Comparative Genomic Analysis of Campylobacter Plasmids Identified in Food Isolates
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
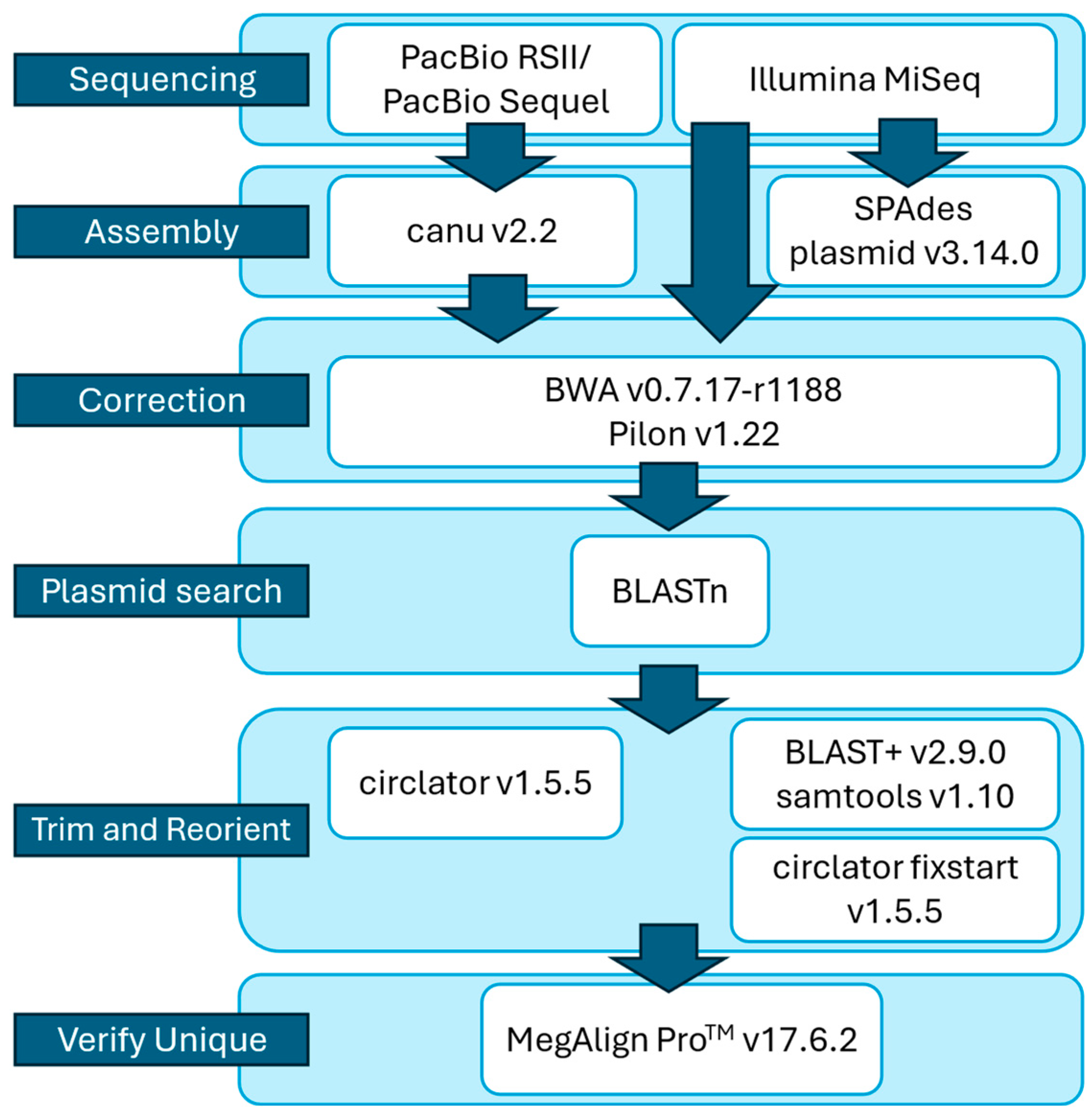
2.2. Genome Sequencing, Assembly, and Plasmid Identification
2.3. Plasmid Pangenome and Phylogeny
| Strain and Species | Source | Plasmid Name | Size (bp) | %GC | Accession No. | Reference |
|---|---|---|---|---|---|---|
| C. jejuni YH001 | Veal livers | pCJP001-1 | 46,524 | 29.74 | CP173351 | This work |
| C. jejuni YH001 | Veal livers | pCJP001-2 | 4354 | 30.57 | CP173352 | This work |
| C. jejuni YH002 | Calf livers | pCJP002 | 45,904 | 29.20 | CP020775 | [36] |
| C. jejuni YH016 | Calf livers | pCJP016 | 29,736 | 28.21 | CP157938 | This work |
| C. jejuni YH018 | Calf livers | pCJP018-1 | 46,524 | 29.74 | CP172373 | This work |
| C. jejuni YH018 | Calf livers | pCJP018-2 | 4366 | 30.85 | CP172374 | This work |
| C. jejuni YH019 | Beef livers | pCJP019-1 | 46,275 | 28.99 | CP172369 | This work |
| C. jejuni YH019 | Beef livers | pCJP019-2 | 30,011 | 28.18 | CP172370 | This work |
| C. jejuni YH019 | Beef livers | pCJP019-3 | 4367 | 30.82 | CP172371 | This work |
| C. jejuni YH020 | Veal livers | pCJP020 | 37,426 | 27.78 | CP172367 | This work |
| C. jejuni YH024 | Calf livers | pCJP024-1 | 45,034 | 29.55 | CP172359 | This work |
| C. jejuni YH024 | Calf livers | pCJP024-2 | 4366 | 30.85 | CP172360 | This work |
| C. jejuni YH025 | Calf livers | pCJP025 | 41,594 | 28.51 | CP172357 | This work |
| C. jejuni YH026 | Calf livers | pCJP026 | 44,973 | 29.12 | CP172355 | This work |
| C. jejuni YH027 | Calf livers | pCJP027 | 46,515 | 29.72 | CP172353 | This work |
| C. jejuni YH029 | Beef livers | pCJP029 | 16,920 | 28.27 | CP172350 | This work |
| C. jejuni (S33Cj) YH010 | Chicken thighs | pCJS010 (pCjS33) | 40,686 | 28.49 | CP131443 | [37] |
| C. jejuni (S36Cj) YH011 | Chicken thighs | pCJS011 (pCjS36) | 86,827 | 26.03 | CP131441 | [37] |
| C. jejuni YH014 | Chicken livers | pCJS014-1 | 47,468 | 30.28 | CP172377 | This work |
| C. jejuni YH014 | Chicken livers | pCJS014-2 | 43,660 | 29.00 | CP172378 | This work |
