Screening and Comparative Genomics of Probiotic Lactic Acid Bacteria from Bee Bread of Apis Cerana: Influence of Stevia and Stevioside on Bacterial Cell Growth and the Potential of Fermented Stevia as an Antidiabetic, Antioxidant, and Antifungal Agent
Abstract
:1. Introduction
2. Materials and Methods
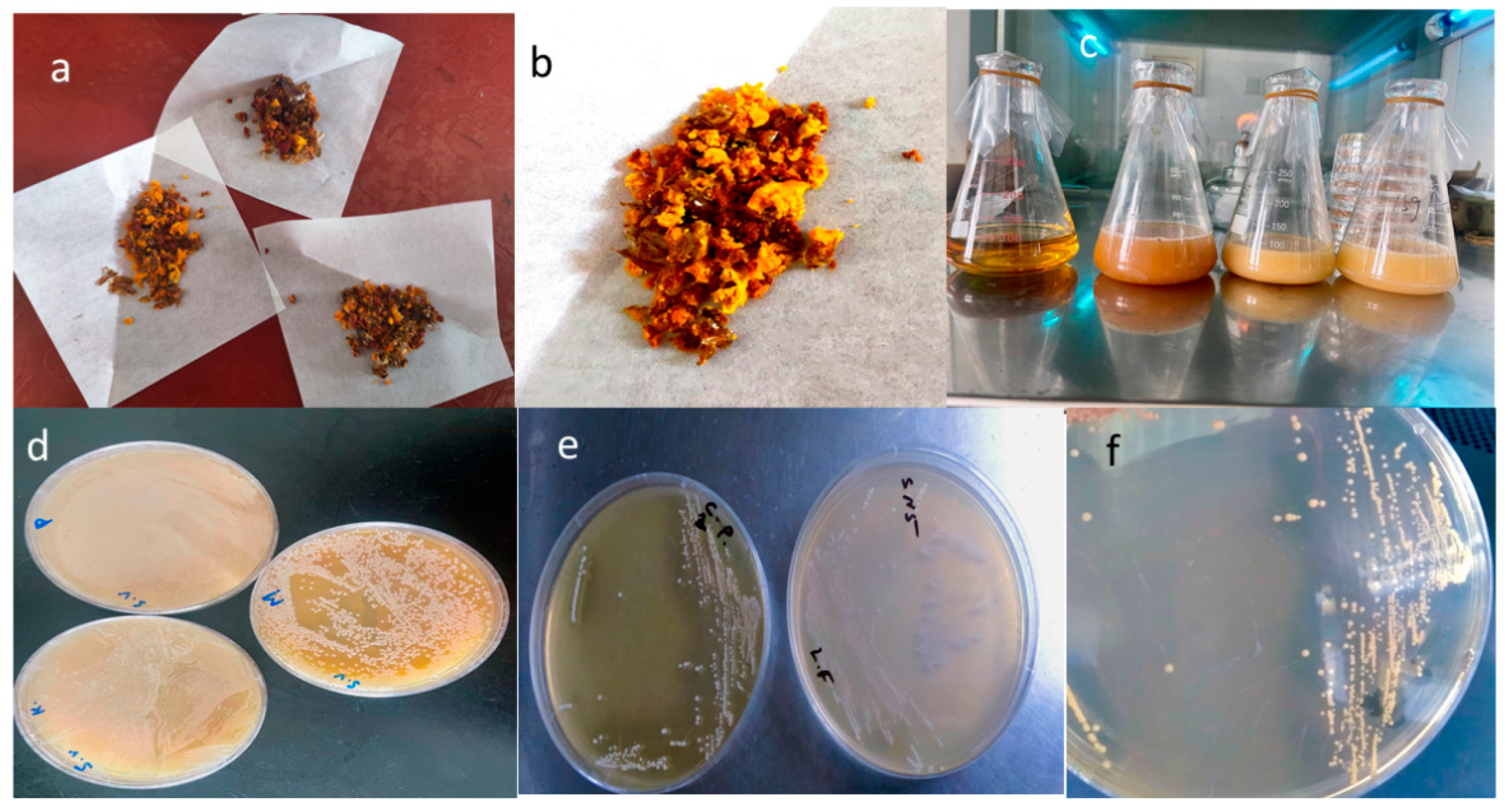
2.1. Sampling of Bee Bread
2.2. Isolation of Lactic Acid Bacteria from Bee Bread
2.3. Identification of Strains
2.4. Morphologic and Biochemical Characterization
2.5. Molecular Identification Using 16S rRNA Gene Sequencing
2.6. Whole Genome Sequencing, Assembly, and Quality Control
2.6.1. Gene Prediction Functional Annotation of the Genome
2.6.2. Genotype Analysis of Carbohydrate Metabolism
2.6.3. Secondary Metabolite Gene Cluster Analysis
2.6.4. Repetitive Sequence and Pathogen-Host Interaction Analysis
2.6.5. Genotype Analysis of Antibiotic Resistance
2.6.6. Cytochrome P450 and VFDB
2.6.7. TCDB Transporter Proteins
2.6.8. Identification of CRISPR-Cas Systems and Prophages
2.7. Evaluation of In Vitro Probiotic Ability of Isolated Strains
2.7.1. Acid and Bile Salts Tolerance
2.7.2. Simulated Human GI Tract Conditions
2.7.3. Analysis of Antagonistic Properties and Cell Surface Hydrophobicity
2.7.4. Adhesion to Intestinal Cells
