Genomic Insights into Probiotic Lactococcus lactis T-21, a Wild Plant-Associated Lactic Acid Bacterium, and Its Preliminary Clinical Safety for Human Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Genomic Sequencing and Comparative Genomic Analysis
| Strain | Origin | Genome Size (Mb) | Proteins | Plasmids * | Complete/ Draft | Genbank Accession | Reference |
|---|---|---|---|---|---|---|---|
| subsp. lactis | |||||||
| T-21 | Cranberry | 2.46 | 2344 | ND | Complete | AP038894 | [8] |
| KF147 | Mung bean sprouts | 2.60 | 2534 | 1 | Complete | CP001834, CP001835 | [4] |
| NCDO 2118 | Frozen peas | 2.55 | 2491 | 1 | Complete | CP009054, CP009055 | [24] |
| G50 | Napier grass | 2.35 | 2239 | ND | Complete | CP025500 | [25] |
| CAB701 | Cabbage | 2.52 | 2440 | 1 | Complete | CP129879, CP129880 | [26] |
| A12 | Wheat sourdough | 2.60 | 2677 | 4 | Complete | LT599049-LT599053 | [27] |
| 14B4 | Almond drupe | 2.58 | 2583 | 1 | Complete | CP028160, CP028161 | [28] |
| IO-1 | Drain water | 2.42 | 2291 | ND | Complete | AP012281 | [29] |
| JCM 5805 | Dairy starter | 2.53 | 2626 | NA | Draft | BBSI00000000 | [30] |
| IL1403 | Dairy starter | 2.37 | 2404 | ND | Complete | AE005176 | [31] |
| KLDS 4.0325 | Koumiss | 2.59 | 2612 | 6 | Complete | CP006766, CP006767, CP007042, CP007043, CP029291-CP029293 | [32] |
| subsp. cremoris | |||||||
| MG1363 | Dairy starter | 2.53 | 2647 | ND | Complete | AM406671 | [33] |
2.2. Phenotypic and Metabolic Characterisation
2.3. Safety and Clinical Trial Design
3. Results and Discussion
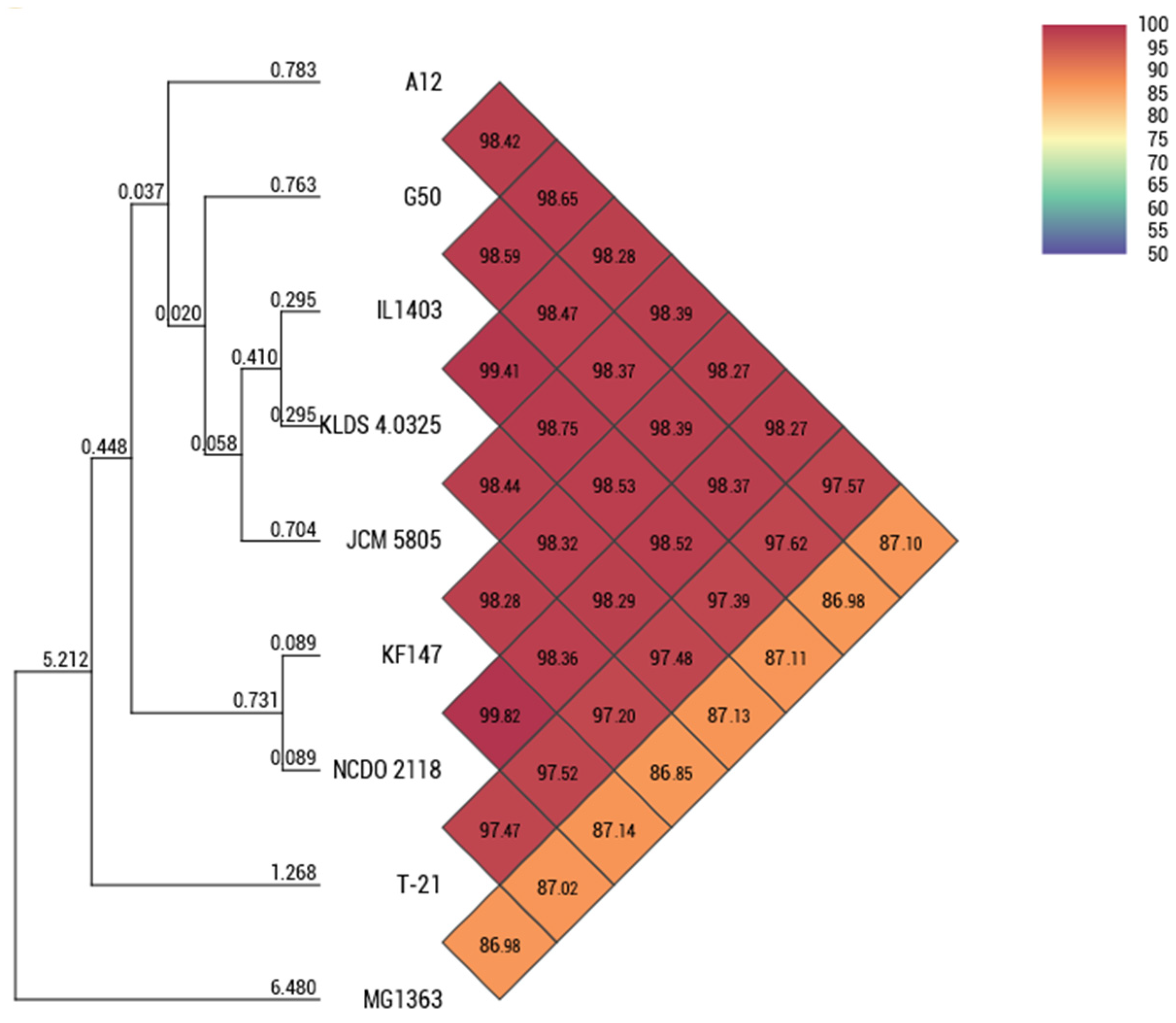
3.1. Genomic Architecture and Strain Diversity
3.2. Genomic Insights into Metabolic Capabilities and Phenotypic Divergence
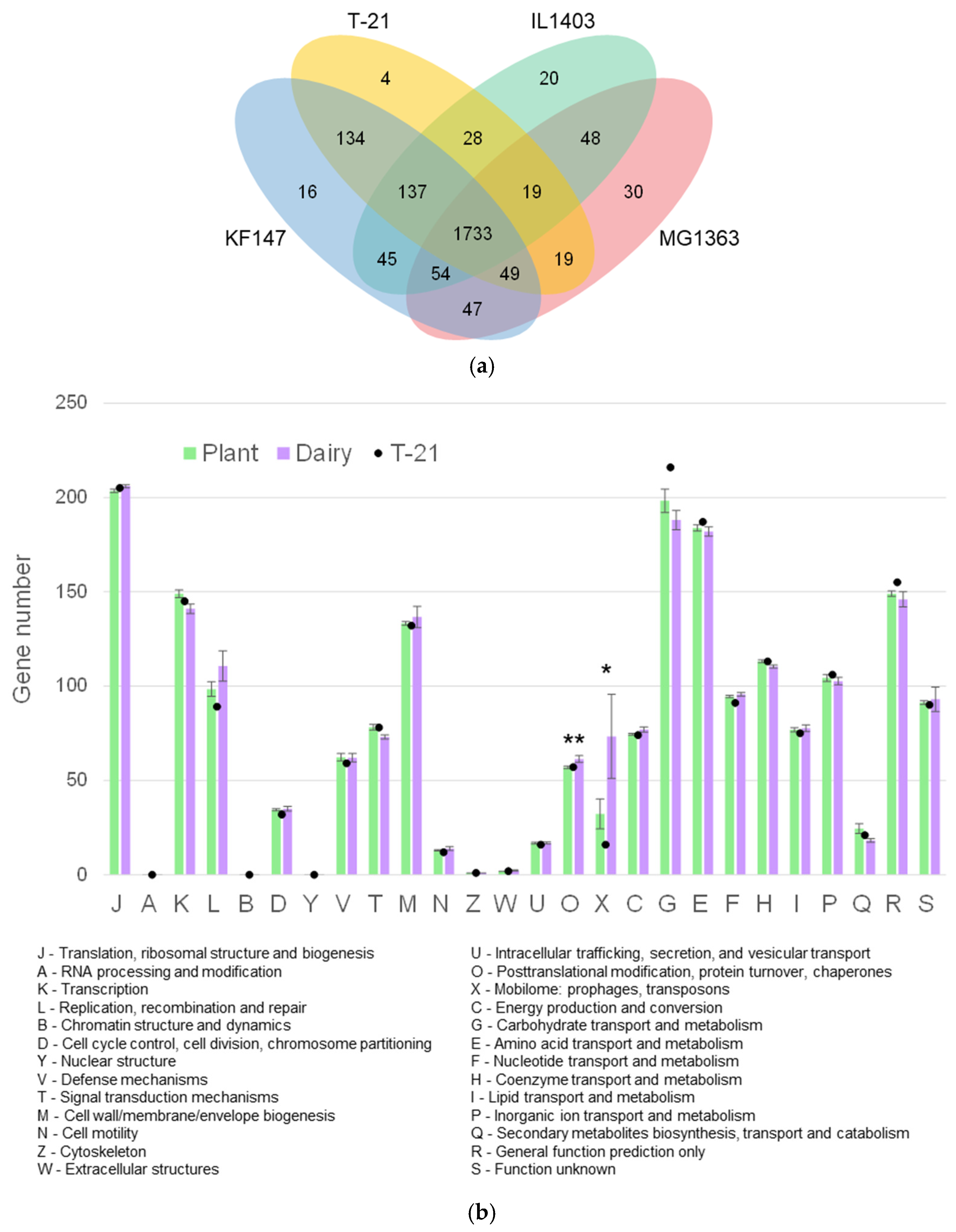
3.2.1. Functional Genomic Overview
3.2.2. Genetic Determinants of EPS Biosynthesis
3.2.3. Enhanced Fermentative Potential and Phenotypic Adaptation
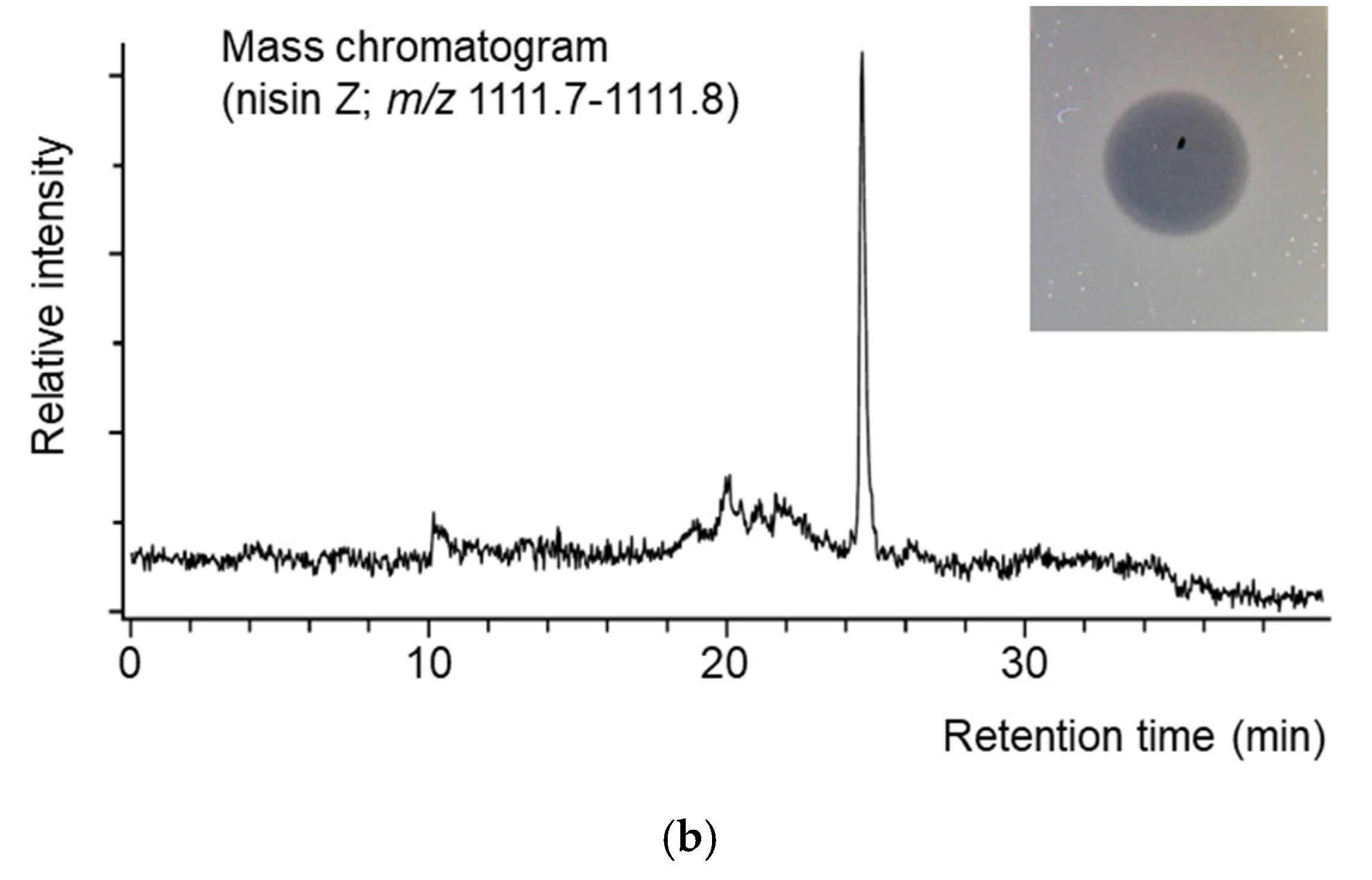
3.2.4. Nisin Production and Antimicrobial Activity
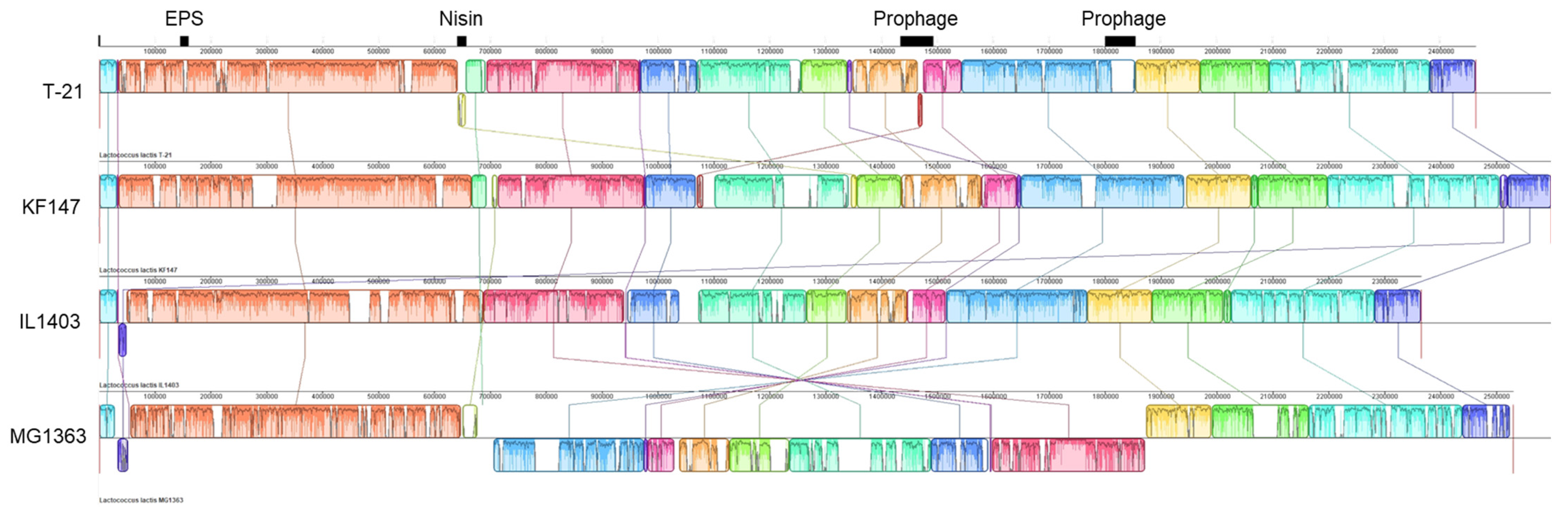
3.3. Prophages: Genomic Features and Considerations