| C. jejuni YH021 | Chicken breasts | pCJS021 | 43,177 | 28.96 | CP172365 | This work |
| C. jejuni YH022 | Chicken thighs | pCJS022 | 48,862 | 28.64 | CP172363 | This work |
| C. coli YH502 | Chicken drumsticks | pCOS502 | 125,964 | 28.11 | CP018901 | [38] |
| C. coli YH503 | Chicken drumsticks | pCOS503-1 | 108,453 | 26.15 | CP025282 | [12] |
| C. coli YH503 | Chicken drumsticks | pCOS503-2 | 5401 | 32.85 | CP173353 | This work |
| C. coli YH504 | Chicken drumsticks | pCOS504 | 110,357 | 26.02 | CP091645 | [12] |
| C. coli YH504 | Chicken drumsticks | pCOS504-2 | 5401 | 32.85 | CP173354 | This work |
| C. coli YH506 | Chicken wings | pCOS506 | 5402 | 30.53 | CP172398 | This work |
| C. coli YH507 | Chicken livers | pCOS507-1 | 150,434 | 27.53 | CP172393 | This work |
| C. coli YH507 | Chicken livers | pCOS507-2 | 37,224 | 25.96 | CP172394 | This work |
| C. coli YH507 | Chicken livers | pCOS507-3 | 29,068 | 29.33 | CP172395 | This work |
| C. coli YH510 | Chicken livers | pCOS510-1 | 117,204 | 28.20 | CP172388 | This work |
| C. coli YH510 | Chicken livers | pCOS510-2 | 38,174 | 25.80 | CP172389 | This work |
| C. coli YH511 | Chicken livers | pCOS511 | 30,429 | 27.88 | CP172386 | This work |
| C. jejuni RM1246-ERRC | Human | pRM1246_ERRC | 45,197 | 29.14 | CP022471 | [39] |
| C. jejuni RM3194 | Human | pRM3194 | 81,079 | 25.99 | CP014345 | [40] |
| C. jejuni 81-176 | Human | pTet * | 45,025 | 29.09 | CP000549 | N/A |
| C. jejuni 81-176 | Human | pVir * | 37,473 | 25.89 | CP000550 | N/A |
| C. coli CVM N17C336 | Chicken breasts | pN17C336-1 * | 146,302 | 27.99 | CP169431 | N/A |
| C. coli CVM N17C264 | Chicken breasts | pN17C264-2 * | 39,356 | 26.18 | CP169460 | N/A |
| C. jejuni NADC 20827 | Turkey | p20827S * | 4366 | 30.83 | CP045047 | [41] |
| C. coli CC20JX12 | Meat | pCC20JX12-5K * | 5363 | 31.51 | CP109816 | N/A |
| C. coli 2014D-0261 | Not reported | p2014D0261-1 * | 52,384 | 28.41 | CP059367 | N/A |
| C. jejuni AR-0413 | Not reported | pAR-0413-2 * | 25,131 | 28.47 | CP044172 | N/A |
| C. jejuni PNUSC002710 | Not reported | pNUSAC002710-2 * | 28,157 | 28.03 | CP132117 | N/A |
| C. coli XK3140 | Chicken liver | pCCDM140S * | 26,812 | 29.28 | MH634990 | [9] |