2.7.5. Flow Cytometry and Plate Count
2.7.6. Antibacterial Activity
2.7.7. Hemolytic Activity of Isolated Strains
2.7.8. Nitrite Degradation Capacity of Strains
2.8. Fermentation Capacity of Strains
2.9. Fermentation of Stevia and Stevioside by Isolated Strains of LAB
2.9.1. Media and Growth Conditions
2.9.2. Fermentation of Isolates with Stevia and Steviosides
2.9.3. Determination of Free Amino Acids During Fermentation
2.9.4. Analytical Determinations
2.10. Antioxidant Activity of Isolates
2.10.1. Assay for DPPH Radical Scavenging
2.10.2. ABTS Radical Scavenging Assay
2.11. Antidiabetic Activity by Inhibiting Carbohydrate Hydrolyzing Enzymes
2.11.1. The Culture Supernatant (CS) and Intact Cells (ICs) Preparation
2.11.2. α-Glucosidase Inhibitory
2.11.3. α-Amylase Inhibitory
2.12. Resistance Against Mycotoxigenic Fungi
2.13. Statistical Analysis
3. Results
3.1. Identification of Isolated Strains of LAB spp.
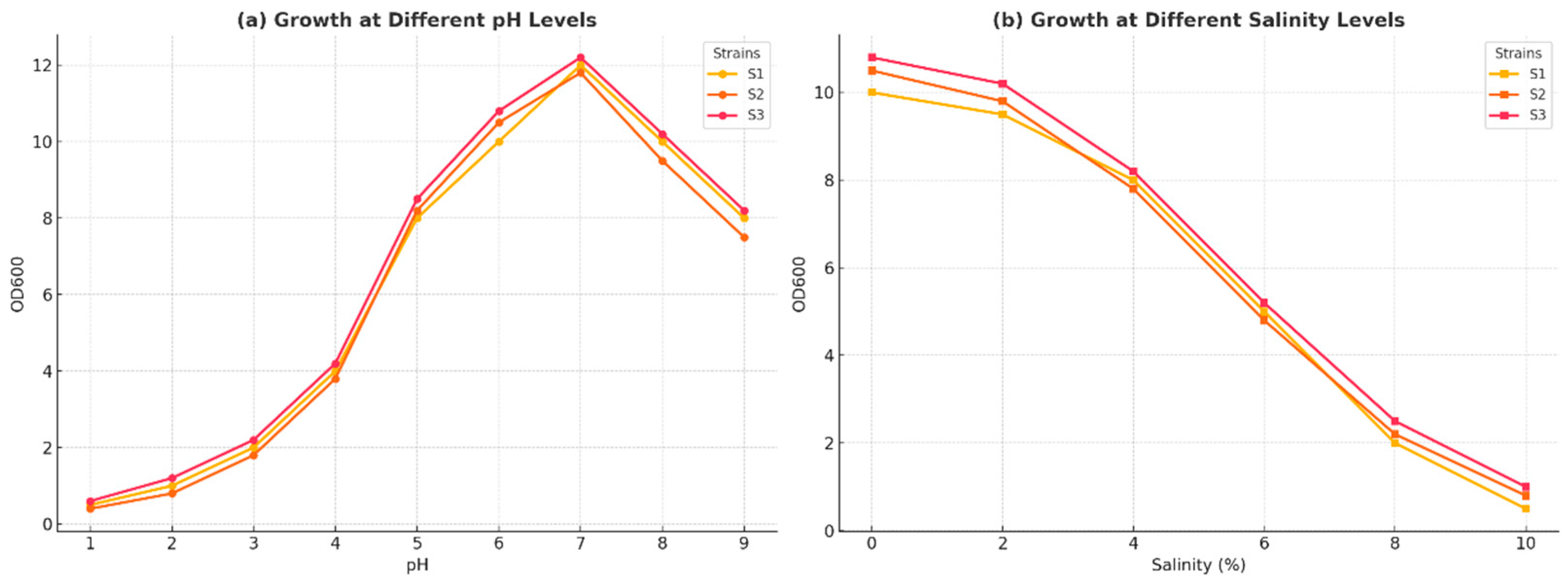
3.2. Physiological and Biochemical Analysis of Strains
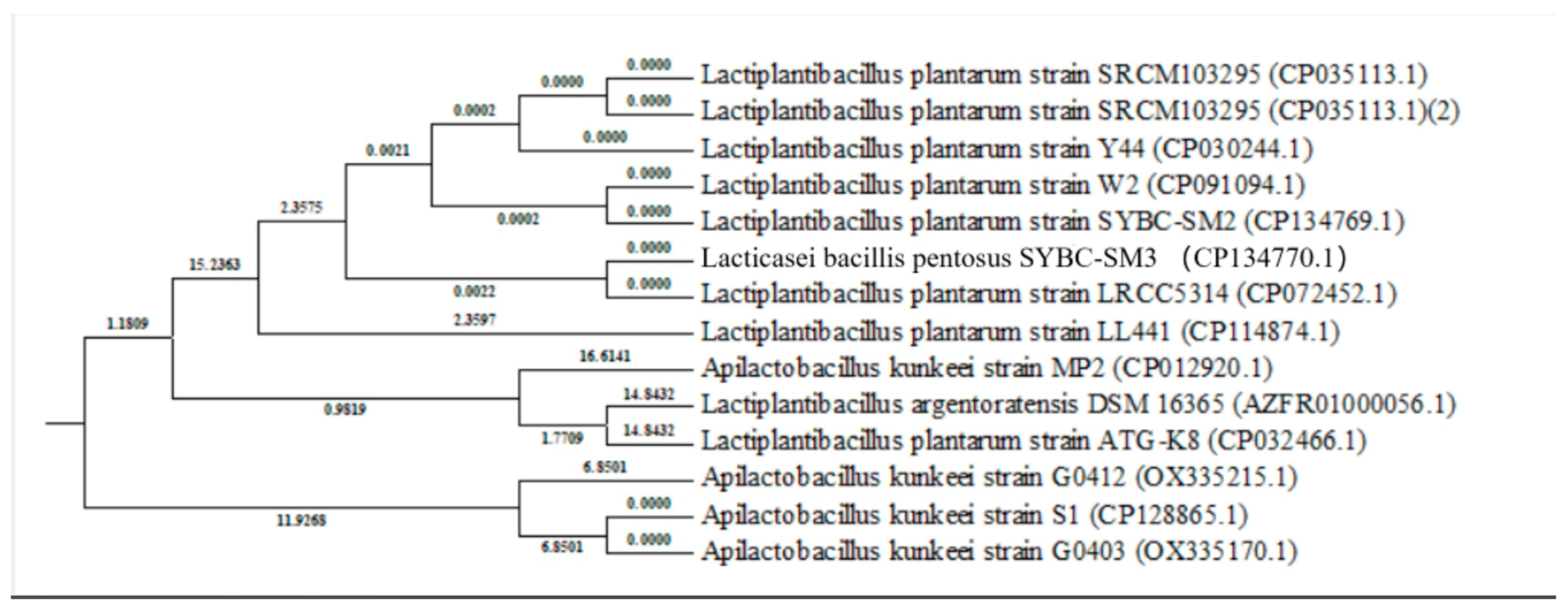
Phylogenetic Analysis
3.3. Comparative Genome Analysis
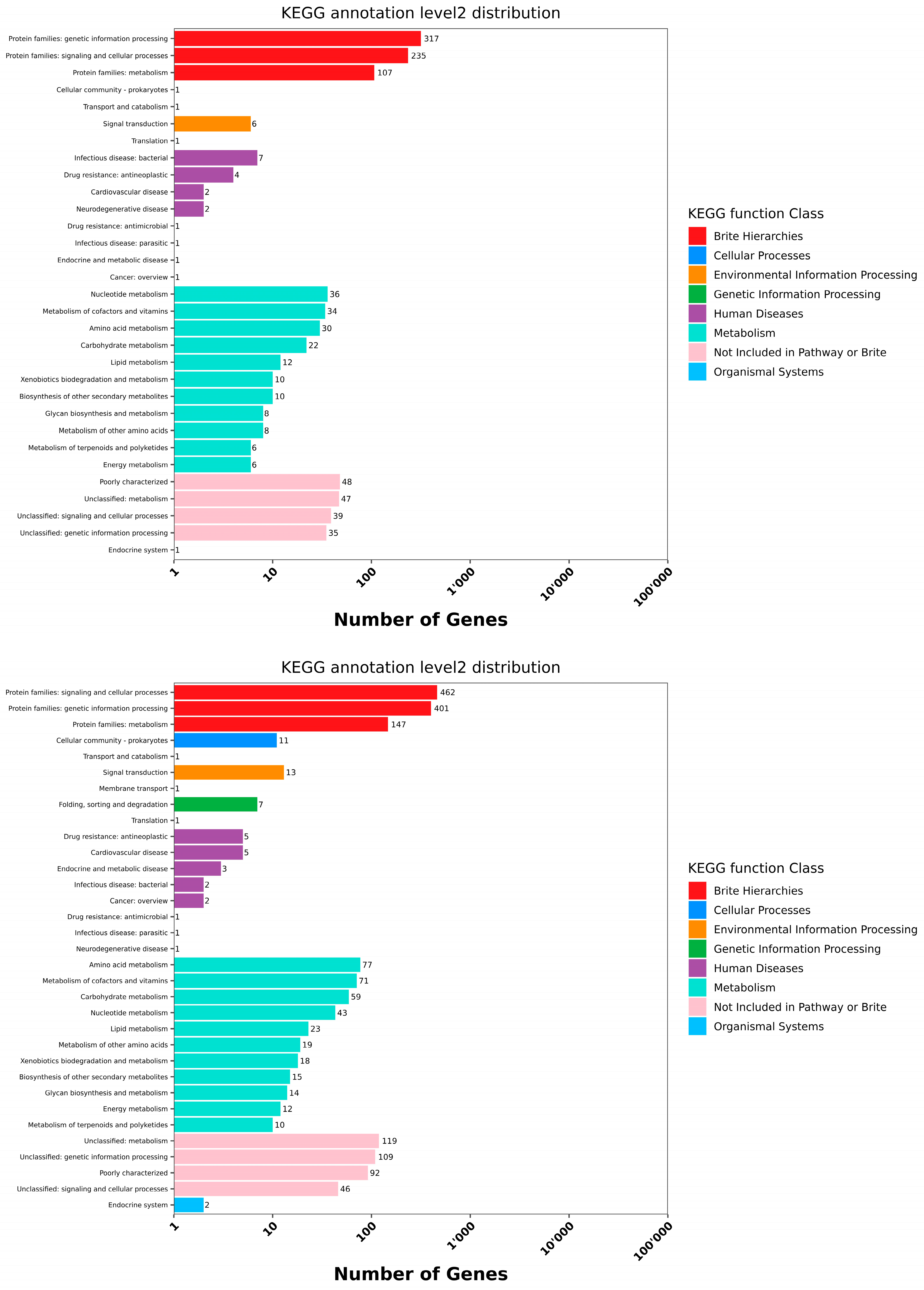
Genome Prediction and Functional Annotation
- Cellular processes: S1 (235), S2 (467), and S3 (520).
