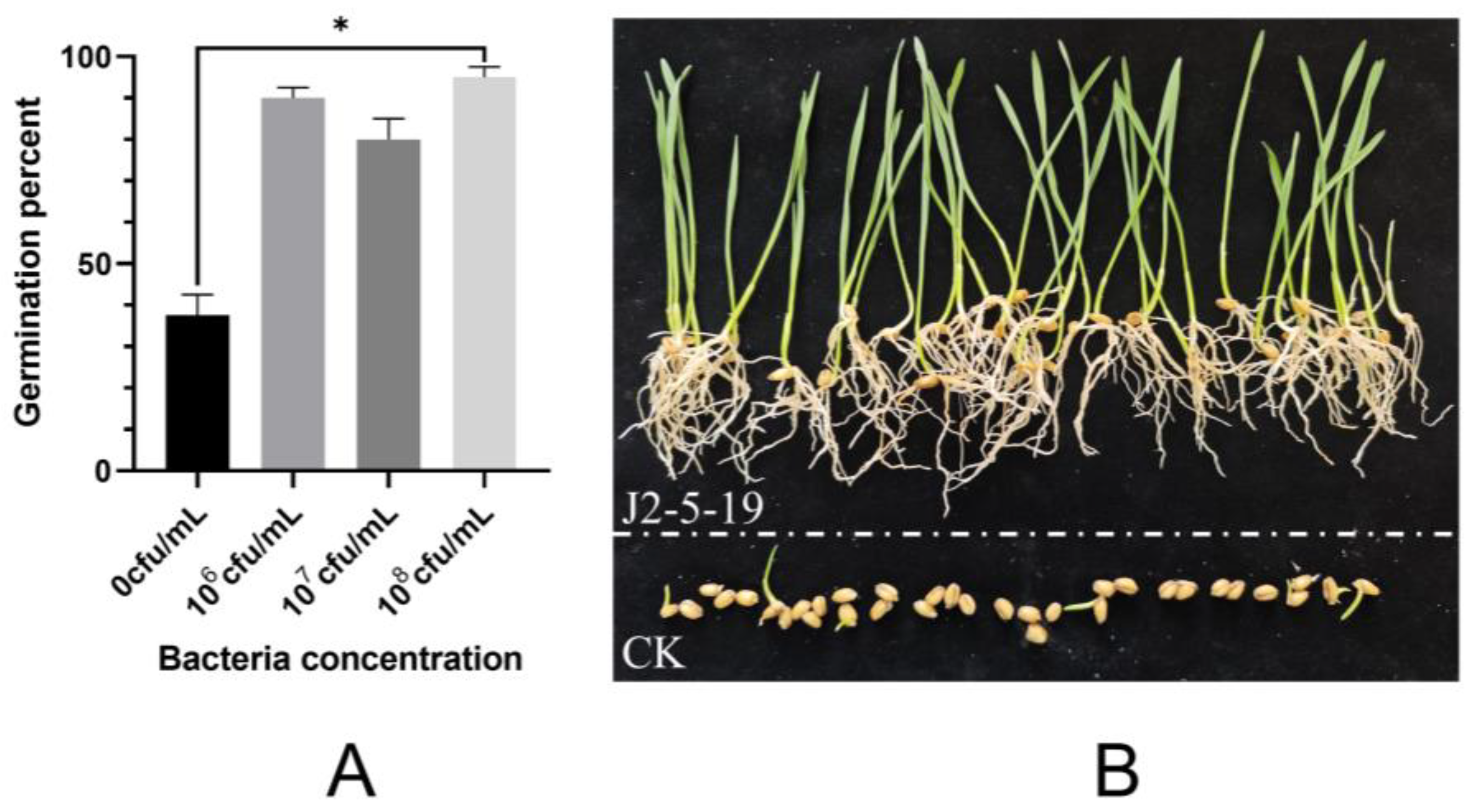
2.3.3. Germination Experiment of Wheat Seeds Under Salt Stress
2.3.4. Corn Pot Experiment Under Salt Stress
2.4. Genome Sequencing and Analysis
2.4.1. Genome Extraction
2.4.2. Genome Sequencing and Assembly
2.4.3. Comparative Genome Analysis
2.4.4. Gene Function Annotation
2.5. Statistical Analysis
3. Results and Discussion
3.1. Morphological Characteristics of the Strain
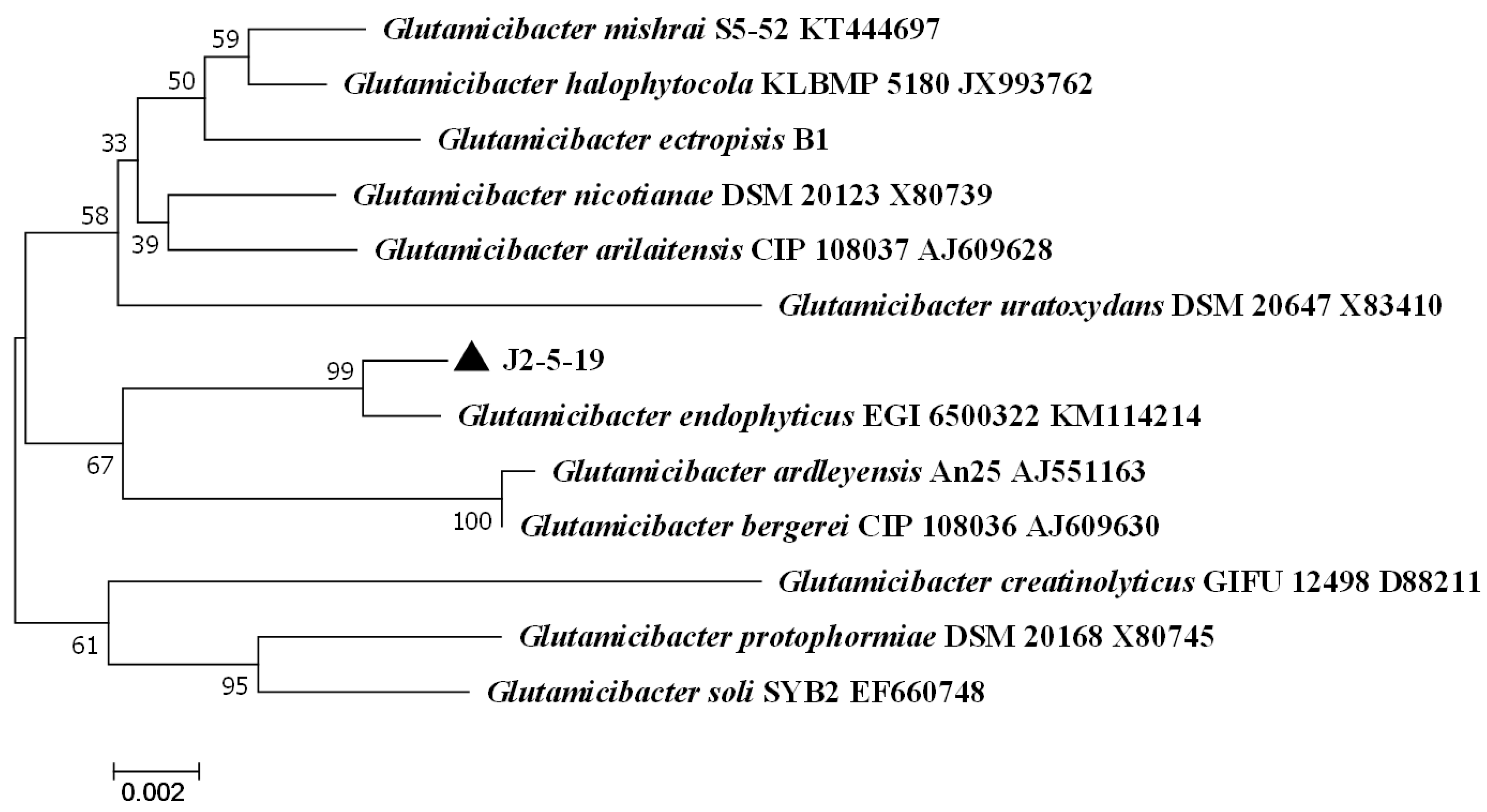
3.2. 16S rRNA Gene Sequencing
3.3. Biochemical Characteristics
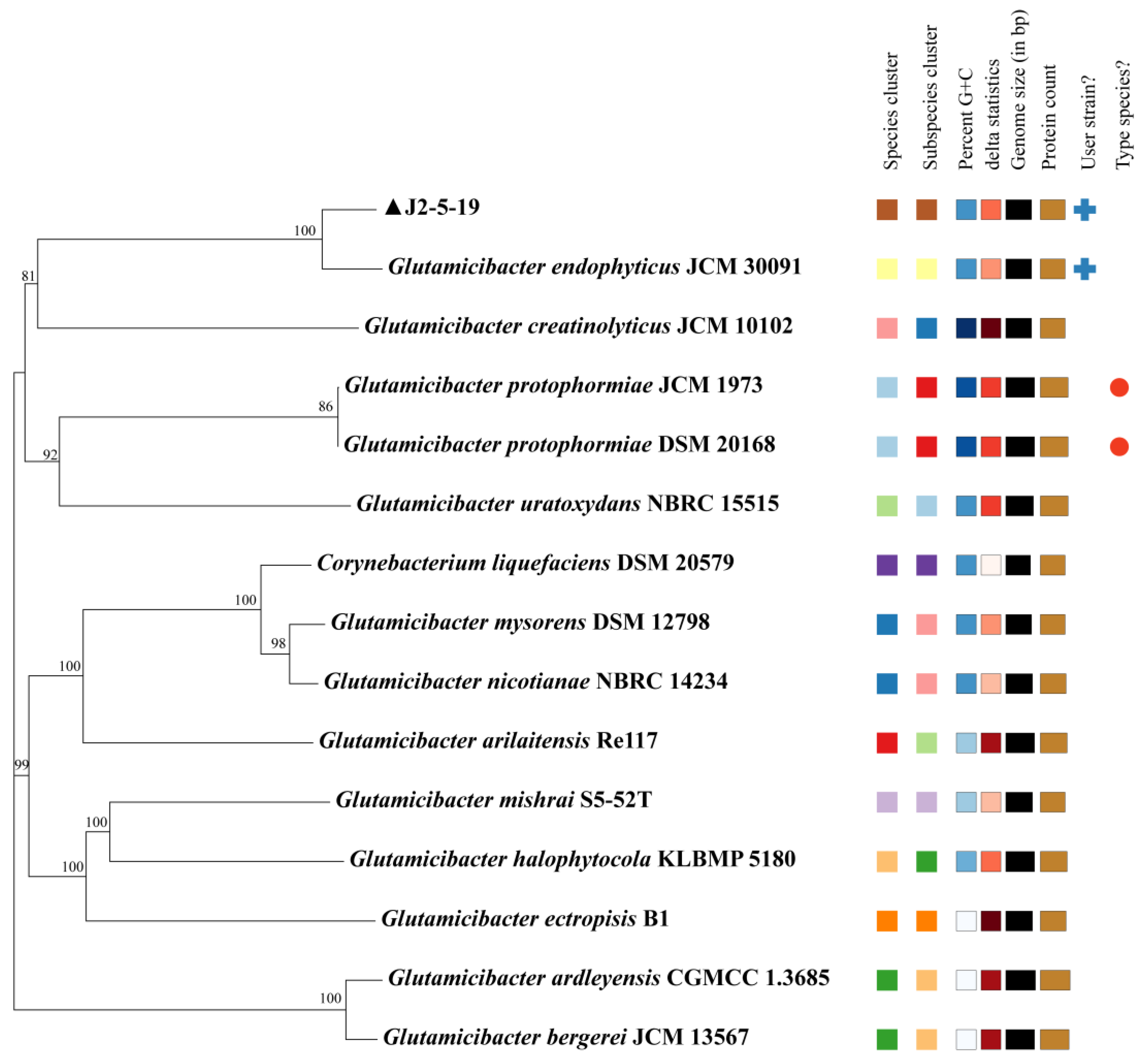
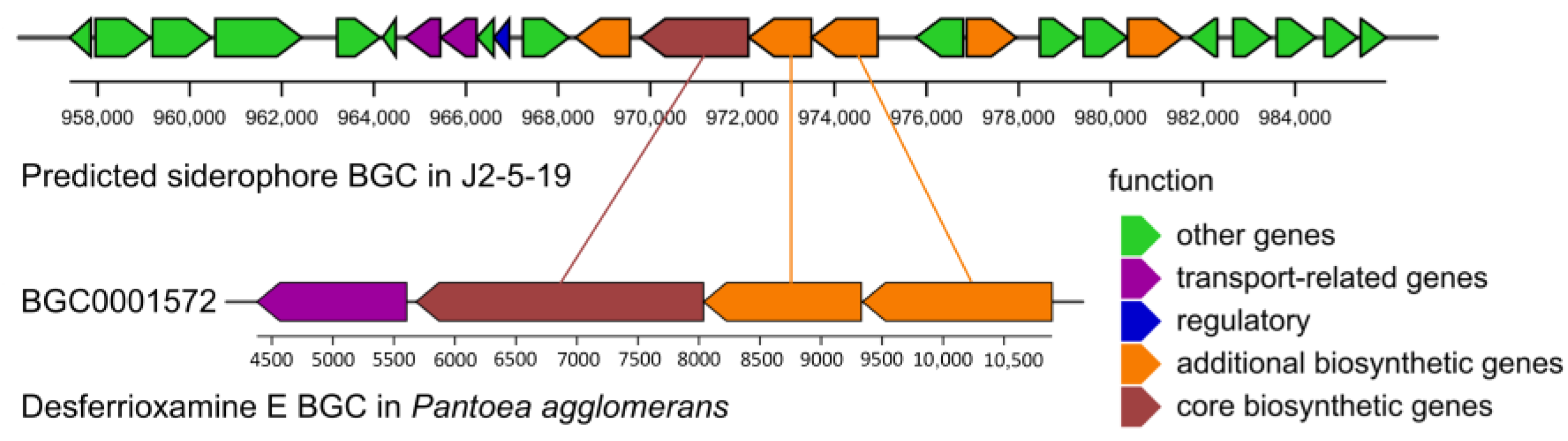
3.4. Genomic Architecture and Comparative Genomic Analysis
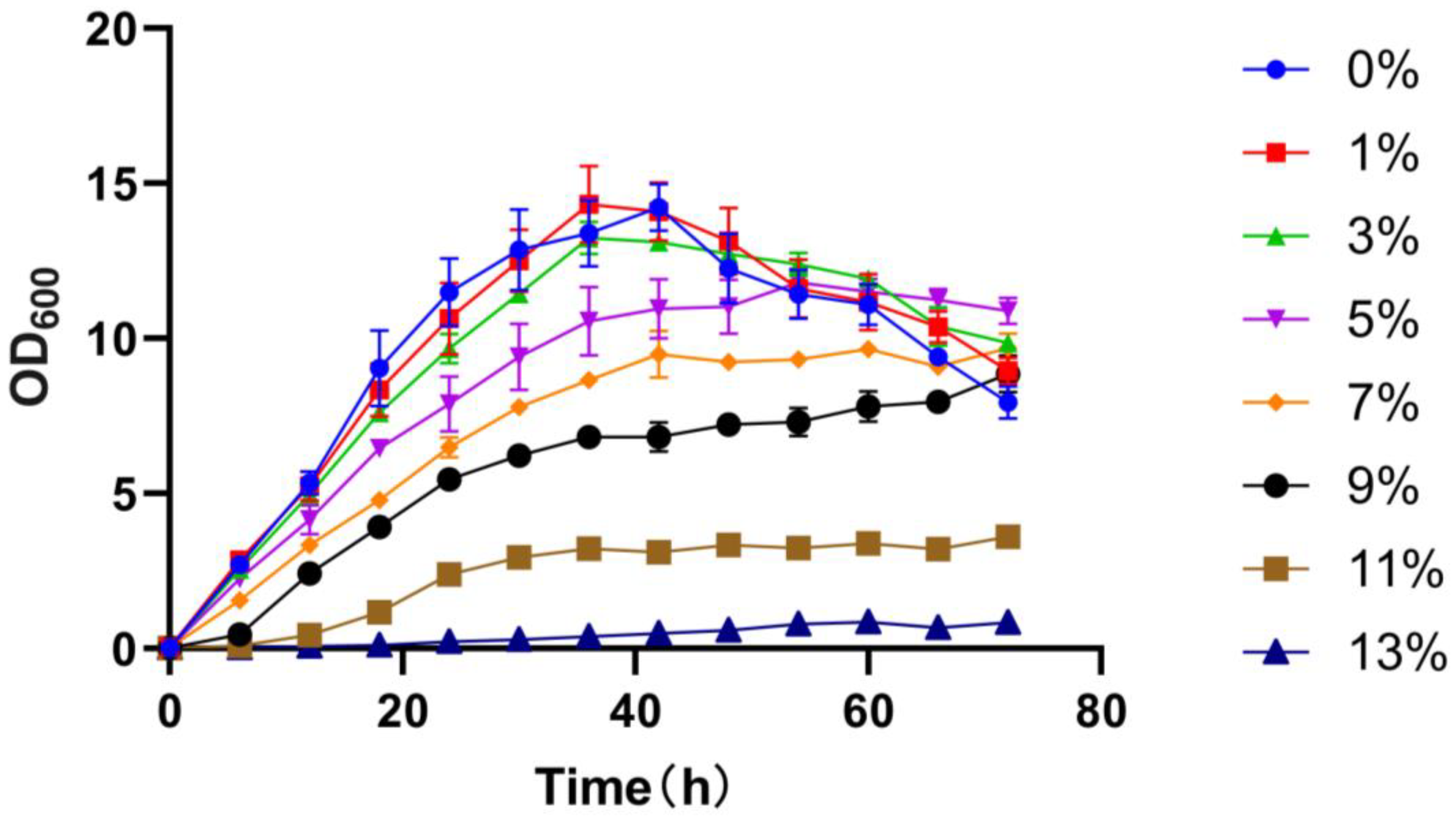
3.5. Salt Tolerance and Mechanism of Salt Resistance in the Strain
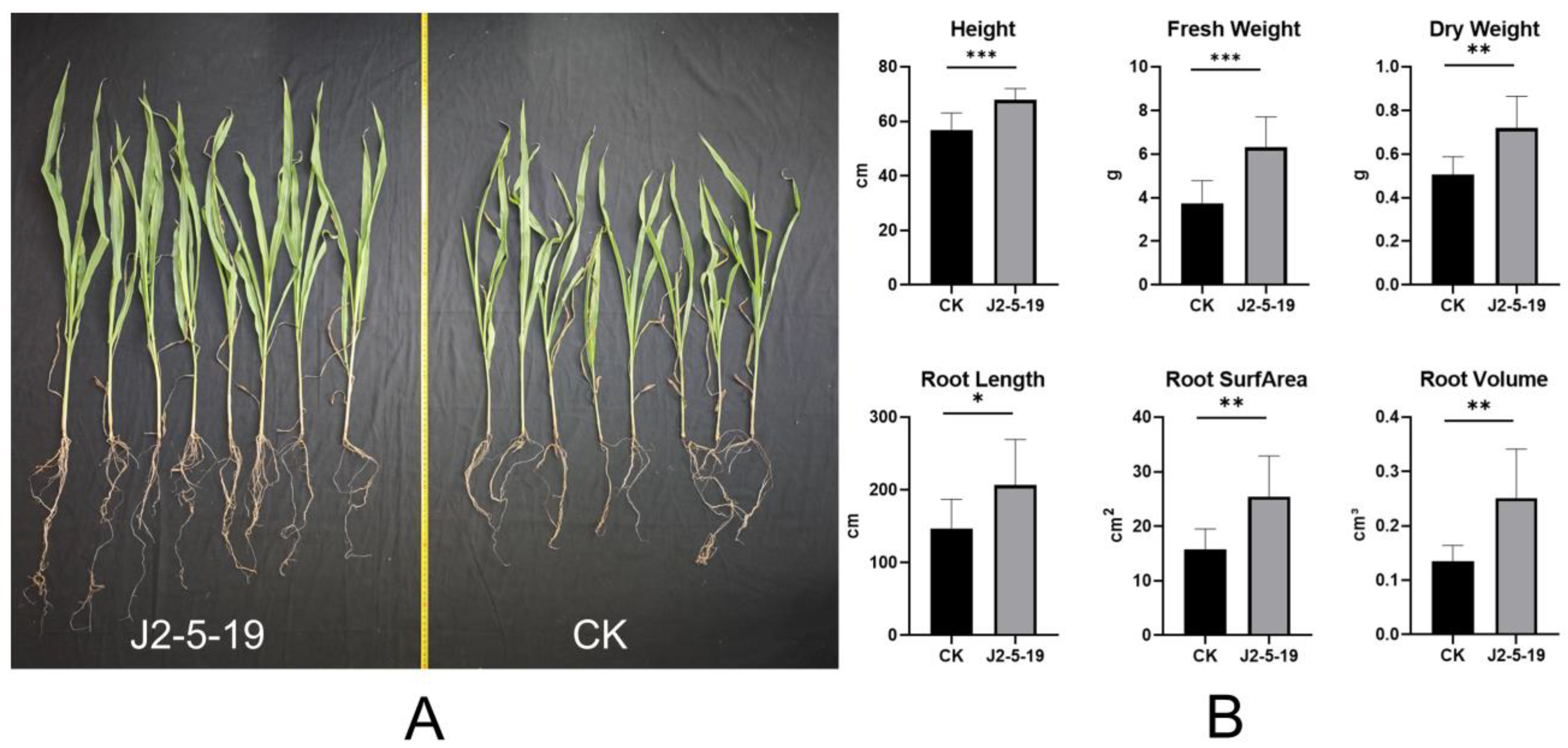
3.6. Growth-Promoting Ability of the Strain and Genomic Interpretation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, A. Soil salinity: A global threat to sustainable development. Soil Use Manag. 2022, 38, 39–67. [Google Scholar] [CrossRef]
- Etesami, H.; Beattie, G.A. Mining Halophytes for Plant Growth-Promoting Halotolerant Bacteria to Enhance the Salinity Tolerance of Non-halophytic Crops. Front. Microbiol. 2018, 9, 148. [Google Scholar] [CrossRef] [PubMed]
- Meena, K.K.; Bitla, U.; Sorty, A.M.; Kumar, S.; Kumar, S.; Wakchaure, G.C.; Singh, D.P.; Stougaard, P.; Suprasanna, P. Understanding the Interaction and Potential of Halophytes and Associated Microbiome for Bio-saline Agriculture. J. Plant Growth Regul. 2023, 42, 6601–6619. [Google Scholar] [CrossRef]
- Wang, H.-F.; Li, L.; Zhang, Y.-G.; Hozzein, W.N.; Zhou, X.-K.; Liu, W.-H.; Duan, Y.-Q.; Li, W.-J. Arthrobacter endophyticus sp. nov., an endophytic actinobacterium isolated from root of Salsola affinis C. A. Mey. Int. J. Syst. Evol. Microbiol. 2015, 65, 2154–2160. [Google Scholar] [CrossRef]
- Busse, H.-J. Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov., Paenarthrobacter gen. nov. and Pseudarthrobacter gen. nov., and emended description of Arthrobacter roseus. Int. J. Syst. Evol. Microbiol. 2016, 66, 9–37. [Google Scholar] [CrossRef] [PubMed]
- Busse, H.-J.; Schumann, P. Reclassification of Arthrobacter endophyticus (Wang et al. 2015) as Glutamicibacter endophyticus comb. nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 1057–1059. [Google Scholar] [CrossRef]
- Sadeghi, H.; Arjmand, S.; Ranaei Siadat, S.O.; Fooladi, J.; Ebrahimipour, G. A novel thermostable alkaline histamine oxidase from Glutamicibacter sp. N1A3101, induced by histamine and its analogue betahistine. AMB Express 2020, 10, 176. [Google Scholar] [CrossRef]