3.4. Safety Assessment and Clinical Trial Outcomes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smid, E.J.; Kleerebezem, M. Production of aroma compounds in lactic fermentations. Annu. Rev. Food Sci. Technol. 2014, 5, 313–326. [Google Scholar] [CrossRef]
- van Hylckama Vlieg, J.E.T.; Rademaker, J.L.W.; Bachmann, H.; Molenaar, D.; Kelly, W.J.; Siezen, R.J. Natural diversity and adaptive responses of Lactococcus lactis. Curr. Opin. Biotechnol. 2006, 17, 183–190. [Google Scholar] [CrossRef]
- Kelly, W.J.; Ward, L.J.H.; Leahy, S.C. Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures. Genome Biol. Evol. 2010, 2, 729–744. [Google Scholar] [CrossRef]
- Siezen, R.J.; Bayjanov, J.; Renckens, B.; Wels, M.; van Hijum, S.A.F.T.; Molenaar, D.; van Hylckama Vlieg, J.E.T. Complete genome sequence of Lactococcus lactis subsp. lactis KF147, a plant-associated lactic acid bacterium. J. Bacteriol. 2010, 192, 2649–2650. [Google Scholar] [CrossRef]
- Bachmann, H.; Starrenburg, M.J.C.; Molenaar, D.; Kleerebezem, M.; van Hylckama Vlieg, J.E.T. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 2012, 22, 115–124. [Google Scholar] [CrossRef]
- Kelleher, P.; Bottacini, F.; Mahony, J.; Kilcawley, K.N.; van Sinderen, D. Comparative and functional genomics of the Lactococcus lactis taxon; insights into evolution and niche adaptation. BMC Genom. 2017, 18, 267. [Google Scholar] [CrossRef] [PubMed]
- Wels, M.; Siezen, R.; van Hijum, S.; Kelly, W.J.; Bachmann, H. Comparative genome analysis of Lactococcus lactis indicates niche adaptation and resolves genotype/phenotype disparity. Front. Microbiol. 2019, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Uehara, K.; Sunada, Y.; Kondo, S.; Matsuo, S. Effects of food containing Lactococcus lactis strain T21 on the improvement of skin condition: A randomized, double-blind, placebo-controlled, parallel-group study. Biosci. Microbiota Food Health 2024, 43, 381–390. [Google Scholar] [CrossRef]