- Signaling and genetic processing: S1 (317), S2 (401), and S3 (411).
- Protein family metabolism: S1 (107), S2 (147), and S3 (155).
- Amino acid metabolism: S1 (30), S2 (77), and S3 (76).
- Carbohydrate metabolism: S1 (22), S2 (59), and S3 (94).
3.4. Evaluation of Probiotic Ability of Isolates
3.4.1. Tolerance to Simulated Gastric Juice and Bile Salts
3.4.2. Hydrophobicity and Autoaggregation
3.4.3. Adhesion to Intestinal Cells and Viable Cell Count
3.4.4. Nitrite Degradation
3.4.5. Antibacterial and Sensitivity Testing
3.4.6. Hemolytic Activity
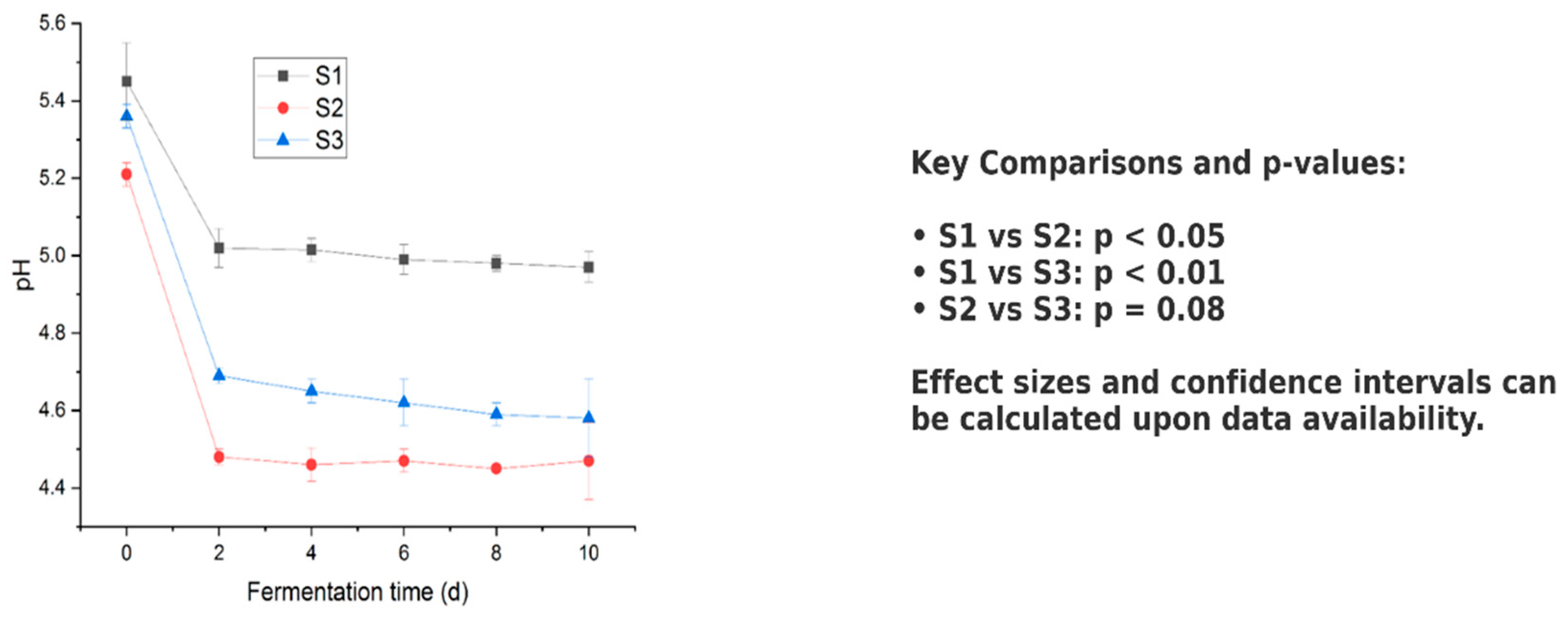
3.5. Fermentation Profile
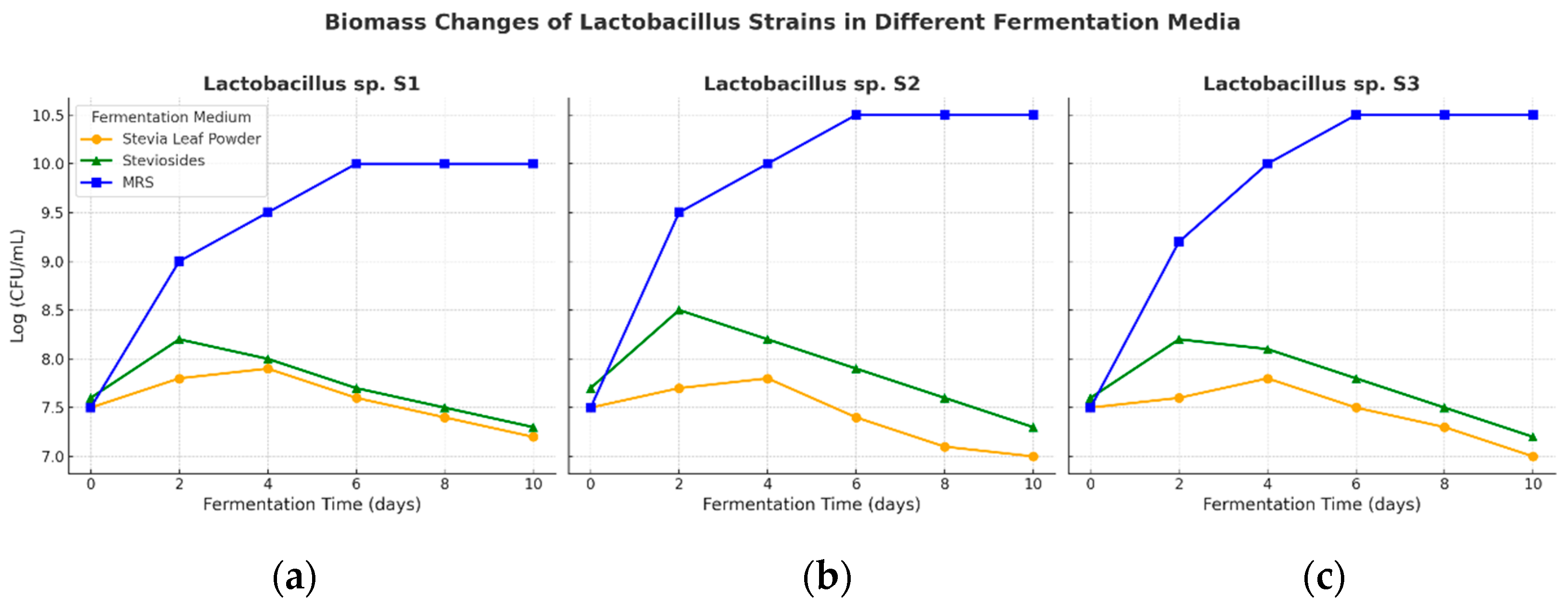
3.6. Fermentation of Stevia Leaf and Stevioside
3.7. Concentration of Essential Amino Acids
3.8. Antidiabetic Effect of Fermented Stevia and Steviosides
3.9. Antioxidant Activity
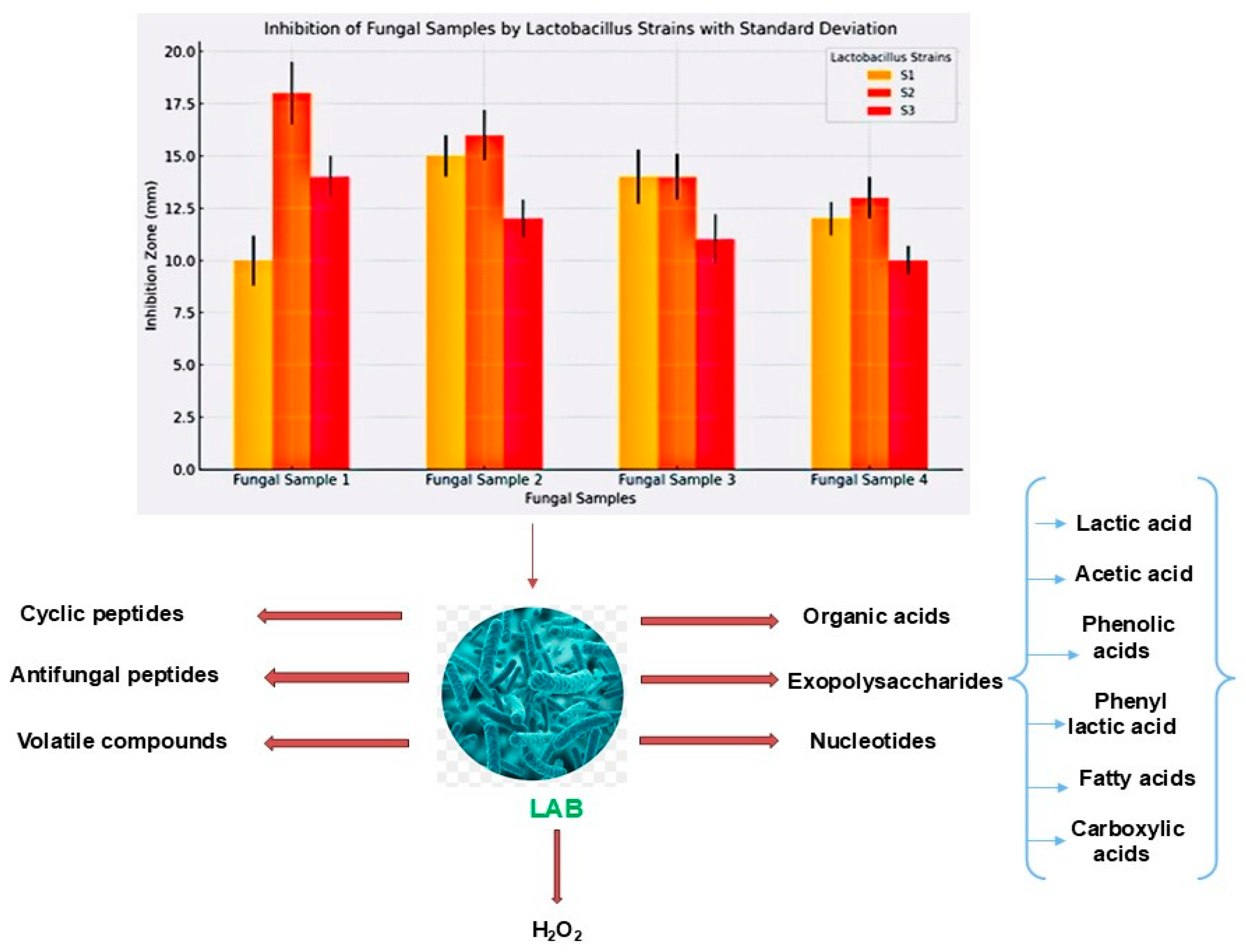
3.10. Inhibition of Fungal Growth
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nowak, A.; Szczuka, D.; Górczyńska, A.; Motyl, I.; Kręgiel, D. Characterization of Apis Mellifera Gastrointestinal Microbiota and Lactic Acid Bacteria for Honeybee Protection—A Review. Cells 2021, 10, 701. [Google Scholar] [CrossRef] [PubMed]
- Asama, T.; Arima, T.; Gomi, T.; Keishi, T.; Tani, H.; Kimura, Y.; Tatefuji, T.; Hashimoto, K. Lactobacillus kunkeei YB38 from Honeybee Products Enhances IgA Production in Healthy Adults. J. Appl. Microbiol. 2015, 119, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Torres-Moreno, R.; Humberto, S.H.-S.; Méndez-Tenorio, A.; Palmeros-Sánchez, B.; Melgar-Lalanne, G. Characterization and Identification of Lactic Acid Bacteria from Mexican Stingless Bees (Apidae: Meliponini). IOP Conf. Ser. Earth Environ. Sci. 2021, 858, 012010. [Google Scholar] [CrossRef]
- Engel, P.; Kwong, W.K.; McFrederick, Q.; Anderson, K.E.; Barribeau, S.M.; Chandler, J.A.; Cornman, R.S.; Dainat, J.; De Miranda, J.R.; Doublet, V. The Bee Microbiome: Impact on Bee Health and Model for Evolution and Ecology of Host-Microbe Interactions. MBio 2016, 7, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Perreau, J.; Powell, J.E.; Han, B.; Zhang, Z.; Kwong, W.K.; Tringe, S.G.; Moran, N.A. Division of Labor in Honey Bee Gut Microbiota for Plant Polysaccharide Digestion. Proc. Natl. Acad. Sci. USA 2019, 116, 25909–25916. [Google Scholar] [CrossRef]
- Suphaphimol, N.; Attasopa, K.; Pakwan, C.; Chantawannakul, P.; Disayathanoowat, T. Cultured-Dependent and Cultured-Independent Study of Bacteria Associated with Thai Commercial Stingless Bee Lepidotrigona Terminata. J. Apic. Res. 2020, 60, 341–348. [Google Scholar] [CrossRef]
- Morais, M.; Moreira, L.; Feás, X.; Estevinho, L.M. Honeybee-Collected Pollen from Five Portuguese Natural Parks: Palynological Origin, Phenolic Content, Antioxidant Properties and Antimicrobial Activity. Food Chem. Toxicol. 2011, 49, 1096–1101. [Google Scholar] [CrossRef]
- Fratini, F.; Cilia, G.; Mancini, S.; Felicioli, A. Royal Jelly: An Ancient Remedy with Remarkable Antibacterial Properties. Microbiol. Res. 2016, 192, 130–141. [Google Scholar] [CrossRef]
- Liu, X.; Qian, M.; Shen, Y.; Qin, X.; Huang, H.; Yang, H.; He, Y.; Bai, W. A High-Throughput Sequencing Approach to the Preliminary Analysis of Bacterial Communities Associated with Changes in Amino Acid Nitrogen, Organic Acid and Reducing Sugar Contents during Soy Sauce Fermentation. Food Chem. 2021, 349, 129131. [Google Scholar] [CrossRef]
- Xia, W.; Liu, P.; Liu, J. Advance in Chitosan Hydrolysis by Non-Specific Cellulases. Bioresour. Technol. 2008, 99, 6751–6762. [Google Scholar] [CrossRef]
- Tuksitha, L.; Chen, Y.-L.S.; Chen, Y.-L.; Wong, K.-Y.; Peng, C.-C. Antioxidant and Antibacterial Capacity of Stingless Bee Honey from Borneo (Sarawak). J. Asia. Pac. Entomol. 2018, 21, 563–570. [Google Scholar] [CrossRef]
- DeGrandi-Hoffman, G.; Chen, Y.; Rivera, R.; Carroll, M.; Chambers, M.; Hidalgo, G.; de Jong, E.W. Honey Bee Colonies Provided with Natural Forage Have Lower Pathogen Loads and Higher Overwinter Survival than Those Fed Protein Supplements. Apidologie 2016, 47, 186–196. [Google Scholar] [CrossRef]
- De Vere, N.; Jones, L.E.; Gilmore, T.; Moscrop, J.; Lowe, A.; Smith, D.; Hegarty, M.J.; Creer, S.; Ford, C.R. Using DNA Metabarcoding to Investigate Honey Bee Foraging Reveals Limited Flower Use despite High Floral Availability. Sci. Rep. 2017, 7, 42838. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Engel, P.; Koch, H.; Moran, N.A. Genomics and Host Specialization of Honey Bee and Bumble Bee Gut Symbionts. Proc. Natl. Acad. Sci. USA 2014, 111, 11509–11514. [Google Scholar] [CrossRef]