- Khan, T.; Alzahrani, O.M.; Sohail, M.; Hasan, K.A.; Gulzar, S.; Rehman, A.U.; Mahmoud, S.F.; Alswat, A.S.; Abdel-Gawad, S.A. Enzyme Profiling and Identification of Endophytic and Rhizospheric Bacteria Isolated from Arthrocnemum macrostachyum. Microorganisms 2022, 10, 2112. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.-Y.; Narsing Rao, M.P.; Wang, H.-F.; Fang, B.-Z.; Liu, Y.-H.; Li, L.; Xiao, M.; Li, W.-J. Transcriptomic analysis of two endophytes involved in enhancing salt stress ability of Arabidopsis thaliana. Sci. Total Environ. 2019, 686, 107–117. [Google Scholar] [CrossRef]
- Sharma, S.; Kulkarni, J.; Jha, B. Halotolerant Rhizobacteria Promote Growth and Enhance Salinity Tolerance in Peanut. Front. Microbiol. 2016, 7, 1600. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Asaf, S.; Khan, A.L.; Adhikari, A.; Jan, R.; Ali, S.; Imran, M.; Kim, K.M.; Lee, I.J. Halotolerant Rhizobacterial Strains Mitigate the Adverse Effects of NaCl Stress in Soybean Seedlings. Biomed. Res. Int. 2019, 2019, 15. [Google Scholar] [CrossRef] [PubMed]
- Ullah, S.; Bano, A. Isolation of plant-growth-promoting rhizobacteria from rhizospheric soil of halophytes and their impact on maize (Zea mays L.) under induced soil salinity. Can. J. Microbiol. 2015, 61, 307–313. [Google Scholar] [CrossRef]
- Amini Hajiabadi, A.; Mosleh Arani, A.; Ghasemi, S.; Rad, M.H.; Etesami, H.; Shabazi Manshadi, S.; Dolati, A. Mining the rhizosphere of halophytic rangeland plants for halotolerant bacteria to improve growth and yield of salinity-stressed wheat. Plant Physiol. Biochem. 2021, 163, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Bharti, N.; Barnawal, D.; Awasthi, A.; Yadav, A.; Kalra, A. Plant growth promoting rhizobacteria alleviate salinity induced negative effects on growth, oil content and physiological status in Mentha arvensis. Acta Physiol. Plant. 2014, 36, 45–60. [Google Scholar] [CrossRef]
- Slatni, T.; Ben Slimene, I.; Harzalli, Z.; Taamalli, W.; Smaoui, A.; Abdelly, C.; Elkahoui, S. Enhancing quinoa (Chenopodium quinoa) growth in saline environments through salt-tolerant rhizobacteria from halophyte biotope. Physiol. Plant. 2024, 176, e14466. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gao, M.; Zhang, W.; Liu, Y.; Wang, S.; Zhang, H.; Li, X.; Yu, S.; Lu, L. Halotolerant Enterobacter asburiae A103 isolated from the halophyte Salix linearistipularis: Genomic analysis and growth-promoting effects on Medicago sativa under alkali stress. Microbiol. Res. 2024, 289, 127909. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.-W.; Gong, Y.; Li, X.-W.; Chen, P.; Ju, X.-Y.; Zhang, C.-M.; Yuan, B.; Lv, Z.-P.; Xing, K.; Qin, S. Enhancement of growth and salt tolerance of tomato seedlings by a natural halotolerant actinobacterium Glutamicibacter halophytocola KLBMP 5180 isolated from a coastal halophyte. Plant Soil 2019, 445, 307–322. [Google Scholar] [CrossRef]