| C. jejuni RM1477 | Human | pRM1477 * | 28,220 | 27.93 | CP071588 | [42] |
2.4. Tetracycline Resistance Testing
3. Results and Discussion
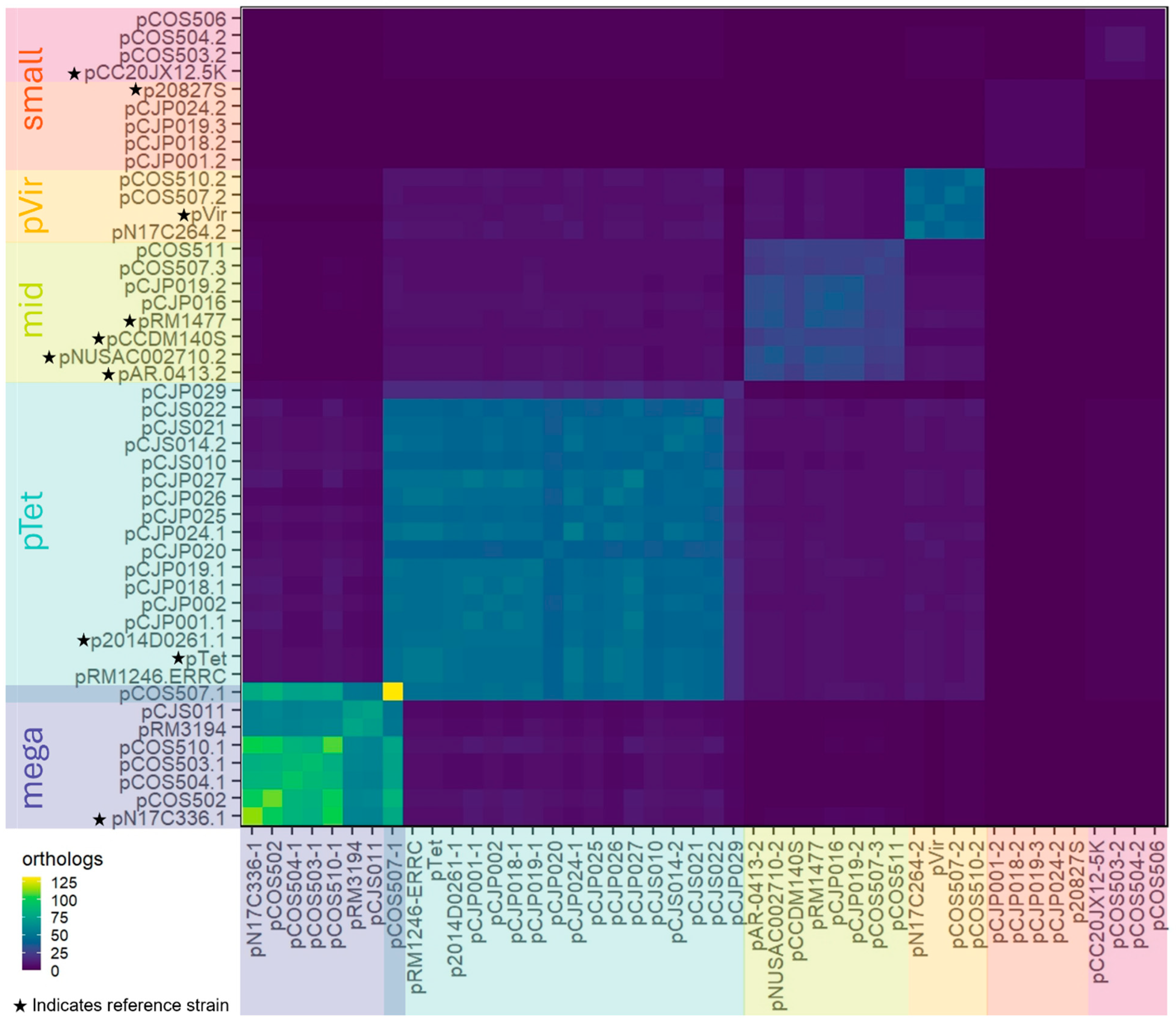
3.1. In Silico Identification of Large and Small Plasmids in Campylobacter Food Isolates
3.2. Pangenomic Analyses of the Conserved Core Genes and Diversity of the Plasmids
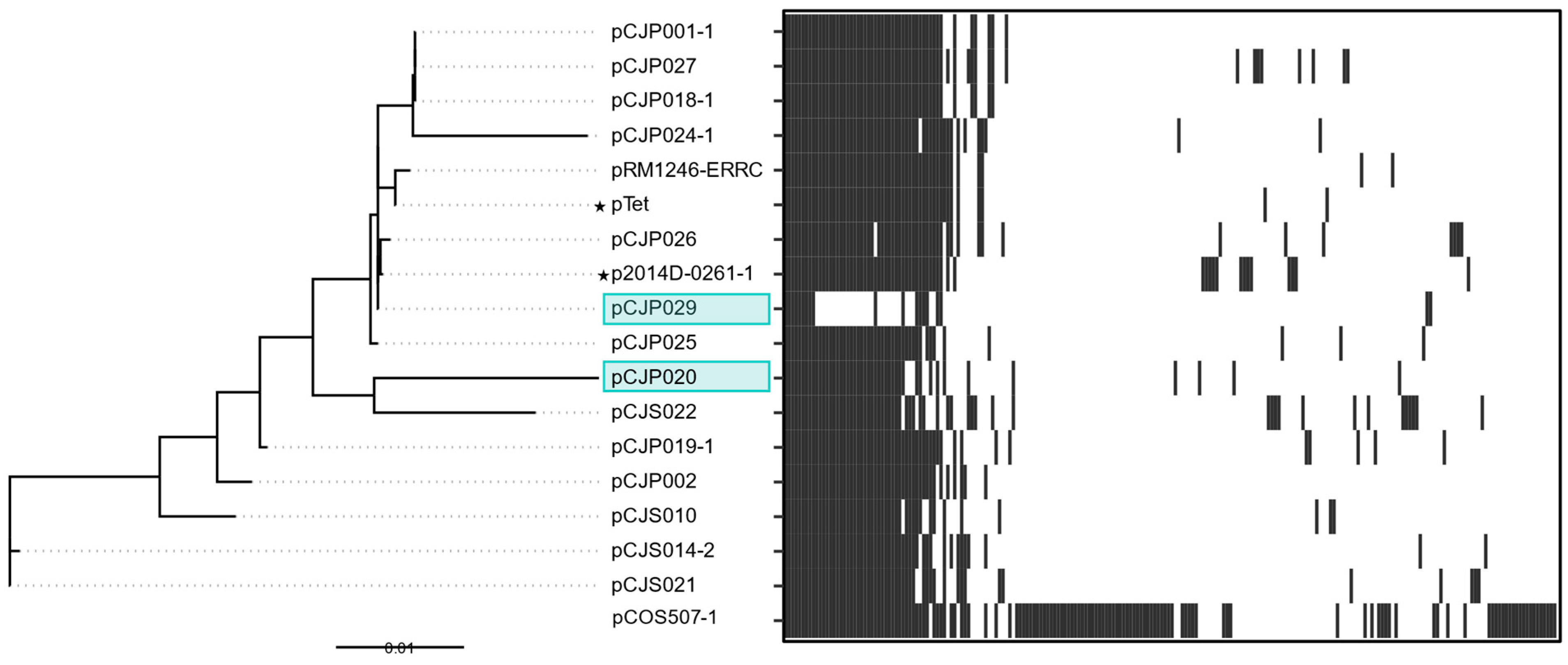
3.2.1. pTet
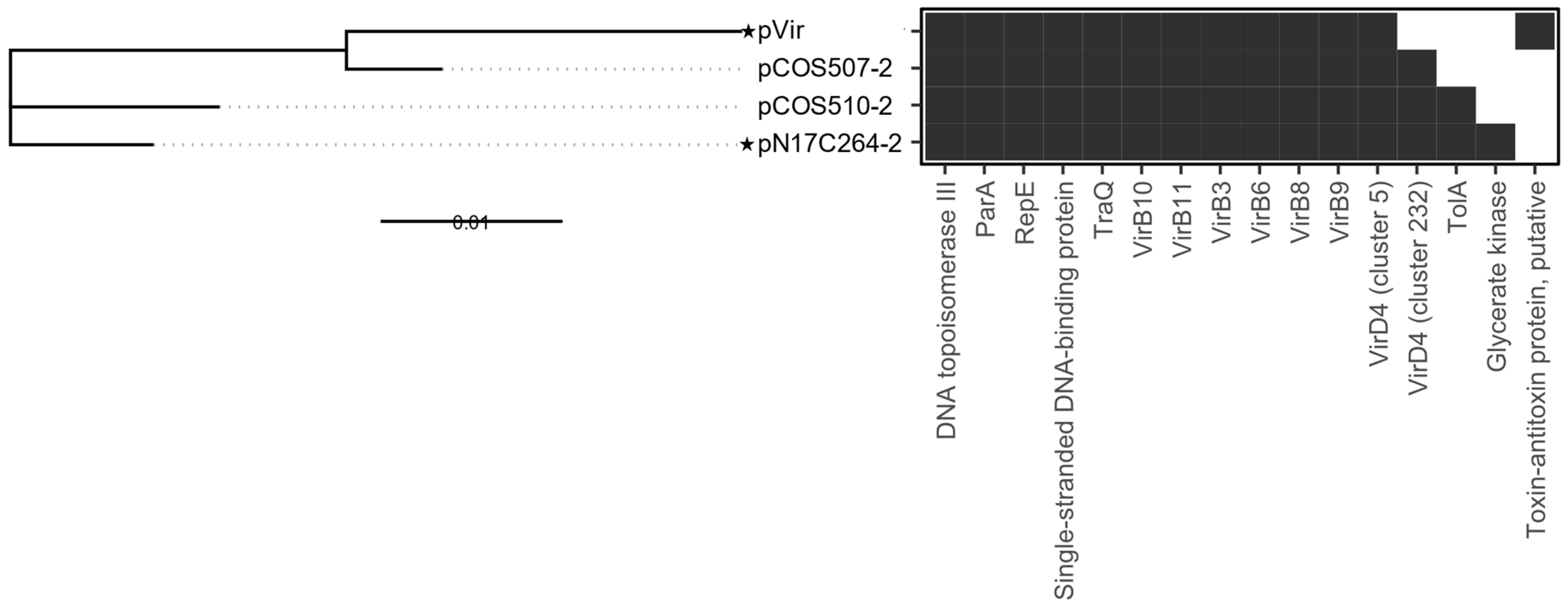
3.2.2. pVir
3.2.3. Small Plasmids (<6 kb)
3.2.4. Mega Plasmids (>80 kb)
3.2.5. Mid Plasmids (~30 kb)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Skarp, C.P.A.; Hänninen, M.-L.; Rautelin, H.I.K. Campylobacteriosis: The Role of Poultry Meat. Clin. Microbiol. Infect. 2016, 22, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Leite, D.; Fernandes, M.; Mena, C.; Gibbs, P.A.; Teixeira, P. Campylobacter spp. as a Foodborne Pathogen: A Review. Front. Microbiol. 2011, 2, 200. [Google Scholar] [CrossRef]
- Tang, Y.; Fang, L.; Xu, C.; Zhang, Q. Antibiotic Resistance Trends and Mechanisms in the Foodborne Pathogen, Campylobacter. Anim. Health. Res. Rev. 2017, 18, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Gabbert, A.D.; Mydosh, J.L.; Talukdar, P.K.; Gloss, L.M.; McDermott, J.E.; Cooper, K.K.; Clair, G.C.; Konkel, M.E. The Missing Pieces: The Role of Secretion Systems in Campylobacter jejuni Virulence. Biomolecules 2023, 13, 135. [Google Scholar] [CrossRef] [PubMed]
- Wysok, B.; Wojtacka, J.; Hänninen, M.-L.; Kivistö, R. Antimicrobial Resistance and Virulence-Associated Markers in Campylobacter Strains from Diarrheic and Non-Diarrheic Humans in Poland. Front. Microbiol. 2020, 11, 1799. [Google Scholar] [CrossRef]
- Bleumink-Pluym, N.M.C.; Van Alphen, L.B.; Bouwman, L.I.; Wösten, M.M.S.M.; Van Putten, J.P.M. Identification of a Functional Type VI Secretion System in Campylobacter jejuni Conferring Capsule Polysaccharide Sensitive Cytotoxicity. PLoS Pathog. 2013, 9, e1003393. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Feye, K.M.; Shi, Z.; Pavlidis, H.O.; Kogut, M.J.; Ashworth, A.; Ricke, S.C. A Historical Review on Antibiotic Resistance of Foodborne Campylobacter. Front. Microbiol. 2019, 10, 1509. [Google Scholar] [CrossRef] [PubMed]