- Aureli, P.; Capurso, L.; Castellazzi, A.M.; Clerici, M.; Giovannini, M.; Morelli, L.; Poli, A.; Pregliasco, F.; Salvini, F.; Zuccotti, G.V. Probiotics and Health: An Evidence-Based Review. Pharmacol. Res. 2011, 63, 366–376. [Google Scholar] [CrossRef]
- Aleklett, K.; Hart, M.; Shade, A. The Microbial Ecology of Flowers: An Emerging Frontier in Phyllosphere Research. Botany 2014, 92, 253–266. [Google Scholar] [CrossRef]
- Olofsson, T.C.; Alsterfjord, M.; Nilson, B.; Butler, È.; Vásquez, A. Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isolated from the honey stomach of the honeybee Apis mellifera. Int. J. Syst. Evol. Microbiol. 2014, 64, 3109–3119. [Google Scholar]
- Torriani, S.; Felis, G.E.; Dellaglio, F. Differentiation of Lactobacillus plantarum, L. Pentosus, and L. Paraplantarum by recA Gene Sequence Analysis and Multiplex PCR Assay with recA Gene-Derived Primers. Appl. Environ. Microbiol. 2001, 67, 3450–3454. [Google Scholar] [CrossRef]
- Torriani, S.; Clementi, F.; Vancanneyt, M.; Hoste, B.; Dellaglio, F.; Kersters, K. Differentiation of Lactobacillus plantarum, L. Pentosus and L. paraplantarum Species by RAPD-PCR and AFLP. Syst. Appl. Microbiol. 2001, 24, 554–560. [Google Scholar]
- Shao, X.; Xu, B.; Zhou, H.; Chen, C.; Li, P. Insight into the Mechanism of Decreasing N-Nitrosodimethylamine by Lactobacillus pentosus R3 in a Model System. Food Control 2021, 121, 107534. [Google Scholar] [CrossRef]
- Davoodi, S.; Behbahani, M.; Shirani, E.; Mohabatkar, H. Influence of Sucrose, Glucose, Stevia Leaf and Stevioside on the Growth and Lactic Acid Production by Lactobacillus plantarum, Lactobacillus brevis and Lactobacillus casei. Iran. J. Sci. Technol. Trans. A Sci. 2016, 40, 275–279. [Google Scholar] [CrossRef]
- Ozcan, T.; Eroglu, E. Effect of Stevia and Inulin Interactions on Fermentation Profile and Short-chain Fatty Acid Production of Lactobacillus acidophilus in Milk and in Vitro Systems. Int. J. Dairy Technol. 2022, 75, 171–181. [Google Scholar] [CrossRef]
- Kunová, G.; Rada, V.; Vidaillac, A.; Lisova, I. Utilisation of Steviol Glycosides from Stevia rebaudiana (Bertoni) by Lactobacilli and Bifidobacteria in in Vitro Conditions. Folia Microbiol. 2014, 59, 251–255. [Google Scholar] [CrossRef]
- Sakr, E.A.E.; Massoud, M.I. Impact of Prebiotic Potential of Stevia Sweeteners-Sugar Used as Synbiotic Preparation on Antimicrobial, Antibiofilm, and Antioxidant Activities. LWT 2021, 144, 111260. [Google Scholar] [CrossRef]
- Poyraz, F.; Yalmanci, D.; İspirli, H.; Dertli, E. Characterization of Bee Bread Produced with Defined Starter Cultures Mimicking the Natural Fermentation Process. Fermentation 2023, 9, 174. [Google Scholar] [CrossRef]
- Guan, Y.; Lv, H.; Wu, G.; Chen, J.; Wang, M.; Zhang, M.; Pang, H.; Duan, Y.; Wang, L.; Tan, Z. Effects of Lactic Acid Bacteria Reducing the Content of Harmful Fungi and Mycotoxins on the Quality of Mixed Fermented Feed. Toxins 2023, 15, 226. [Google Scholar] [CrossRef]
- Tsuda, H.; Kodama, K. Evaluating the Technological Properties of Lactic Acid Bacteria in Wagyu Cattle Milk. J. Dairy Res. 2021, 88, 210–216. [Google Scholar] [CrossRef]
- Li, P.; Banjade, S.; Cheng, H.-C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F. Phase Transitions in the Assembly of Multivalent Signalling Proteins. Nature 2012, 483, 336–340. [Google Scholar] [CrossRef]
- Lim, Y.-S.; Kim, J.; Kang, H. Isolation and Identification of Lactic Acid Bacteria with Probiotic Activities from Kimchi and Their Fermentation Properties in Milk. J. Dairy Sci. Biotechnol. 2019, 37, 115–128. [Google Scholar]
- Altermann, E.; Russell, W.M.; Azcarate-Peril, M.A.; Barrangou, R.; Buck, B.L.; McAuliffe, O.; Souther, N.; Dobson, A.; Duong, T.; Callanan, M. Complete Genome Sequence of the Probiotic Lactic Acid Bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 2005, 102, 3906–3912. [Google Scholar] [CrossRef]
- Voulgari, K.; Hatzikamari, M.; Delepoglou, A.; Georgakopoulos, P.; Litopoulou-Tzanetaki, E.; Tzanetakis, N. Antifungal Activity of Non-Starter Lactic Acid Bacteria Isolates from Dairy Products. Food Control 2010, 21, 136–142. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, L.; Zhang, Y.; Ran, R.; Meng, X.; Liu, S. Research Advances in the Degradation of Aflatoxin by Lactic Acid Bacteria. J. Venom. Anim. Toxins Incl. Trop. Dis. 2023, 29, e20230029. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; Magan, N.; Medina, A. Evaluation of the Risk of Fungal Spoilage When Substituting Sucrose with Commercial Purified Stevia Glycosides in Sweetened Bakery Products. Int. J. Food Microbiol. 2016, 231, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Y.I.; Zhou, T.; Bullerman, L.B. Sourdough Lactic Acid Bacteria as Antifungal and Mycotoxin-Controlling Agents. Food Sci. Technol. Int. 2016, 22, 79–90. [Google Scholar] [CrossRef]