- Goswami, D.; Pithwa, S.; Dhandhukia, P.; Thakker, J.N. Delineating Kocuria turfanensis 2M4 as a credible PGPR: A novel IAA-producing bacteria isolated from saline desert. J. Plant Interact. 2014, 9, 566–576. [Google Scholar] [CrossRef]
- Latif, A.; Ahmad, R.; Ahmed, J.; Shah, M.M.; Ahmad, R.; Hassan, A. Novel halotolerant rhizobacterial strains mitigated the salt stress in-vitro and in-vivo and improved the growth of tomato plants. Sci. Hortic. 2023, 319, 112115. [Google Scholar] [CrossRef]
- Zhu, R.; Cao, Y.; Li, G.; Guo, Y.; Ma, L.; Bu, N.; Hao, L. Paraburkholderia sp. GD17 improves rice seedling tolerance to salinity. Plant Soil 2021, 467, 373–389. [Google Scholar] [CrossRef]
- Pandey, S.; Gupta, S. Diversity analysis of ACC deaminase producing bacteria associated with rhizosphere of coconut tree (Cocos nucifera L.) grown in Lakshadweep islands of India and their ability to promote plant growth under saline conditions. J. Biotechnol. 2020, 324, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Bakelli, A.; Amrani, S.; Bouri, M.; Kalayci, S.; Sahin, F. Endophytic bacteria of common tamarisk (Tamarix gallica L.) and alkali seepweed (Suaeda fruticosa (L.) forssk.) as potential biocontrol and plant growth-promoting agents in arid environments. Appl. Ecol. Environ. Res. 2022, 20, 3073–3098. [Google Scholar] [CrossRef]
- Zhou, D.; Yin, Z.; Li, X.; Cui, Y.; Cheng, Q.; Du, B.; Liu, K.; Wang, C.; Ding, Y. Complete Genome Sequence of Pseudomonas chloritidismutans 6L11 with Plant Growth–Promoting and Salt-Tolerant Properties. Mol. Plant-Microbe Interact. 2022, 35, 870–874. [Google Scholar] [CrossRef]
- Gong, Y.; Chen, L.-J.; Pan, S.-Y.; Li, X.-W.; Xu, M.-J.; Zhang, C.-M.; Xing, K.; Qin, S. Antifungal potential evaluation and alleviation of salt stress in tomato seedlings by a halotolerant plant growth-promoting actinomycete Streptomyces sp. KLBMP5084. Rhizosphere 2020, 16, 100262. [Google Scholar] [CrossRef]