- Luangtongkum, T.; Jeon, B.; Han, J.; Plummer, P.; Logue, C.M.; Zhang, Q. Antibiotic Resistance in Campylobacter: Emergence, Transmission and Persistence. Future Microbiol. 2009, 4, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Marasini, D.; Karki, A.B.; Buchheim, M.A.; Fakhr, M.K. Phylogenetic Relatedness Among Plasmids Harbored by Campylobacter jejuni and Campylobacter coli Isolated from Retail Meats. Front. Microbiol. 2018, 9, 2167. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Capobianco, J.; Armstrong, C.M.; Chen, C.-Y.; Counihan, K.; Lee, J.; Reed, S.; Tilman, S. Detection and Isolation of Campylobacter spp. from Raw Meat. JoVE 2024, 204, 66462. [Google Scholar] [CrossRef]
- He, Y.; Yao, X.; Gunther, N.W.; Xie, Y.; Tu, S.-I.; Shi, X. Simultaneous Detection and Differentiation of Campylobacter jejuni, C. coli, and C. lari in Chickens Using a Multiplex Real-Time PCR Assay. Food Anal. Methods 2010, 3, 321–329. [Google Scholar] [CrossRef]
- He, Y.; Reed, S.; Yan, X.; Zhang, D.; Strobaugh, T.; Capobianco, J.; Gehring, A. Complete Genome Sequences of Multidrug-Resistant Campylobacter coli Strains YH501, YH503, and YH504, from Retail Chicken. Microbiol. Resour. Announc. 2022, 11, e00237-22. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k -Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Hunt, M.; Silva, N.D.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated Circularization of Genome Assemblies Using Long Sequencing Reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucl. Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- MegAlign Pro, DNASTAR. Available online: https://www.dnastar.com/software/lasergene/megalign-pro/ (accessed on 1 December 2024).
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A Modular and Extensible Implementation of the RAST Algorithm for Building Custom Annotation Pipelines and Annotating Batches of Genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024.