- Farhat, L.B.; Aissaoui, N.; Torrijos, R.; Luz, C.; Meca, G.; Abidi, F. Correlation between Metabolites of Lactic Acid Bacteria Isolated from Dairy Traditional Fermented Tunisian Products and Antifungal and Antioxidant Activities. J. Appl. Microbiol. 2022, 133, 3069–3082. [Google Scholar] [CrossRef] [PubMed]
- Lebeer, S.; Vanderleyden, J.; De Keersmaecker, S.C.J. Genes and Molecules of Lactobacilli Supporting Probiotic Action. Microbiol. Mol. Biol. Rev. 2008, 72, 728–764. [Google Scholar] [CrossRef]
- Barta, D.G.; Cornea-Cipcigan, M.; Margaoan, R.; Vodnar, D.C. Biotechnological Processes Simulating the Natural Fermentation Process of Bee Bread and Therapeutic Properties—An Overview. Front. Nutr. 2022, 9, 871896. [Google Scholar] [CrossRef]
- Toropov, V.; Demyanova, E.; Shalaeva, O.; Sitkin, S.; Vakhitov, T. Whole-Genome Sequencing of Lactobacillus helveticus D75 and D76 Confirms Safety and Probiotic Potential. Microorganisms 2020, 8, 329. [Google Scholar] [CrossRef]
- Qureshi, N.; Gu, Q.; Li, P. Whole Genome Sequence Analysis and in Vitro Probiotic Characteristics of a Lactobacillus Strain Lactobacillus paracasei ZFM54. J. Appl. Microbiol. 2020, 129, 422–433. [Google Scholar] [CrossRef]
- Filannino, P.; Di Cagno, R.; Tlais, A.Z.A.; Cantatore, V.; Gobbetti, M. Fructose-Rich Niches Traced the Evolution of Lactic Acid Bacteria toward Fructophilic Species. Crit. Rev. Microbiol. 2019, 45, 65–81. [Google Scholar] [CrossRef]
- Syed Yaacob, S.N.; Huyop, F.; Kamarulzaman Raja Ibrahim, R.; Wahab, R.A. Identification of Lactobacillus spp. and Fructobacillus spp. Isolated from Fresh Heterotrigona itama Honey and Their Antagonistic Activities Against Clinical Pathogenic Bacteria. J. Apic. Res. 2018, 57, 395–405. [Google Scholar] [CrossRef]
- Poornachandra Rao, K.; Chennappa, G.; Suraj, U.; Nagaraja, H.; Charith Raj, A.P.; Sreenivasa, M.Y. Probiotic Potential of Lactobacillus Strains Isolated from Sorghum-Based Traditional Fermented Food. Probiotics Antimicrob. Proteins 2015, 7, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Ben-Miled, H.; Semmar, N.; Castellanos, M.S.; Ben-Mahrez, K.; Benoit-Biancamano, M.-O.; Réjiba, S. Effect of Honey Bee Forage Plants in Tunisia on Diversity and Antibacterial Potential of Lactic Acid Bacteria and Bifidobacteria from Apis mellifera intermissa and Its Products. Arch. Microbiol. 2023, 205, 295. [Google Scholar] [CrossRef] [PubMed]
- Gilliam, M. Microbiology of Pollen and Bee Bread: The Yeasts. Apidologie 1979, 10, 43–53. [Google Scholar] [CrossRef]
- Raveendran, M.; Lee, A.J.; Sharma, R.; Wälti, C.; Actis, P. Rational Design of DNA Nanostructures for Single Molecule Biosensing. Nat. Commun. 2020, 11, 4384. [Google Scholar] [CrossRef]
- Basharat, S.; Meng, T.; Zhai, L.; Hussain, A.; Aqeel, S.M.; Khan, S.; Shah, O.U.; Liao, X. Bacterial Diversity of Stingless Bee Honey in Yunnan, China: Isolation and Genome Sequencing of a Novel Acid-Resistant Lactobacillus pentosus (SYBC-MI) with Probiotic and L. tryptophan Producing Potential via Millet Fermentation. Front. Bioeng. Biotechnol. 2023, 11, 1272308. [Google Scholar] [CrossRef]
- Chin, C.-S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figureueroa-Balderas, R.; Morales-Cruz, A. Phased Diploid Genome Assembly with Single-Molecule Real-Time Sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef]
- Sergey, K.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k-Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar]
- Bradford, E.L.; Wax, N.; Bueren, E.K.; Walke, J.B.; Fell, R.; Belden, L.K.; Haak, D.C. Comparative Genomics of Lactobacillaceae from the Gut of Honey Bees, Apis mellifera, from the Eastern United States. G3 2022, 12, jkac286. [Google Scholar] [CrossRef]
- Asenjo, F.; Olmos, A.; Henríquez-Piskulich, P.; Polanco, V.; Aldea, P.; Ugalde, J.A.; Trombert, A.N. Genome Sequencing and Analysis of the First Complete Genome of Lactobacillus kunkeei Strain MP2, an Apis mellifera Gut Isolate. PeerJ 2016, 4, e1950. [Google Scholar] [CrossRef]
- Botes, M.; van Reenen, C.A.; Dicks, L.M.T. Evaluation of Enterococcus Mundtii ST4SA and Lactobacillus plantarum 423 as Probiotics by Using a Gastro-Intestinal Model with Infant Milk Formulations as Substrate. Int. J. Food Microbiol. 2008, 128, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Kant, R.; Blom, J.; Palva, A.; Siezen, R.J.; de Vos, W.M. Comparative Genomics of Lactobacillus. Microb. Biotechnol. 2011, 4, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Bujnakova, D.; Kmet, V. Aggregation of Animal LAB with O157 Enterohemorrhagic Escherichia coli. J. Vet. Med. Ser. B 2002, 49, 152–154. [Google Scholar] [CrossRef]