- Fukao, M.; Oshima, K.; Morita, H.; Toh, H.; Suda, W.; Kim, S.-W.; Suzuki, S.; Yakabe, T.; Hattori, M.; Yajima, N. Genomic analysis by deep sequencing of the probiotic Lactobacillus brevis KB290 harboring nine plasmids reveals genomic stability. PLoS ONE 2013, 8, e60521. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, Y.; Fujisawa, T.; Kaminuma, E.; Nakamura, Y.; Arita, M. DFAST and DAGA: Web-based integrated genome annotation tools and resources. Biosci. Microbiota Food Health 2016, 35, 173–184. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, Y.; Ikeda-Ohtsubo, W.; Nagata, Y.; Tsuda, M. GenomeMatcher: A graphical user interface for DNA sequence comparison. BMC Bioinform. 2008, 9, 376. [Google Scholar] [CrossRef]
- Sun, J.; Lu, F.; Luo, Y.; Bie, L.; Xu, L.; Wang, Y. OrthoVenn3: An integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 2023, 51, W397–W403. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015, 43, D261–D269. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: Faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef]
- Oliveira, L.C.; Saraiva, T.D.L.; Soares, S.C.; Ramos, R.T.J.; Sá, P.H.C.G.; Carneiro, A.R.; Miranda, F.; Freire, M.; Renan, W.; Júnior, A.F.O.; et al. Genome sequence of Lactococcus lactis subsp. lactis NCDO 2118, a GABA-producing strain. Genome Announc. 2014, 2, e00980-14. [Google Scholar] [CrossRef]
- Nakano, K.; Minami, M.; Shinzato, M.; Shimoji, M.; Ashimine, N.; Shiroma, A.; Ohki, S.; Nakanishi, T.; Tamotsu, H.; Teruya, K.; et al. Complete genome sequence of Lactococcus lactis subsp. lactis G50 with immunostimulating activity, isolated from Napier Grass. Genome Announc. 2018, 6, e00069-18. [Google Scholar] [CrossRef]
- Jeong, H.; Kim, S.; Hwang, U.-S.; Choi, H.; Park, Y.-S. Immunostimulatory activity of Lactococcus lactis subsp. lactis CAB701 isolated from Jeju Cabbage. Microorganisms 2023, 11, 1718. [Google Scholar] [CrossRef] [PubMed]
- Passerini, D.; Coddeville, M.; Le Bourgeois, P.; Loubière, P.; Ritzenthaler, P.; Fontagné-Faucher, C.; Daveran-Mingot, M.-L.; Cocaign-Bousquet, M. The carbohydrate metabolism signature of Lactococcus lactis strain A12 reveals its sourdough ecosystem origin. Appl. Environ. Microbiol. 2013, 79, 5844–5852. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.D.; Huynh, S.; Parker, C.T.; Han, R.; Hnasko, R.; Gorski, L.; McGarvey, J.A. Complete genome sequence of Lactococcus lactis subsp. lactis strain 14B4, which inhibits the growth of Salmonella enterica serotype Poona in vitro. Microbiol. Resour. Announc. 2018, 7, e01364-18. [Google Scholar] [CrossRef] [PubMed]