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Simon, G.; Noam, R.; Robert, R.; Pedro, A.C.; Marco, S.; Scherer, C. Viridis(Lite)—Colorblind-Friendly Color Maps for R; rOpenSci: Berkeley, CA, USA, 2024. [Google Scholar]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; Van Nimwegen, E. Automated Reconstruction of Whole-Genome Phylogenies from Short-Sequence Reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.M.; Harrison, A.O.; McAllister, S.M.; Polson, S.W.; Wommack, K.E. Iroki: Automatic Customization and Visualization of Phylogenetic Trees. PeerJ. 2020, 26, e8584. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H.E. MOB-Suite: Software Tools for Clustering, Reconstruction and Typing of Plasmids from Draft Assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- Robertson, J.; Bessonov, K.; Schonfeld, J.; Nash, J.H.E. Universal Whole-Sequence-Based Plasmid Typing and Its Utility to Prediction of Host Range and Epidemiological Surveillance. Microb. Genom. 2020, 6, e000435. [Google Scholar] [CrossRef] [PubMed]
- Ghatak, S.; Armstrong, C.M.; Reed, S.; He, Y. Comparative Methylome Analysis of Campylobacter jejuni Strain YH002 Reveals a Putative Novel Motif and Diverse Epigenetic Regulations of Virulence Genes. Front. Microbiol. 2020, 11, 610395. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Kanrar, S.; Reed, S.; Lee, J.; Capobianco, J. Whole Genome Sequences, De Novo Assembly, and Annotation of Antibiotic Resistant Campylobacter jejuni Strains S27, S33, and S36 Newly Isolated from Chicken Meat. Microorganisms 2024, 12, 159. [Google Scholar] [CrossRef] [PubMed]
- Ghatak, S.; He, Y.; Reed, S.; Strobaugh, T.; Irwin, P. Whole Genome Sequencing and Analysis of Campylobacter coli YH502 from Retail Chicken Reveals a Plasmid-Borne Type VI Secretion System. Genom. Data 2017, 11, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Gunther, N.W.; Reichenberger, E.R. Complete Genome Sequence of Campylobacter jejuni RM1246-ERRC, Which Exhibits Resistance to Quaternary Ammonium Compounds. Genome Announc. 2017, 5, e00978-17. [Google Scholar] [CrossRef]