- Katiku, M.M.; Matofari, J.W.; Nduko, J.M. Preliminary Evaluation of Probiotic Properties and Safety Profile of Lactiplantibacillus plantarum Isolated from Spontaneously Fermented Milk, Amabere Amaruranu. Heliyon 2022, 8, e10342. [Google Scholar] [CrossRef]
- Andrade-Velásquez, A.; Hernández Sánchez, H.; Dorantes-Álvarez, L.; Palmeros-Sánchez, B.; Torres-Moreno, R.; Hernández-Rodríguez, D.; Melgar-Lalanne, G. Honey Characterization and Identification of Fructophilic Lactic Acid Bacteria of Fresh Samples from Melipona beecheii, Scaptotrigona pectoralis, Plebeia llorentei, and Plebeia jatiformis Hives. Front. Sustain. Food Syst. 2023, 7, 1113920. [Google Scholar] [CrossRef]
- Baharudin, M.M.A.; Ngalimat, M.S.; Mohd Shariff, F.; Balia Yusof, Z.N.; Karim, M.; Baharum, S.N.; Sabri, S. Antimicrobial Activities of Bacillus Velezensis Strains Isolated from Stingless Bee Products against Methicillin-Resistant Staphylococcus aureus. PLoS ONE 2021, 16, e0251514. [Google Scholar] [CrossRef]
- Kačániová, M.; Terentjeva, M.; Žiarovská, J.; Kowalczewski, P.Ł. In Vitro Antagonistic Effect of Gut Bacteriota Isolated from Indigenous Honey Bees and Essential Oils against Paenibacillus larvae. Int. J. Mol. Sci. 2020, 21, 6736. [Google Scholar] [CrossRef]
- Verma, M.; Singh, S.B. In Vitro Evaluation of Antidiabetic, Antioxidant and Probiotic Attributes of Lactobacillus casei Strain NCDC 357. Indian J. 2022, 24, 288–297. [Google Scholar] [CrossRef]
- Sallan, S.; Yılmaz Oral, Z.F.; Kaya, M. A Review on the Role of Lactic Acid Bacteria in the Formation and Reduction of Volatile Nitrosamines in Fermented Sausages. Foods 2023, 12, 702. [Google Scholar] [CrossRef]
- Mao, Y.; Zhang, X.; Xu, Z. Identification of Antibacterial Substances of Lactobacillus plantarum DY-6 for Bacteriostatic Action. Food Sci. Nutr. 2020, 8, 2854–2863. [Google Scholar] [CrossRef]
- Zhao, W.-W.; Xu, J.-J.; Chen, H.-Y. Photoelectrochemical DNA Biosensors. Chem. Rev. 2014, 114, 7421–7441. [Google Scholar] [CrossRef] [PubMed]
- Deniņa, I.; Semjonovs, P.; Fomina, A.; Treimane, R.; Linde, R. The Influence of Stevia Glycosides on the Growth of Lactobacillus reuteri Strains. Lett. Appl. Microbiol. 2014, 58, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Banwo, K.; Asogwa, F.C.; Ogunremi, O.R.; Adesulu-Dahunsi, A.; Sanni, A. Nutritional Profile and Antioxidant Capacities of Fermented Millet and Sorghum Gruels Using Lactic Acid Bacteria and Yeasts. Food Biotechnol. 2021, 35, 199–220. [Google Scholar] [CrossRef]
- Zhao, M.; Huang, D.; Zhang, X.; Koffas, M.A.G.; Zhou, J.; Deng, Y. Metabolic Engineering of Escherichia coli for Producing Adipic Acid through the Reverse Adipate-Degradation Pathway. Metab. Eng. 2018, 47, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Mu, G.; Gao, Y.; Tuo, Y.; Li, H.; Zhang, Y.; Qian, F.; Jiang, S. Assessing and Comparing Antioxidant Activities of Lactobacilli Strains by Using Different Chemical and Cellular Antioxidant Methods. J. Dairy Sci. 2018, 101, 10792–10806. [Google Scholar] [CrossRef]
- Li, S.; Zhao, Y.; Zhang, L.; Zhang, X.; Huang, L.; Li, D.; Niu, C.; Yang, Z.; Wang, Q. Antioxidant Activity of Lactobacillus plantarum Strains Isolated from Traditional Chinese Fermented Foods. Food Chem. 2012, 135, 1914–1919. [Google Scholar] [CrossRef]
- Pieniz, S.; Andreazza, R.; Anghinoni, T.; Camargo, F.; Brandelli, A. Probiotic Potential, Antimicrobial and Antioxidant Activities of Enterococcus Durans Strain LAB18s. Food Control 2014, 37, 251–256. [Google Scholar] [CrossRef]
- Zhong, Z.; Ji, X.; Xing, R.; Liu, S.; Guo, Z.; Chen, X.; Li, P. The Preparation and Antioxidant Activity of the Sulfanilamide Derivatives of Chitosan and Chitosan Sulfates. Bioorg. Med. Chem. 2007, 15, 3775–3782. [Google Scholar] [CrossRef]
- Chai, L.-J.; Lan, T.; Cheng, Z.; Zhang, J.; Deng, Y.; Wang, Y.; Li, Y.; Wang, F.; Piao, M. Stevia rebaudiana Leaves Fermented by Lactobacillus plantarum Exhibit Resistance to Microorganisms and Cancer Cell Lines in Vitro: A Potential Sausage Preservative. Food Chem. 2024, 432, 137187. [Google Scholar] [CrossRef]
- Laslo, É.; György, É.; András, C.D. Bioprotective Potential of Lactic Acid Bacteria. Acta Univ. Sapientiae Aliment. 2020, 13, 118–130. [Google Scholar] [CrossRef]
- Hurtado, A.; Othman, N.B.; Chammem, N.; Hamdi, M.; Ferrer, S.; Reguant, C.; Bordons, A.; Rozès, N. Characterization of Lactobacillus Isolates from Fermented Olives and Their Bacteriocin Gene Profiles. Food Microbiol. 2011, 28, 1514–1518. [Google Scholar] [CrossRef] [PubMed]
- Morais, P.B.; Calaça, P.S.S.T.; Rosa, C.A. Microorganisms Associated with Stingless Bees. In Pot-Honey: A Legacy of Stingless Bees; Springer: Berlin/Heidelberg, Germany, 2012; pp. 173–186. [Google Scholar]