- John, J.E.; Maheswari, M.; Kalaiselvi, T.; Prasanthrajan, M.; Poornachandhra, C.; Rakesh, S.S.; Gopalakrishnan, B.; Davamani, V.; Kokiladevi, E.; Ranjith, S. Biomining Sesuvium portulacastrum for halotolerant PGPR and endophytes for promotion of salt tolerance in Vigna mungo L. Front. Microbiol. 2023, 14, 1085787. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.F.; Han, C.; Liu, T.; Wang, Y.M.; Sun, P.; Pang, Q.Y.; Zhang, X.C.; Xiang, W.S.; Zhao, J.W. Integrated physiological, biochemical and transcriptomic analyses reveal the mechanism of salt tolerance induced by a halotolerant Serratia sp. NTN6 in maize. Environ. Exp. Bot. 2024, 221, 16. [Google Scholar] [CrossRef]
- Alexander, A.; Singh, V.K.; Mishra, A.; Jha, B. Plant growth promoting rhizobacterium Stenotrophomonas maltophilia BJ01 augments endurance against N2 starvation by modulating physiology and biochemical activities of Arachis hypogea. PLoS ONE 2019, 14, e0222405. [Google Scholar] [CrossRef]
- Acuña, J.J.; Rilling, J.I.; Inostroza, N.G.; Zhang, Q.; Wick, L.Y.; Sessitsch, A.; Jorquera, M.A. Variovorax sp. strain P1R9 applied individually or as part of bacterial consortia enhances wheat germination under salt stress conditions. Sci. Rep. 2024, 14, 2070. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, S.N.; Gibbons, N.E. Effect of some metal ions on the growth of Halobacterium cutirubrum. Can. J. Microbiol. 1960, 6, 165–169. [Google Scholar] [CrossRef]
- Dong, X.; Cai, M. Manual for Systematic Identification of Common Bacteria; Science Press: Beijing, China, 2001. [Google Scholar]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.; Xu, J.; Acosta, K.; Poulev, A.; Lebeis, S.; Lam, E. Bacterial Production of Indole Related Compounds Reveals Their Role in Association Between Duckweeds and Endophytes. Front. Chem. 2018, 6, 265. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021, 50, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xiao, X.; Wang, P.; Zeng, X.; Wang, F. Arthrobacter ardleyensis sp. nov., isolated from Antarctic lake sediment and deep-sea sediment. Arch. Microbiol. 2005, 183, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Irlinger, F.; Bimet, F.; Delettre, J.; Lefèvre, M.; Grimont, P.A.D. Arthrobacter bergerei sp. nov. and Arthrobacter arilaitensis sp. nov., novel coryneform species isolated from the surfaces of cheeses. Int. J. Syst. Evol. Microbiol. 2005, 55, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Padan, E.; Tzubery, T.; Herz, K.; Kozachkov, L.; Rimon, A.; Galili, L. NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+antiporter. Biochim. Biophys. Acta BBA Bioenerg. 2004, 1658, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.; Moir, A. The Bacillus cereus GerN and GerT Protein Homologs Have Distinct Roles in Spore Germination and Outgrowth, Respectively. J. Bacteriol. 2008, 190, 6148–6152. [Google Scholar] [CrossRef]