- Gunther, N.W.; Reichenberger, E.R.; Bono, J.L. Complete Genome Sequence of UV-Resistant Campylobacter jejuni RM3194, Including an 81.08-Kilobase Plasmid. Genome Announc. 2016, 4, e00305-16. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, D.W.; Maki, J.J.; Looft, T.; Ricker, N.; Sylte, M.J. Complete Genome Sequence of Campylobacter jejuni Strain NADC 20827, Isolated from Commercial Turkeys. Microbiol. Resour. Announc. 2020, 9, e01403-19. [Google Scholar] [CrossRef] [PubMed]
- Heikema, A.P.; Strepis, N.; Horst-Kreft, D.; Huynh, S.; Zomer, A.; Kelly, D.J.; Cooper, K.K.; Parker, C.T. Biomolecule Sulphation and Novel Methylations Related to Guillain-Barré Syndrome-Associated Campylobacter jejuni Serotype HS:19. Microb. Genom. 2021, 7, 000660. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Ott, R.; Pohl, S.; Burghard, S.; Weig, M.; Groß, U. Identification and Characterization of a Major Subgroup of Conjugative Campylobacter jejuni Plasmids. J. Infect. 2005, 50, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.K. Use of Antibiotics as Feed Additives: A Burning Question. Front. Microbiol. 2014, 5, 334. [Google Scholar] [CrossRef]
- Baquero, F. Low-Level Antibacterial Resistance: A Gateway to Clinical Resistance. Drug Resist. Updates 2001, 4, 93–105. [Google Scholar] [CrossRef]
- Bacon, D.J.; Alm, R.A.; Burr, D.H.; Hu, L.; Kopecko, D.J.; Ewing, C.P.; Trust, T.J.; Guerry, P. Involvement of a Plasmid in Virulence of Campylobacter jejuni 81-176. Infect. Immun. 2000, 68, 4384–4390. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P. Plasmid Incompatibility. Microbiol. Rev. 1987, 51, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Kamruzzaman, M.; Wu, A.Y.; Iredell, J.R. Biological Functions of Type II Toxin-Antitoxin Systems in Bacteria. Microorganisms 2021, 9, 1276. [Google Scholar] [CrossRef] [PubMed]


| Strain | Plasmids | Tetracycline MIC (µg/mL) | tetO Location |
|---|---|---|---|
| C. jejuni YH014 | pCJS014-2 | >64 | pTet |
| C. jejuni YH019 | pCJP019-1, pCJP019-2, pCJP019-3 | >64 | pTet and chromosome |
| C. jejuni YH020 | pCJP020 | >64 | chromosome |
| C. jejuni YH025 | pCJP025 | >64 | pTet |
| C. coli YH507 | pCOS507-1, pCOS507-2, pCOS507-3 | >64 | pTet |
| C. jejuni YH001 | pCJP001-1, pCJP001-2 | 64 | pTet |
| C. jejuni YH002 | pCJP002 | 64 | pTet and chromosome |
| C. jejuni YH018 | pCJP018-1 | 64 | pTet |