- Shimizu-Kadota, M.; Kato, H.; Shiwa, Y.; Oshima, K.; Machii, M.; Araya-Kojima, T.; Zendo, T.; Hattori, M.; Sonomoto, K.; Yoshikawa, H. Genomic features of Lactococcus lactis IO-1, a lactic acid bacterium that utilizes xylose and produces high levels of L-lactic acid. Biosci. Biotechnol. Biochem. 2013, 77, 1804–1808. [Google Scholar] [CrossRef]
- Fujii, T.; Tomita, Y.; Ikushima, S.; Horie, A.; Fujiwara, D. Draft genome sequence of Lactococcus lactis subsp. lactis JCM 5805T, a strain that induces plasmacytoid dendritic cell activation. Genome Announc. 2015, 3, e00113-15. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, A.; Wincker, P.; Mauger, S.; Jaillon, O.; Malarme, K.; Weissenbach, J.; Ehrlich, S.D.; Sorokin, A. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001, 11, 731–753. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, Y.; Huo, G. Complete genome sequence of Lactococcus lactis subsp. lactis KLDS4.0325. Genome Announc. 2013, 1, e00962-13. [Google Scholar] [CrossRef]
- Wegmann, U.; O’Connell-Motherway, M.; Zomer, A.; Buist, G.; Shearman, C.; Canchaya, C.; Ventura, M.; Goesmann, A.; Gasson, M.J.; Kuipers, O.P.; et al. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 2007, 189, 3256–3270. [Google Scholar] [CrossRef]
- Zendo, T.; Nakayama, J.; Fujita, K.; Sonomoto, K. Bacteriocin detection by liquid chromatography/mass spectrometry for rapid identification. J. Appl. Microbiol. 2008, 104, 499–507. [Google Scholar] [CrossRef]
- Zendo, T.; Ohashi, C.; Maeno, S.; Piao, X.; Salminen, S.; Sonomoto, K.; Endo, A. Kunkecin A, a new nisin variant bacteriocin produced by the fructophilic lactic acid bacterium Apilactobacillus kunkeei FF30-6 isolated from honey bees. Front. Microbiol. 2020, 11, 571903. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- ProBiotix Health Ltd. Notice to US Food and Drug Administration of the Conclusion That the Intended Use of Lactobacillus plantarum ECGC 13110402 (LPLDL®) Is Generally Recognized as Safe; GRAS Notice No 000847; ProBiotix Health Ltd.: Wakefield, UK, 2019. [Google Scholar]
