Genome-Wide Survey of Donor Chromosomal Genes Involved in Trans-Kingdom Conjugation via the RP4-T4SS Machinery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Yeast, and Growth Media
2.2. Donor and Recipient Cell Cultures
2.3. Trans-Kingdom Conjugation
2.4. Bacterial Conjugation
2.5. Complementation Analysis
2.6. Data Analysis
3. Results
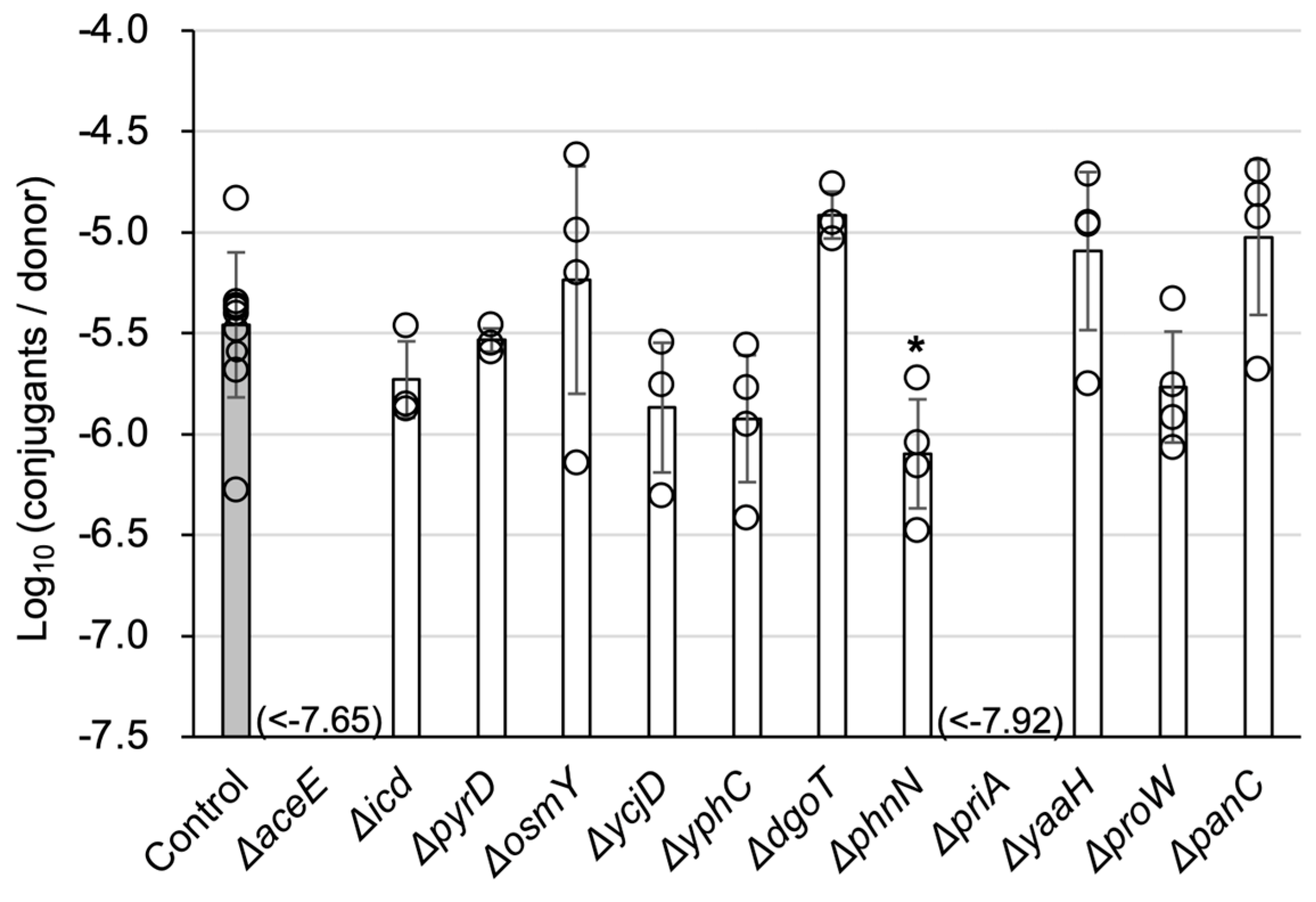
3.1. Screening of Chromosomal Mutants Defective in TKC
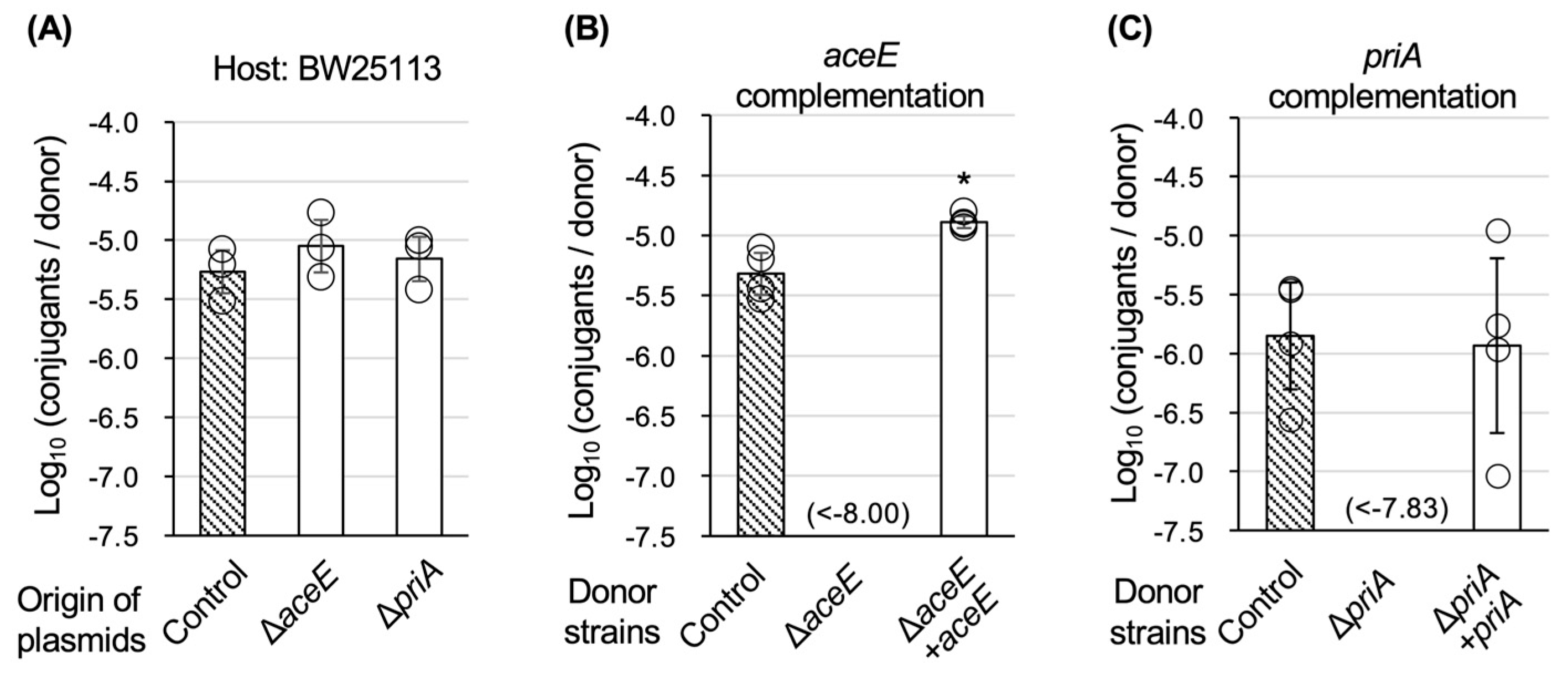
3.2. Confirmation of TKC Defectiveness in ∆aceE and ∆priA Mutants
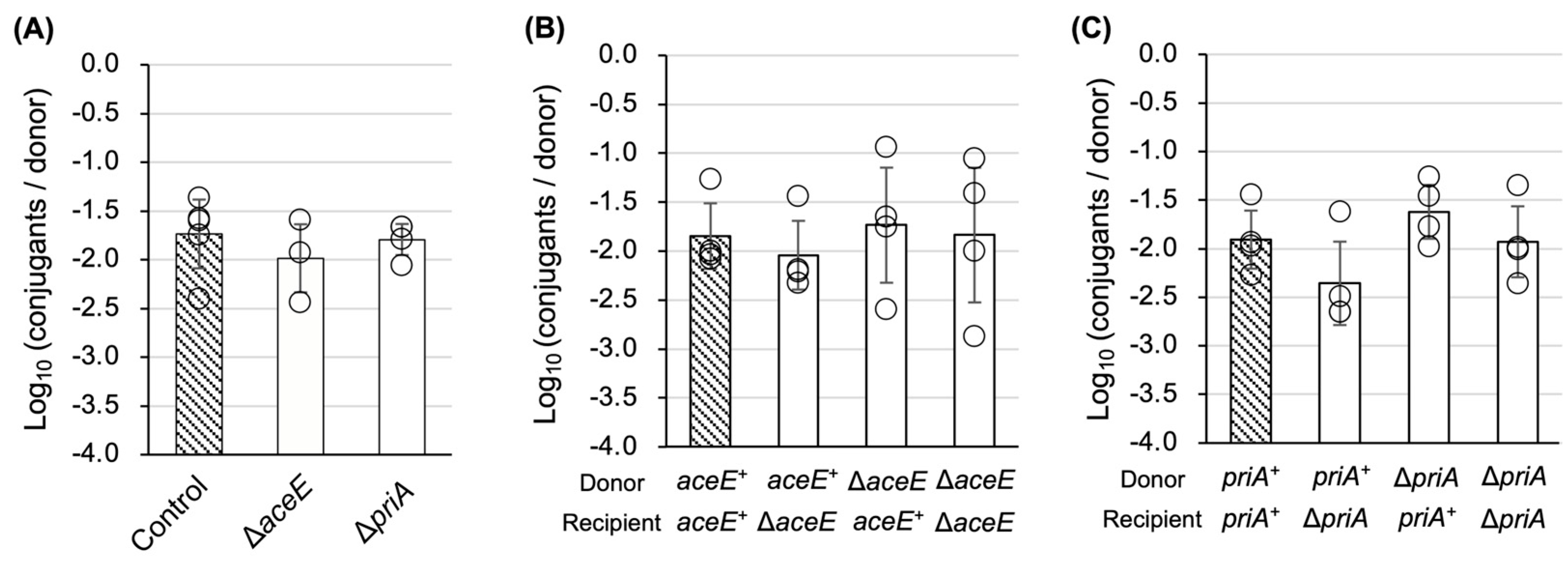
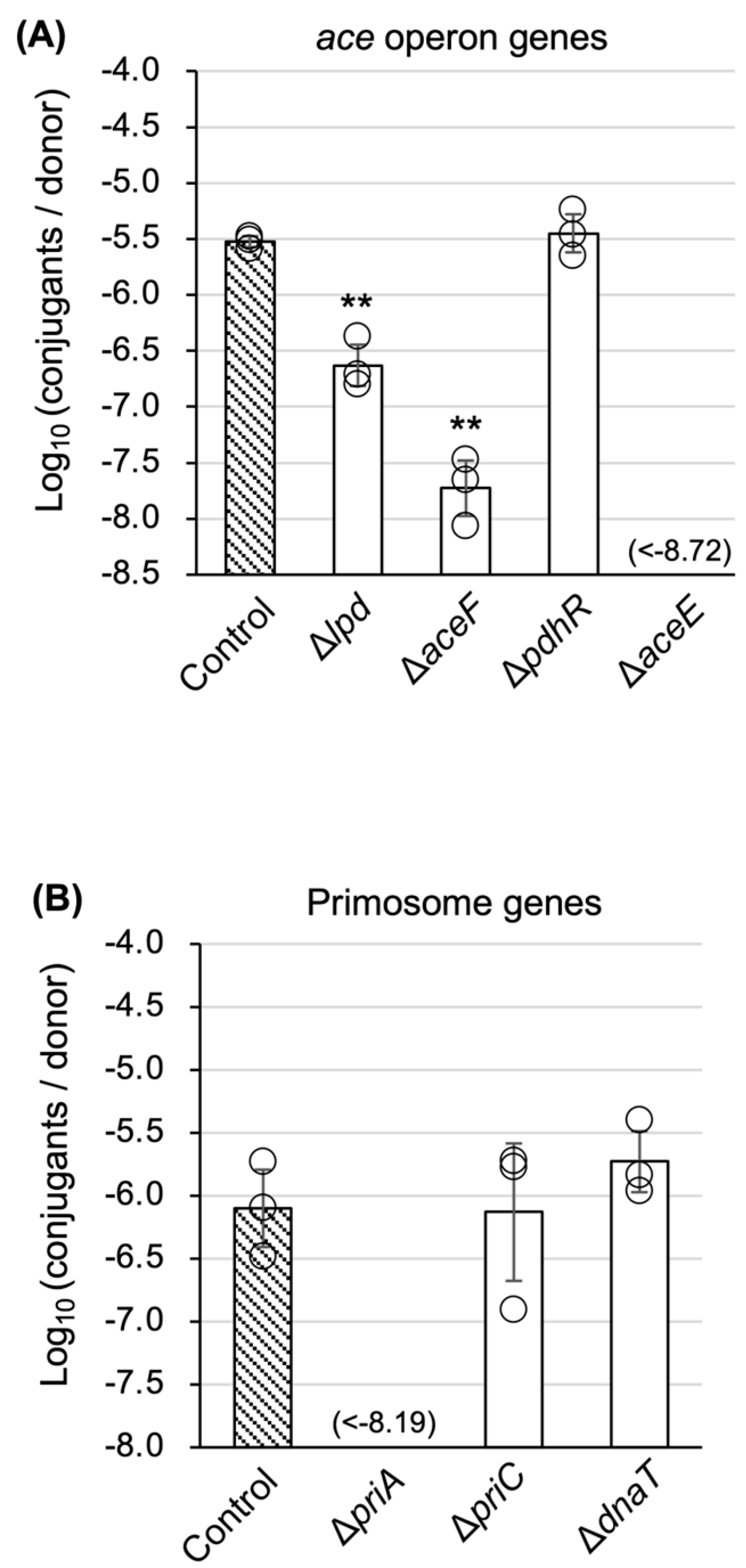