| C. jejuni YH022 | pCJS022 | 64 | pTet |
| C. jejuni YH024 | pCJP024-1, pCJP024-2 | 64 | pTet |
| C. jejuni YH027 | pCJP027 | 64 | pTet |
| C. jejuni YH029 | pCJP029 | 64 | chromosome |
| C. jejuni YH026 | pCJP026 | 32 | pTet |
| C. jejuni YH010 | pCJS010 | 4 | pTet |
| C. jejuni YH021 | pCJS021 | 4 | pTet |
| C. coli YH503 | pCOS503-1, pCOS503-2 | 0.5 | none |
| C. coli YH504 | pCOS504-1, pCOS504-1 | 0.25 | none |
| C. coli YH510 | pCOS510-1, pCOS510-2 | 0.25 | none |
| C. coli YH511 | pCOS511 | 0.25 | none |
| C. jejuni YH011 | pCJS011 | 0.12 | none |
| C. coli YH506 | pCOS506 | 0.12 | none |
| C. coli YH502 | pCOS502 | 0.06 | none |
| Plasmid Name | Replicon Type | Relaxase Type | Predicted Mobility | Cluster |
|---|---|---|---|---|
| pCOS502 | - | MOBQ | mobilizable | mega |
| pCOS510-1 | - | MOBQ | mobilizable | mega |
| pN17C336-1 * | - | MOBQ | mobilizable | mega |
| pCJS011 (pCjS36) | - | - | non-mobilizable | mega |
| pCOS503-1 | - | - | non-mobilizable | mega |
| pCOS504 | - | - | non-mobilizable | mega |
| pRM3194 | - | - | non-mobilizable | mega |
| pCOS507-1 | rep_cluster_475 | MOBP | conjugative | mega/pTet |
| pAR-0413-2 | - | - | - | mid |
| pCCDM140S | - | - | - | mid |
| pNUSAC002710-2 | - | - | - | mid |
| pRM1477 | - | - | - | mid |
| pCJP016 | - | MOBP | conjugative | mid |
| pCJP019-2 | - | MOBP | conjugative | mid |
| pCOS507-3 | - | MOBP | conjugative | mid |
| pCOS511 | - | MOBP | conjugative | mid |
| p2014D0261-1 | - | - | - | pTet |
| pCJP001-1 | rep_cluster_475 | MOBP | conjugative | pTet |
| pCJP002 | rep_cluster_475 | MOBP | conjugative | pTet |
| pCJP018 | rep_cluster_475 | MOBP | conjugative | pTet |
| pCJP019-1 | rep_cluster_475 | MOBP | conjugative | pTet |
| pCJP020 | - | MOBP | conjugative | pTet |
| pCJP024 | rep_cluster_475 | MOBP | conjugative | pTet |
| pCJP025 | - | MOBP | conjugative | pTet |
| pCJP026 | rep_cluster_475 | MOBP | conjugative | pTet |
| pCJP027 | rep_cluster_475 | MOBP | conjugative | pTet |
| pCJS010 (pCjS33) | - | MOBP | conjugative | pTet |
| pCJS014-2 | - | MOBP | conjugative | pTet |
| pCJS021 | - | MOBP | conjugative | pTet |
| pCJS022 | - | MOBP | conjugative | pTet |