- Fukao, M.; Oki, A.; Segawa, S. Genome-based assessment of safety characteristics of Lacticaseibacillus paracasei NY1301 and genomic differences in closely related strains marketed as probiotics. Biosci. Microbiota Food Health 2024, 43, 145–149. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Zeidan, A.A.; Poulsen, V.K.; Janzen, T.; Buldo, P.; Derkx, P.M.F.; Øregaard, G.; Neves, A.R. Polysaccharide production by lactic acid bacteria: From genes to industrial applications. FEMS Microbiol. Rev. 2017, 41, S168–S200. [Google Scholar] [CrossRef] [PubMed]
- Lammens, W.; Le Roy, K.; Schroeven, L.; Van Laere, A.; Rabijns, A.; Van den Ende, W. Structural insights into glycoside hydrolase family 32 and 68 enzymes: Functional implications. J. Exp. Bot. 2009, 60, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Graebin, N.G.; Schöffer, J.D.N.; Andrades, D.D.; Hertz, P.F.; Ayub, M.A.Z.; Rodrigues, R.C. Immobilization of glycoside hydrolase families GH1, GH13, and GH70: State of the art and perspectives. Molecules 2016, 21, 1074. [Google Scholar] [CrossRef]
- Deo, D.; Davray, D.; Kulkarni, R. A Diverse repertoire of exopolysaccharide biosynthesis gene clusters in Lactobacillus revealed by comparative analysis in 106 sequenced genomes. Microorganisms 2019, 7, 444. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, V.K.; Derkx, P.; Oregaard, G. High-throughput screening for texturing Lactococcus strains. FEMS Microbiol. Lett. 2019, 366, fnz001. [Google Scholar] [CrossRef]
- Boels, I.C.; Beerthuyzen, M.M.; Kosters, M.H.W.; Van Kaauwen, M.P.W.; Kleerebezem, M.; De Vos, W.M. Identification and functional characterization of the Lactococcus lactis Rfb operon, required for dTDP-rhamnose biosynthesis. J. Bacteriol. 2004, 186, 1239–1248. [Google Scholar] [CrossRef]