- Waditee, R.; Hibino, T.; Tanaka, Y.; Nakamura, T.; Incharoensakdi, A.; Takabe, T. Halotolerant Cyanobacterium Aphanothece halophytica Contains an Na+/H+ Antiporter, Homologous to Eukaryotic Ones, with Novel Ion Specificity Affected by C-terminal Tail*. J. Biol. Chem. 2001, 276, 36931–36938. [Google Scholar] [CrossRef]
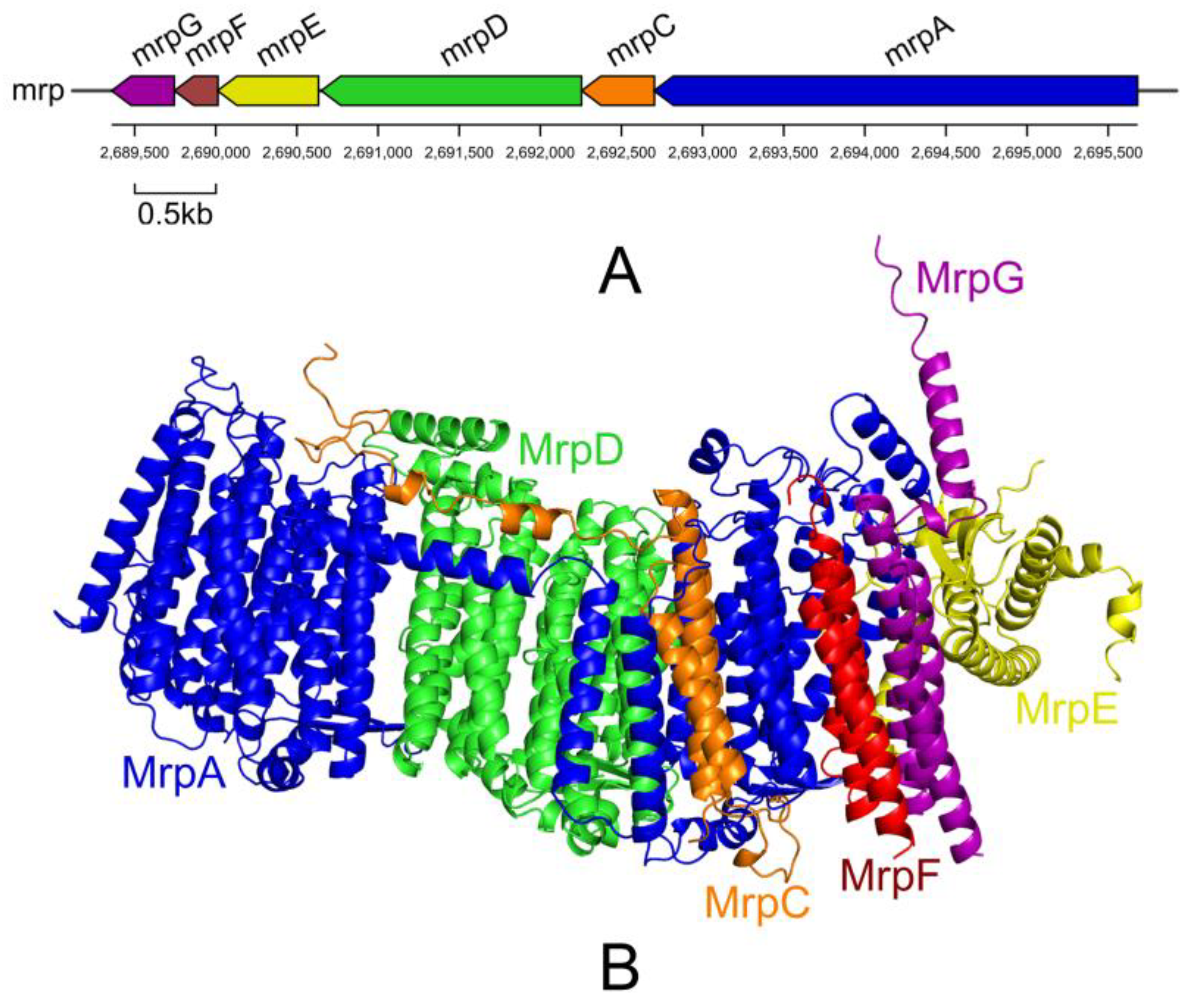
- Swartz, T.H.; Ikewada, S.; Ishikawa, O.; Ito, M.; Krulwich, T.A. The Mrp system: A giant among monovalent cation/proton antiporters? Extremophiles 2005, 9, 345–354. [Google Scholar] [CrossRef]
- Ito, M.; Morino, M.; Krulwich, T.A. Mrp Antiporters Have Important Roles in Diverse Bacteria and Archaea. Front. Microbiol. 2017, 8, 2325. [Google Scholar] [CrossRef]
- Hamann, A.; Bossemeyer, D.; Bakker, E.P. Physical mapping of the K+ transport trkA gene of Escherichia coli and overproduction of the TrkA protein. J. Bacteriol. 1987, 169, 3138–3145. [Google Scholar] [CrossRef]
- Nakamura, T.; Yuda, R.; Unemoto, T.; Bakker, E.P. KtrAB, a New Type of Bacterial K+-Uptake System from Vibrio alginolyticus. J. Bacteriol. 1998, 180, 3491–3494. [Google Scholar] [CrossRef] [PubMed]
- Gundlach, J.; Herzberg, C.; Kaever, V.; Gunka, K.; Hoffmann, T.; Weiß, M.; Gibhardt, J.; Thürmer, A.; Hertel, D.; Daniel, R.; et al. Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci. Signal. 2017, 10, eaal3011. [Google Scholar] [CrossRef]
- Dinnbier, U.; Limpinsel, E.; Schmid, R.; Bakker, E.P. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 1988, 150, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef] [PubMed]
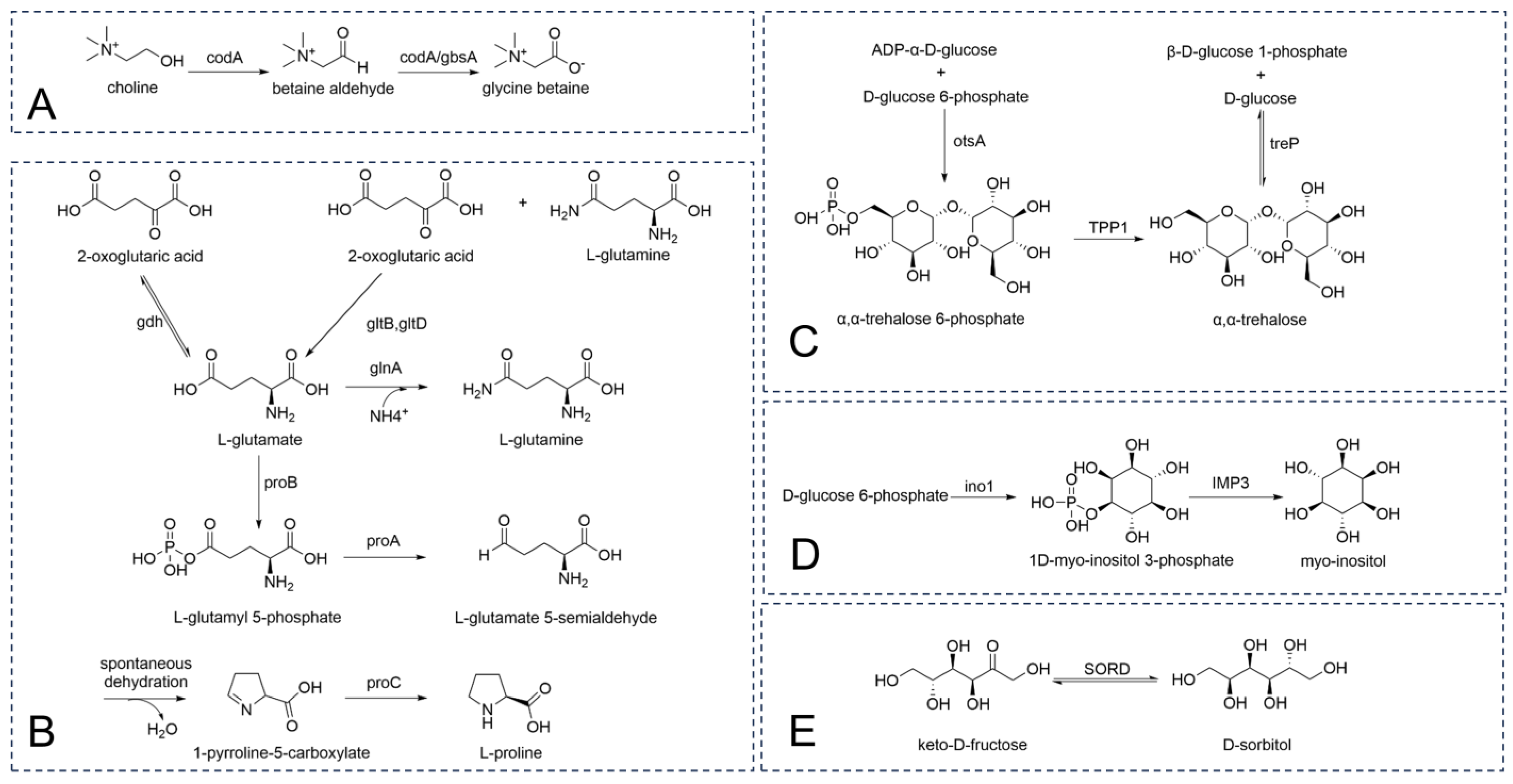
- Rand, T.; Halkier, T.; Hansen, O.C. Structural Characterization and Mapping of the Covalently Linked FAD Cofactor in Choline Oxidase from Arthrobacter globiformis. Biochemistry 2003, 42, 7188–7194. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Chen, N.; Shi, M.; Xian, M.; Song, Y.; Liu, J. The metabolism and biotechnological application of betaine in microorganism. Appl. Microbiol. Biotechnol. 2016, 100, 3865–3876. [Google Scholar] [CrossRef] [PubMed]
- Saum, S.H.; Sydow, J.F.; Palm, P.; Pfeiffer, F.; Oesterhelt, D.; Müller, V. Biochemical and Molecular Characterization of the Biosynthesis of Glutamine and Glutamate, Two Major Compatible Solutes in the Moderately Halophilic Bacterium Halobacillus halophilus. J. Bacteriol. 2006, 188, 6808–6815. [Google Scholar] [CrossRef] [PubMed]