| pRM1246_ERRC | rep_cluster_475 | MOBP | conjugative | pTet |
| pTet * | rep_cluster_475 | MOBP | conjugative | pTet |
| pCJP029 | rep_cluster_475 | MOBP | mobilizable | pTet |
| pCOS507-2 | rep_cluster_1502 | MOBP | conjugative | pVir |
| pCOS510-2 | rep_cluster_1502 | MOBP | conjugative | pVir |
| pN17C264-2 * | rep_cluster_1502 | MOBP | conjugative | pVir |
| pVir * | rep_cluster_1502 | MOBP | conjugative | pVir |
| p20827S * | rep_cluster_795 | MOBP | mobilizable | small |
| pCJP001-2 | rep_cluster_795 | MOBP | mobilizable | small |
| pCJP018-2 | rep_cluster_795 | MOBP | mobilizable | small |
| pCJP019-3 | rep_cluster_795 | MOBP | mobilizable | small |
| pCJP024-2 | rep_cluster_795 | MOBP | mobilizable | small |
| pCC20JX12-5K * | rep_cluster_896 | - | non-mobilizable | small |
| pCOS503-2 | rep_cluster_896 | - | non-mobilizable | small |
| pCOS504-2 | rep_cluster_896 | - | non-mobilizable | small |
| pCOS506 | - | - | non-mobilizable | small |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Dykes, G.E.; Kanrar, S.; Liu, Y.; Gunther, N.W., IV; Counihan, K.L.; Lee, J.; Capobianco, J.A. Comparative Genomic Analysis of Campylobacter Plasmids Identified in Food Isolates. Microorganisms 2025, 13, 206. https://doi.org/10.3390/microorganisms13010206
He Y, Dykes GE, Kanrar S, Liu Y, Gunther NW IV, Counihan KL, Lee J, Capobianco JA. Comparative Genomic Analysis of Campylobacter Plasmids Identified in Food Isolates. Microorganisms. 2025; 13(1):206. https://doi.org/10.3390/microorganisms13010206
Chicago/Turabian StyleHe, Yiping, Gretchen Elizabeth Dykes, Siddhartha Kanrar, Yanhong Liu, Nereus W. Gunther, IV, Katrina L. Counihan, Joe Lee, and Joseph A. Capobianco. 2025. "Comparative Genomic Analysis of Campylobacter Plasmids Identified in Food Isolates" Microorganisms 13, no. 1: 206. https://doi.org/10.3390/microorganisms13010206
APA StyleHe, Y., Dykes, G. E., Kanrar, S., Liu, Y., Gunther, N. W., IV, Counihan, K. L., Lee, J., & Capobianco, J. A. (2025). Comparative Genomic Analysis of Campylobacter Plasmids Identified in Food Isolates. Microorganisms, 13(1), 206. https://doi.org/10.3390/microorganisms13010206