- Siezen, R.J.; Starrenburg, M.J.C.; Boekhorst, J.; Renckens, B.; Molenaar, D.; van Hylckama Vlieg, J.E.T. Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl. Environ. Microbiol. 2008, 74, 424–436. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Kasper, D.L. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat. Rev. Immunol. 2006, 6, 849–858. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.M.; Ross, R.P.; Fitzgerald, G.F.; Caplice, N.M.; Stanton, C. Sugar-coated: Exopolysaccharide producing lactic acid bacteria for food and human health applications. Food Funct. 2015, 6, 679–693. [Google Scholar] [CrossRef] [PubMed]
- Caggianiello, G.; Kleerebezem, M.; Spano, G. Exopolysaccharides produced by lactic acid bacteria: From health-promoting benefits to stress tolerance mechanisms. Appl. Microbiol. Biotechnol. 2016, 100, 3877–3886. [Google Scholar] [CrossRef]
- Kuipers, O.P.; Beerthuyzen, M.M.; Siezen, R.J.; De Vos, W.M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 1993, 216, 281–291. [Google Scholar] [CrossRef]
- Romero, D.A.; Magill, D.; Millen, A.; Horvath, P.; Fremaux, C. Dairy lactococcal and streptococcal phage-host interactions: An industrial perspective in an evolving phage landscape. FEMS Microbiol. Rev. 2020, 44, 909–932. [Google Scholar] [CrossRef] [PubMed]
- Aucouturier, A.; Chain, F.; Langella, P.; Bidnenko, E. Characterization of a prophage-free derivative strain of Lactococcus lactis ssp. lactis IL1403 reveals the importance of prophages for phenotypic plasticity of the host. Front. Microbiol. 2018, 9, 2032. [Google Scholar] [CrossRef] [PubMed]
- Kullar, R.; Goldstein, E.J.C.; Johnson, S.; McFarland, L.V. Lactobacillus bacteremia and probiotics: A review. Microorganisms 2023, 11, 896. [Google Scholar] [CrossRef]
- Barlow, S.; Chesson, A.; Collins, J.D.; Dybing, E.; Flynn, A.; Fruijtier-Pölloth, C.; Hardy, A.; Knaap, A.; Kuiper, H.; Le Neindre, P.; et al. Introduction of a qualified presumption of safety (QPS) approach for assessment of selected microorganisms referred to EFSA-opinion of the scientific committee. EFSA J. 2007, 5, 587. [Google Scholar]