- Iorizzo, M.; Letizia, F.; Ganassi, S.; Testa, B.; Petrarca, S.; Albanese, G.; Di Criscio, D.; De Cristofaro, A. Functional Properties and Antimicrobial Activity from Lactic Acid Bacteria as Resources to Improve the Health and Welfare of Honey Bees. Insects 2022, 13, 308. [Google Scholar] [CrossRef] [PubMed]
- Mesquita, A.; Silveira, L.; Filho, I.; Lima, V.; Filho, V.; Araujo, A.; Silva, T.; Araújo, K.; Macedo, L. Metabolism and Physiology of Lactobacilli: A Review. J. Environ. Anal. Prog. 2017, 2, 115. [Google Scholar] [CrossRef]
- Rangberg, A.; Mathiesen, G.; Amdam, G.V.; Diep, D.B. The Paratransgenic Potential of Lactobacillus kunkeei in the Honey Bee Apis mellifera. Benef. Microbes 2015, 6, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Braghini, F.; Biluca, F.C.; Schulz, M.; Gonzaga, L.V.; Costa, A.C.O.; Fett, R. Stingless Bee Honey: A Precious but Unregulated Product-Reality and Expectations. Food Rev. Int. 2022, 38, 683–712. [Google Scholar] [CrossRef]
- Elzeini, H.M.; Ali, A.R.A.A.; Nasr, N.F.; Hassan, M.; Hassan, A.A.M.; Elenany, Y.E. Probiotic Capability of Novel Lactic Acid Bacteria Isolated from Worker Honey Bees Gut Microbiota. FEMS Microbiol. Lett. 2021, 368, fnab030. [Google Scholar] [CrossRef]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune System Stimulation by the Native Gut Microbiota of Honey Bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef]
- Vásquez, A.; Olofsson, T.C. The Lactic Acid Bacteria Involved in the Production of Bee Pollen and Bee Bread. J. Apic. Res. 2009, 48, 189–195. [Google Scholar] [CrossRef]
- Borges, D.; Guzman-Novoa, E.; Goodwin, P.H. Effects of Prebiotics and Probiotics on Honey Bees (Apis mellifera) Infected with the Microsporidian Parasite Nosema Ceranae. Microorganisms 2021, 9, 481. [Google Scholar] [CrossRef]
- Shi, W.; Syrenne, R.; Sun, J.; Yuan, J.S. Molecular Approaches to Study the Insect Gut Symbiotic Microbiota at the ‘Omics’ Age. Insect Sci. 2010, 17, 199–219. [Google Scholar] [CrossRef]
- Gomez-Perez, A.; Kyryakov, P.; MT, B.; Asbah, N.; Noohi, F.; Iouk, T.; VI, T. Empirical Validation of a Hypothesis of the Hormetic Selective Forces Driving the Evolution of Longevity Regulation Mechanisms. Front. Genet. 2016, 7, 216. [Google Scholar] [CrossRef] [PubMed]
- Mohan, A.; Quek, S.-Y.; Gutierrez-Maddox, N.; Gao, Y.; Shu, Q. Effect of Honey in Improving the Gut Microbial Balance. Food Qual. Saf. 2017, 1, 107–115. [Google Scholar] [CrossRef]
- Rodrigues, N.P.A.; Garcia, E.F.; de Souza, E.L. Selection of Lactic Acid Bacteria with Promising Probiotic Aptitudes from Fruit and Ability to Survive in Different Food Matrices. Braz. J. Microbiol. 2021, 52, 2257–2269. [Google Scholar] [CrossRef]
- Yang, S.; Zhai, L.; Huang, L.; Meng, D.; Li, J.; Hao, Z.; Guan, Z.; Cai, Y.; Liao, X. Mining of Alkaline Proteases from Bacillus Altitudinis W3 for Desensitization of Milk Proteins: Their Heterologous Expression, Purification, and Characterization. Int. J. Biol. Macromol. 2020, 153, 1220–1230. [Google Scholar] [CrossRef]
- Hill, D.; Sugrue, I.; Tobin, C.; Hill, C.; Stanton, C.; Ross, R.P. The Lactobacillus casei Group: History and Health Related Applications. Front. Microbiol. 2018, 9, 2107. [Google Scholar] [CrossRef] [PubMed]
- Tuo, Y.; Yu, H.; Ai, L.; Wu, Z.; Guo, B.; Chen, W. Aggregation and Adhesion Properties of 22 Lactobacillus Strains. J. Dairy Sci. 2013, 96, 4252–4257. [Google Scholar] [CrossRef] [PubMed]
- Cheek, N.N. Taking Perspective the next Time around. Commentary on: “Perceived Perspective Taking: When Others Walk in Our Shoes”. Front. Psychol. 2015, 6, 434. [Google Scholar] [CrossRef]
- Won, G.; Choi, S.-I.; Park, N.; Kim, J.-E.; Kang, C.-H.; Kim, G.-H. In Vitro Antidiabetic, Antioxidant Activity, and Probiotic Activities of Lactiplantibacillus plantarum and Lacticaseibacillus paracasei Strains. Curr. Microbiol. 2021, 78, 3181–3191. [Google Scholar] [CrossRef]
- Heller, K.J. Probiotic Bacteria in Fermented Foods: Product Characteristics and Starter Organisms. Am. J. Clin. Nutr. 2001, 73, 374s–379s. [Google Scholar] [CrossRef]
- Anderson, K.E.; Ricigliano, V.A. Honey Bee Gut Dysbiosis: A Novel Context of Disease Ecology. Curr. Opin. Insect Sci. 2017, 22, 125–132. [Google Scholar] [CrossRef]
- Wang, G.; Si, Q.; Yang, S.; Jiao, T.; Zhu, H.; Tian, P.; Wang, L.; Li, X.; Gong, L.; Zhao, J. Lactic Acid Bacteria Reduce Diabetes Symptoms in Mice by Alleviating Gut Microbiota Dysbiosis and Inflammation in Different Manners. Food Funct. 2020, 11, 5898–5914. [Google Scholar] [CrossRef] [PubMed]