- Rexer, H.U.; Schäberle, T.; Wohlleben, W.; Engels, A. Investigation of the functional properties and regulation of three glutamine synthetase-like genes in Streptomyces coelicolor A3(2). Arch. Microbiol. 2006, 186, 447–458. [Google Scholar] [CrossRef]
- Csonka, L.N.; Leisinger, T. Biosynthesis of Proline. EcoSal Plus 2007, 2. [Google Scholar] [CrossRef] [PubMed]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef]
- Habibur Rahman Pramanik, M.; Imai, R. Functional Identification of a Trehalose 6-phosphate Phosphatase Gene that is Involved in Transient Induction of Trehalose Biosynthesis during Chilling Stress in Rice. Plant Mol. Biol. 2005, 58, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Koliwer-Brandl, H.; Syson, K.; van de Weerd, R.; Chandra, G.; Appelmelk, B.; Alber, M.; Ioerger, T.R.; Jacobs, W.R., Jr.; Geurtsen, J.; Bornemann, S.; et al. Metabolic Network for the Biosynthesis of Intra- and Extracellular α-Glucans Required for Virulence of Mycobacterium tuberculosis. PLoS Pathog. 2016, 12, e1005768. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.S.; Fukuda, T.; Sena, C.B.C.; Yamaryo-Botte, Y.; McConville, M.J.; Kinoshita, T. Inositol lipid metabolism in mycobacteria: Biosynthesis and regulatory mechanisms. Biochim. Biophys. Acta (BBA)–Gen. Subj. 2011, 1810, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Karacaoğlan, V.; Özer, I. Steady-state kinetic properties of sorbitol dehydrogenase from chicken liver. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2005, 140, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Choquet, G.; Jehan, N.; Pissavin, C.; Blanco, C.; Jebbar, M. OusB, a Broad-Specificity ABC-Type Transporter from Erwinia chrysanthemi, Mediates Uptake of Glycine Betaine and Choline with a High Affinity. Appl. Environ. Microbiol. 2005, 71, 3389–3398. [Google Scholar] [CrossRef]
- Kempf, B.; Bremer, E. OpuA, an Osmotically Regulated Binding Protein-dependent Transport System for the Osmoprotectant Glycine Betaine in Bacillus subtilis(*). J. Biol. Chem. 1995, 270, 16701–16713. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.-K.; Gort, S.; Maloy, S. A cryptic proline permease in Salmonella typhimurium. Microbiology 1997, 143, 2903–2911. [Google Scholar] [CrossRef]
- Peter, H.; Weil, B.; Burkovski, A.; Krämer, R.; Morbach, S. Corynebacterium glutamicum Is Equipped with Four Secondary Carriers for Compatible Solutes: Identification, Sequencing, and Characterization of the Proline/Ectoine Uptake System, ProP, and the Ectoine/Proline/Glycine Betaine Carrier, EctP. J. Bacteriol. 1998, 180, 6005–6012. [Google Scholar] [CrossRef] [PubMed]