- Aquilina, G.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; de Knecht, J.; Dierick, N.A.; Gralak, M.A.; Gropp, J.; Ingrid, H.; Hogstrand, C.; et al. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 274040. [Google Scholar]


| Ingredients: | |
| Skimmed milk powder, glucose–fructose syrup, sugar, yeast extract, seven flavourings | |
| Nutritional facts (value for daily dose, 180 g) | |
| Energy (kcal) | 116 |
| Protein (g) | 5.3 |
| Fat (g) | 0.2 |
| Carbohydrate (g) | 24.0 |
| Sodium (mg) | 87 |
| Carbohydrate | T-21 (Cranberry) | KF147 * (Mung Bean Sprouts) | JCM 5805 (Dairy Starter) | IL-1403 * (Dairy Starter) | Carbohydrate Type |
|---|---|---|---|---|---|
| Mono/oligosaccharides | |||||
| L-Arabinose | + | + | − | − | Pentose |
| D-Ribose | +/− | + | + | + | Pentose |
| D-Xylose | + | + | + | − | Pentose |
| D-Lyxose | +/− | − | − | − | Pentose |
| D-Galactose | + | + | + | + | Hexose |
| D-Glucose | + | + | + | + | Hexose |
| D-Fructose | + | + | + | + | Hexose |
| D-Mannose | + | + | + | + | Hexose |
| N-Acetylglucosamine | + | + | + | + | Hexose |
| Arbutin | + | + | + | + | Aryl-monosaccharide |
| Salicin | + | + | + | + | Aryl-monosaccharide |
| Amygdalin | +/− | + | +/− | +/− | Aryl-disaccharide |
| D-Cellobiose | + | + | + | + | Disaccharide |
| D-Maltose | + | + | + | + | Disaccharide |
| D-Lactose | +/− | + | + | +/− | Disaccharide |
| D-Sucrose | + | + | − | − | Disaccharide |
| D-Trehalose | + | + | + | + | Disaccharide |
| D-Trehalose | + | + | + | + | Disaccharide |
| Gentiobiose | +/− | + | +/− | + | Disaccharide |
| D-Melibiose | − | + | − | − | Disaccharide |
| D-Raffinose | − | + | − | − | Trisaccharide |
| Gluconate | +/− | +/− | − | − | Sugar acid |
| Polyos | |||||
| Glycerol | − | − | +/− | − | |
| D-Mannitol | +/− | + | − | − | |
| Polysaccharides | |||||
| Starch | +/− | + | +/− | + |
| Group | Number of Participants | Number of Adverse Events | Symptoms Reported | Severity | Relevance to Test Beverages |
|---|---|---|---|---|---|
| T-21 group | 8 | 14 | Headache (2 cases) | Mild | Not associated |
| Nasal mucus and sore throat (1 case) | |||||
| Headache and stiff shoulders (2 cases) | |||||