- Matthews, A.; Grimaldi, A.; Walker, M.; Bartowsky, E.; Grbin, P.; Jiranek, V. Lactic Acid Bacteria as a Potential Source of Enzymes for Use in Vinification. Appl. Environ. Microbiol. 2004, 70, 5715–5731. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Kim, H.; Lee, J.Y.; Won, G.; Choi, S.-I.; Kim, G.-H.; Kang, C.-H. The Antioxidant, Anti-Diabetic, and Anti-Adipogenesis Potential and Probiotic Properties of Lactic Acid Bacteria Isolated from Human and Fermented Foods. Fermentation 2021, 7, 123. [Google Scholar] [CrossRef]
- Huligere, S.S.; Chandana Kumari, V.B.; Alqadi, T.; Kumar, S.; Cull, C.A.; Amachawadi, R.G.; Ramu, R. Isolation and Characterization of Lactic Acid Bacteria with Potential Probiotic Activity and Further Investigation of Their Activity by α-Amylase and α-Glucosidase Inhibitions of Fermented Batters. Front. Microbiol. 2023, 13, 1042263. [Google Scholar] [CrossRef]





| PCR product | 2.5 µL |
| 2×KAPA HiFi Ready Mix | 12.5 µL |
| Forward primer (25 µM) | 0.25 µL |
| Reverse primer (25 µM) | 0.25 µL |
| PCR-grade water | 9.5 µL |
| Total volume | 25 µL |
| Characteristic | Isolated Strains | ||
|---|---|---|---|
| S1 | S2 | S3 | |
| Growth in MRS at: | |||
| 15 °C | − | w | − |
| 45 °C | − | − | − |
| The Optimum growth temperature (°C) | 30 | 30 | 30 |
| pH 4.0 | ++ | ++ | ++ |
| pH 3.5 | + | + | + |
| pH 3.0 | − | + | w |
| pH 2.5 | − | − | w |
| pH 2.0 | − | − | − |
| 6% | + | w | w |
| 8% | w | − | w |
| 10% | − | − | − |
| Others: | |||
| Motility | − | − | − |
| Gram-stain | + | + | + |
| Catalase assay | − | − | − |
| H2S production test | − | − | − |
| Gelatin liquefaction test | − | − | − |
| Indole production test | − | − | − |
| Starch hydrolysis test | + | + | + |
| Acid production from: | |||
| L-arabinose | + | + | + |
| D-cellulose-disaccharide | + | + | + |
| D-galactose | + | + | + |
| D-lactose | ++ | ++ | ++ |
| D-maltose | + | ++ | ++ |
| D-mannose | + | ++ | + |
| D-mannitol | + | + | + |
| D-triose | − | + | − |
| L-rhamnose | − | − | − |
| D-ribose | + | + | + |
| D-sucrose | + | + | + |
| D-xylose | − | − | − |
| L-xylose | − | − | − |
| D-fructose | + | + | + |
| D-sorbitol | + | + | + |
| L-sorbose | − | − | − |
| inositol | − | − | − |
| glycerol | + | + | + |
| Strain | Zone of Inhibition Size (mm) | ||
|---|---|---|---|
| Escherichia coli | Salmonella | Staphylococcus aureus | |
| S1 | 12.67 ± 1.04 | − | 15.08 ± 1.01 |
| S2 | 15.77 ± 0.84 | 9.55 ± 1.12 | 11.04 ± 0.98 |
| S3 | 13.35 ± 1.04 | 11.02 ± 0.54 | 10.68 ± 0.22 |
| Strain | Drug Sensitivity | Auto-Aggregation (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| GEN | S | KAN | ERY | C | AMP | VA | ||
| S1 | S | S | R | R | S | S | S | 17.43 ± 4.16 |
| S2 | S | S | R | R | S | S | S | 28.14 ± 3.85 |
| S3 | S | S | R | R | S | S | S | 32.14 ± 3.48 |
| Isolates | α-Glucosidase Inhibition * (CS) | (IC) | α-Amylase Inhibition * (CS) | (IC) |
|---|---|---|---|---|
| S1 | 49.35 ± 1.97 a | 21.50 ± 2.23 a | 59.35 ± 2.34 b | 32.25 ± 1.97 c |
| S2 | 52.42 ± 2.47 b | 25.57 ± 2.45 b | 58.21 ± 3.24 b | 34.24 ± 2.47 d |
| S3 | 59.45 ± 1.84 c | 26.35 ± 2.06 c | 63.35 ± 3.24 d | 36.12 ± 1.84 d |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basharat, S.; Zhai, L.; Jiang, F.; Asjad, T.; Khan, A.; Liao, X. Screening and Comparative Genomics of Probiotic Lactic Acid Bacteria from Bee Bread of Apis Cerana: Influence of Stevia and Stevioside on Bacterial Cell Growth and the Potential of Fermented Stevia as an Antidiabetic, Antioxidant, and Antifungal Agent. Microorganisms 2025, 13, 216. https://doi.org/10.3390/microorganisms13020216
Basharat S, Zhai L, Jiang F, Asjad T, Khan A, Liao X. Screening and Comparative Genomics of Probiotic Lactic Acid Bacteria from Bee Bread of Apis Cerana: Influence of Stevia and Stevioside on Bacterial Cell Growth and the Potential of Fermented Stevia as an Antidiabetic, Antioxidant, and Antifungal Agent. Microorganisms. 2025; 13(2):216. https://doi.org/10.3390/microorganisms13020216
Chicago/Turabian StyleBasharat, Samra, Lixin Zhai, Fuyao Jiang, Tanzila Asjad, Adil Khan, and Xiangru Liao. 2025. "Screening and Comparative Genomics of Probiotic Lactic Acid Bacteria from Bee Bread of Apis Cerana: Influence of Stevia and Stevioside on Bacterial Cell Growth and the Potential of Fermented Stevia as an Antidiabetic, Antioxidant, and Antifungal Agent" Microorganisms 13, no. 2: 216. https://doi.org/10.3390/microorganisms13020216
APA StyleBasharat, S., Zhai, L., Jiang, F., Asjad, T., Khan, A., & Liao, X. (2025). Screening and Comparative Genomics of Probiotic Lactic Acid Bacteria from Bee Bread of Apis Cerana: Influence of Stevia and Stevioside on Bacterial Cell Growth and the Potential of Fermented Stevia as an Antidiabetic, Antioxidant, and Antifungal Agent. Microorganisms, 13(2), 216. https://doi.org/10.3390/microorganisms13020216