- Moses, S.; Sinner, T.; Zaprasis, A.; Stöveken, N.; Hoffmann, T.; Belitsky, B.R.; Sonenshein, A.L.; Bremer, E. Proline Utilization by Bacillus subtilis: Uptake and Catabolism. J. Bacteriol. 2012, 194, 745–758. [Google Scholar] [CrossRef]
- Kronemeyer, W.; Peekhaus, N.; Krämer, R.; Sahm, H.; Eggeling, L. Structure of the gluABCD cluster encoding the glutamate uptake system of Corynebacterium glutamicum. J. Bacteriol. 1995, 177, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Mikkat, S.; Hagemann, M. Molecular analysis of the ggtBCD gene cluster of Synechocystis sp. strain PCC6803 encoding subunits of an ABC transporter for osmoprotective compounds. Arch. Microbiol. 2000, 174, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Sahintoth, M.; Frillingos, S.; Lengeler, J.W.; Kaback, H.R. Active Transport by the CscB Permease in Escherichia coli K-12. Biochem. Biophys. Res. Commun. 1995, 208, 1116–1123. [Google Scholar] [CrossRef] [PubMed]
- Steger, R.; Weinand, M.; Krämer, R.; Morbach, S. LcoP, an osmoregulated betaine/ectoine uptake system from Corynebacterium glutamicum. FEBS Lett. 2004, 573, 155–160. [Google Scholar] [CrossRef]
- Paulsen, I.T.; Seshadri, R.; Nelson, K.E.; Eisen, J.A.; Heidelberg, J.F.; Read, T.D.; Dodson, R.J.; Umayam, L.; Brinkac, L.M.; Beanan, M.J.; et al. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 2002, 99, 13148–13153. [Google Scholar] [CrossRef] [PubMed]
- Young, J.P.W.; Crossman, L.C.; Johnston, A.W.B.; Thomson, N.R.; Ghazoui, Z.F.; Hull, K.H.; Wexler, M.; Curson, A.R.J.; Todd, J.D.; Poole, P.S.; et al. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 2006, 7, R34. [Google Scholar] [CrossRef] [PubMed]
- Shende, V.V.; Bauman, K.D.; Moore, B.S. The shikimate pathway: Gateway to metabolic diversity. Nat. Prod. Rep. 2024, 41, 604–648. [Google Scholar] [CrossRef] [PubMed]
- Tivendale, N.D.; Davies, N.W.; Molesworth, P.P.; Davidson, S.E.; Smith, J.A.; Lowe, E.K.; Reid, J.B.; Ross, J.J. Reassessing the Role of N-Hydroxytryptamine in Auxin Biosynthesis. Plant Physiol. 2010, 154, 1957–1965. [Google Scholar] [CrossRef] [PubMed]
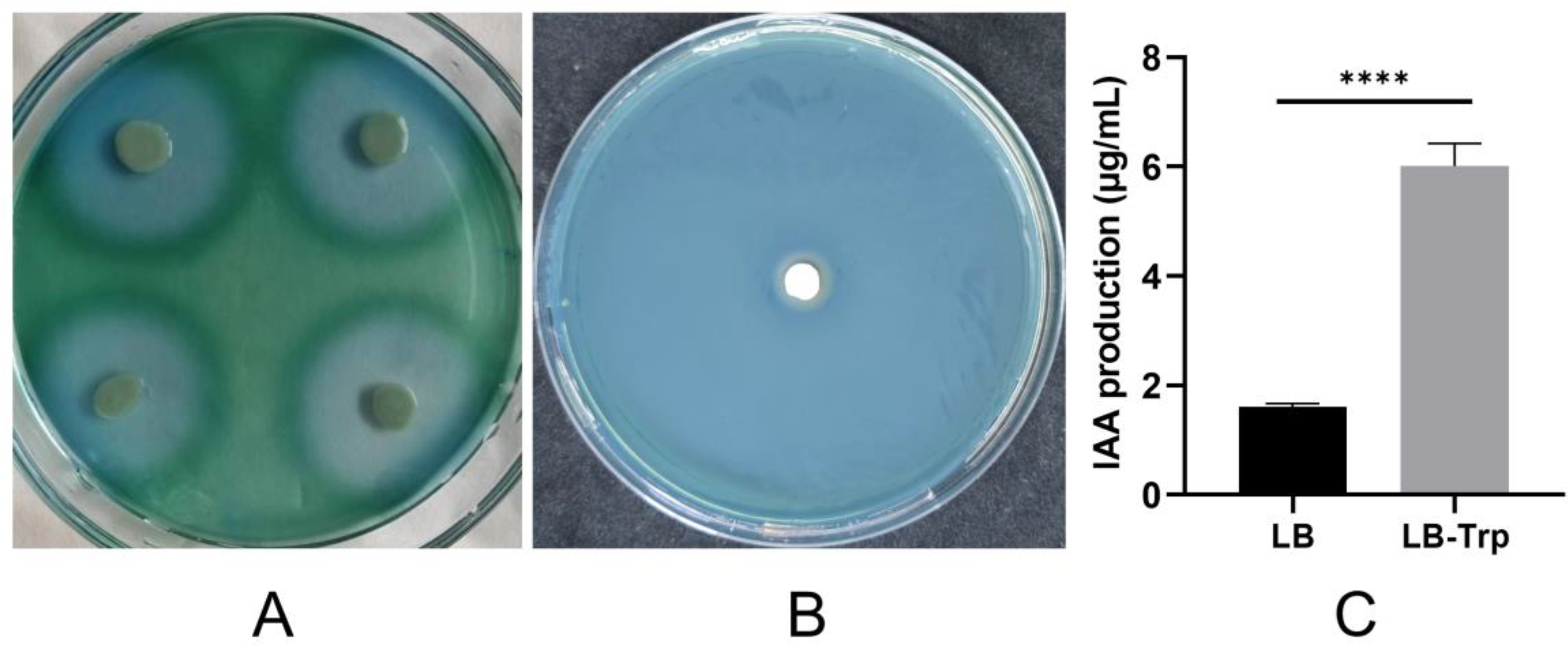
- Duca, D.; Lorv, J.; Patten, C.L.; Rose, D.; Glick, B.R. Indole-3-acetic acid in plant–microbe interactions. Antonie Van Leeuwenhoek 2014, 106, 85–125. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Kim, K.-J. Structural insight into molecular mechanism of cytokinin activating protein from Pseudomonas aeruginosa PAO1. Environ. Microbiol. 2018, 20, 3214–3223. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, S.; Tsuboi, Y.; Oda, S.; Kim, H.G.; Kumagai, H.; Suzuki, H. The Putrescine Importer PuuP of Escherichia coli K-12. J. Bacteriol. 2009, 191, 2776–2782. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, H.; Horinouchi, S.; Beppu, T. Putrescine Oxidase of Micrococcus Rubens: Primary Structure and Escherichia coli. Microbiology 1993, 139, 425–432. [Google Scholar] [CrossRef]