| Stiff shoulders (1 case) | |||||
| Sneezing and runny nose (2 cases) | |||||
| Fever, cough, and phlegm (1 case) | |||||
| Nasal mucus and nasal congestion (1 case) | |||||
| Abdominal pain (2 cases) | |||||
| Sore throat (1 case) | |||||
| Abdominal pain and diarrhoea (1 case) | |||||
| Placebo group | 7 | 10 | Diarrhoea (1 case) | Mild | Not associated |
| Constipation (1 case) | |||||
| Fever and headache (1 case) | |||||
| Headache (3 cases) | |||||
| Sudden sensorineural hearing loss (1 case) | |||||
| Stiff shoulders and headache (1 case) | |||||
| Abdominal pain (1 case) | |||||
| Nasal mucus (1 case) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukao, M.; Tagawa, K.; Sunada, Y.; Uehara, K.; Sugimoto, T.; Zendo, T.; Nakayama, J.; Segawa, S. Genomic Insights into Probiotic Lactococcus lactis T-21, a Wild Plant-Associated Lactic Acid Bacterium, and Its Preliminary Clinical Safety for Human Application. Microorganisms 2025, 13, 388. https://doi.org/10.3390/microorganisms13020388
Fukao M, Tagawa K, Sunada Y, Uehara K, Sugimoto T, Zendo T, Nakayama J, Segawa S. Genomic Insights into Probiotic Lactococcus lactis T-21, a Wild Plant-Associated Lactic Acid Bacterium, and Its Preliminary Clinical Safety for Human Application. Microorganisms. 2025; 13(2):388. https://doi.org/10.3390/microorganisms13020388
Chicago/Turabian StyleFukao, Masanori, Keisuke Tagawa, Yosuke Sunada, Kazuya Uehara, Takuya Sugimoto, Takeshi Zendo, Jiro Nakayama, and Shuichi Segawa. 2025. "Genomic Insights into Probiotic Lactococcus lactis T-21, a Wild Plant-Associated Lactic Acid Bacterium, and Its Preliminary Clinical Safety for Human Application" Microorganisms 13, no. 2: 388. https://doi.org/10.3390/microorganisms13020388
APA StyleFukao, M., Tagawa, K., Sunada, Y., Uehara, K., Sugimoto, T., Zendo, T., Nakayama, J., & Segawa, S. (2025). Genomic Insights into Probiotic Lactococcus lactis T-21, a Wild Plant-Associated Lactic Acid Bacterium, and Its Preliminary Clinical Safety for Human Application. Microorganisms, 13(2), 388. https://doi.org/10.3390/microorganisms13020388