- Nicholson, W.L. The Bacillus subtilis ydjL (bdhA) Gene Encodes Acetoin Reductase/2,3-Butanediol Dehydrogenase. Appl. Environ. Microbiol. 2008, 74, 6832–6838. [Google Scholar] [CrossRef] [PubMed]
- Grundy, F.J.; Waters, D.A.; Takova, T.Y.; Henkin, T.M. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol. Microbiol. 1993, 10, 259–271. [Google Scholar] [CrossRef] [PubMed]
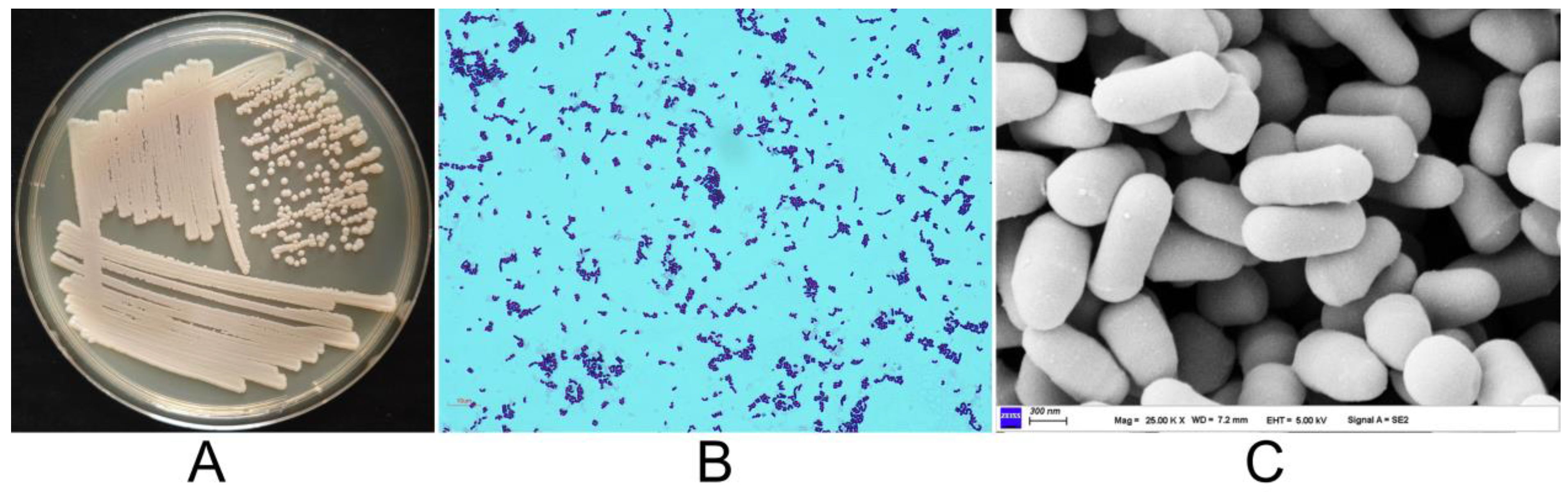


| Characteristics | J2-5-19 | EGI 6500322T |
|---|---|---|
| VP test | − | ND |
| Hydrolysis of | ||
| Tween 60 | − | + |
| Urea | − | + |
| PNPG | + | − |
| Assimilation of | ||
| Mannose | w | + |
| Mannitol | − | + |
| J2-5-19 & G. endophyticus | G. ardleyensis & G. bergerei | |
|---|---|---|
| ANIb | 95.93% | 96.84% |
| ANIm | 96.26% | 97.96% |
| DDH estimate | 67.4% (64.4–70.3%) | 79.7% (76.8–82.4%) |
| Probability that DDH > 70% | 73.15% | 90.36% |
| Difference in GC% | 0.01 | 0.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Sun, S.; Liu, T.; Lei, X.; Liu, R.; Zhang, J.; Dai, S.; Li, J.; Ding, Y. Association Analysis of the Genomic and Functional Characteristics of Halotolerant Glutamicibacter endophyticus J2-5-19 from the Rhizosphere of Suaeda salsa. Microorganisms 2025, 13, 208. https://doi.org/10.3390/microorganisms13010208
Sun L, Sun S, Liu T, Lei X, Liu R, Zhang J, Dai S, Li J, Ding Y. Association Analysis of the Genomic and Functional Characteristics of Halotolerant Glutamicibacter endophyticus J2-5-19 from the Rhizosphere of Suaeda salsa. Microorganisms. 2025; 13(1):208. https://doi.org/10.3390/microorganisms13010208
Chicago/Turabian StyleSun, Longhao, Shanshan Sun, Tianyang Liu, Xinmin Lei, Ruiqi Liu, Junyi Zhang, Shanshan Dai, Jing Li, and Yanqin Ding. 2025. "Association Analysis of the Genomic and Functional Characteristics of Halotolerant Glutamicibacter endophyticus J2-5-19 from the Rhizosphere of Suaeda salsa" Microorganisms 13, no. 1: 208. https://doi.org/10.3390/microorganisms13010208
APA StyleSun, L., Sun, S., Liu, T., Lei, X., Liu, R., Zhang, J., Dai, S., Li, J., & Ding, Y. (2025). Association Analysis of the Genomic and Functional Characteristics of Halotolerant Glutamicibacter endophyticus J2-5-19 from the Rhizosphere of Suaeda salsa. Microorganisms, 13(1), 208. https://doi.org/10.3390/microorganisms13010208