3.3. Characterization of TKC Defectiveness in ∆aceE and ∆priA Mutants
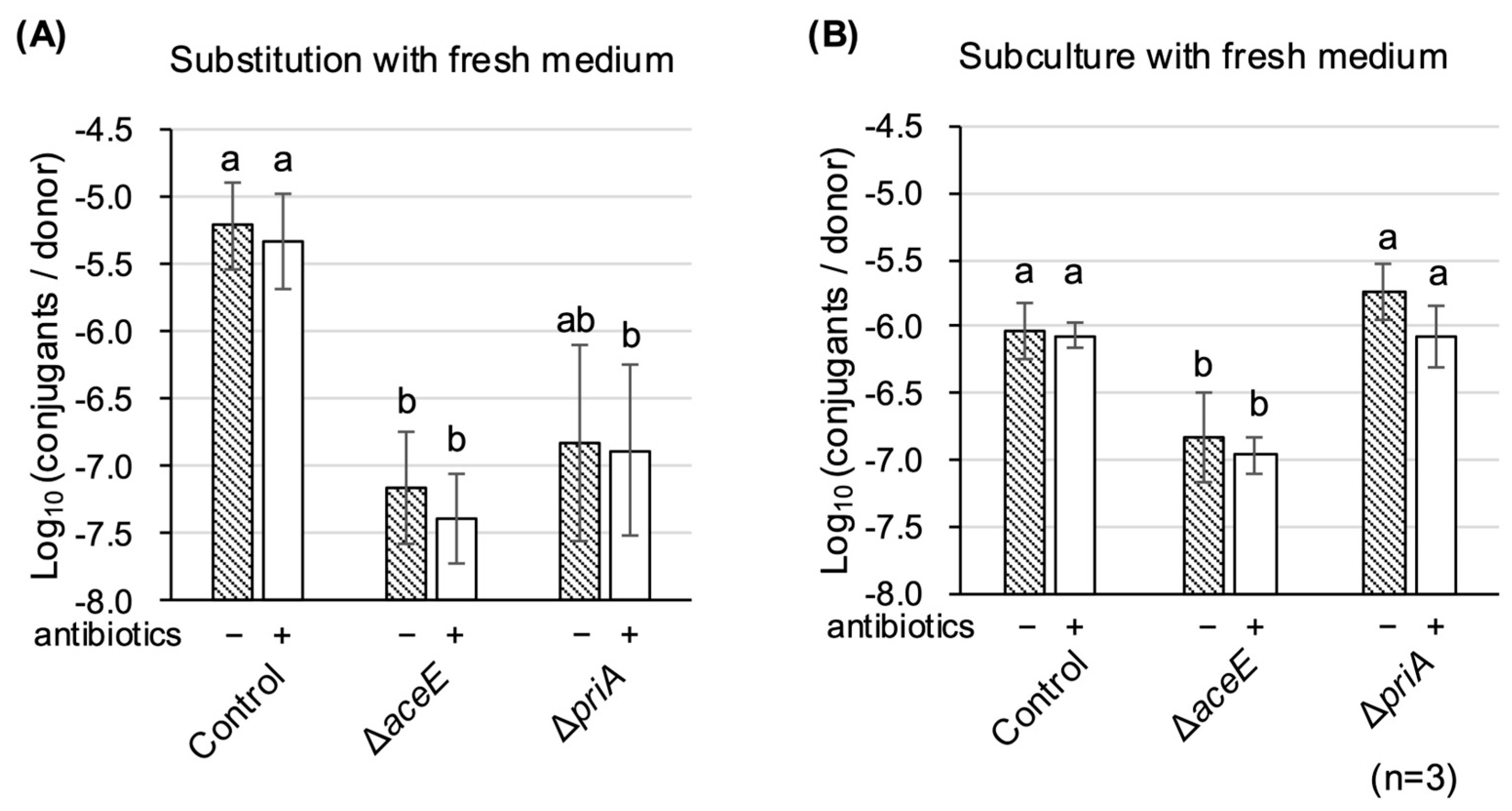
3.4. Culture Medium Substitution Recovers TKC Defectiveness in ∆aceE and ∆priA Mutants
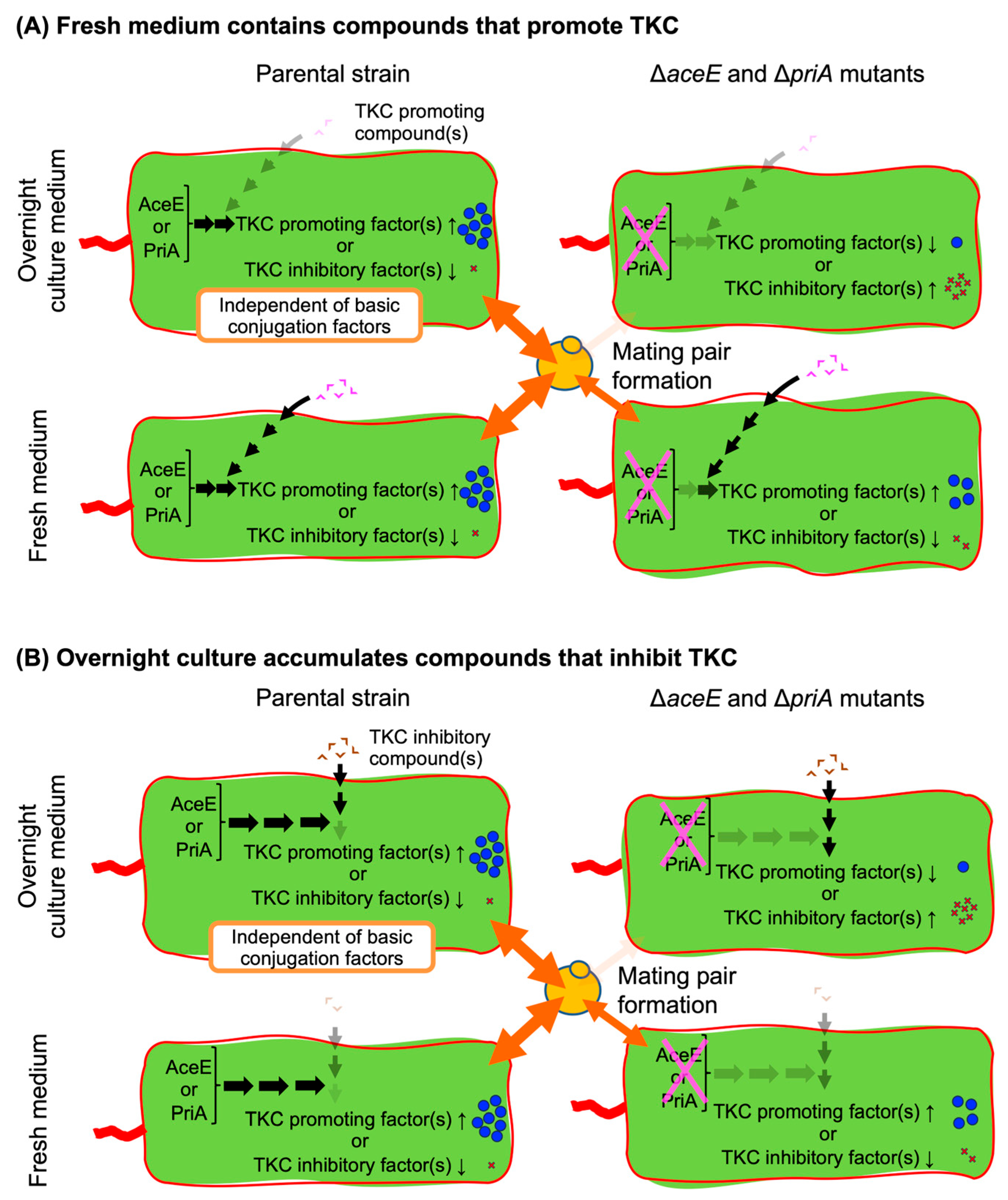
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TKC | Trans-kingdom conjugation |
| NBRP | National BioResource Project |
| SD | Standard deviation |
| TNB | Tris-HCl with NaCl buffer |
| YPD | Yeast-extract/peptone/dextrose |
References
- Furuya, E.Y.; Lowy, F.D. Antimicrobial-resistant bacteria in the community setting. Nat. Rev. Microbiol. 2006, 4, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.D.; Harb, L.; Khara, P.; Zeng, L.; Hu, B.; Christie, P.J. Type IV secretion systems: Advances in structure, function, and activation. Mol. Microbiol. 2021, 115, 436–452. [Google Scholar] [CrossRef] [PubMed]
- Pansegrau, W.; Lanka, E.; Barth, P.T.; Figurski, D.H.; Guiney, D.G.; Haas, D.; Helinski, D.R.; Schwab, H.; Stanisich, V.A.; Thomas, C.M. Complete nucleotide sequence of birmingham IncPα plasmids: Compilation and comparative analysis. J. Mol. Biol. 1994, 239, 623–663. [Google Scholar] [CrossRef] [PubMed]
- Schmidhauser, T.J.; Helinski, D.R. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of Gram-negative bacteria. J. Bacteriol. 1985, 164, 446–455. [Google Scholar] [CrossRef]
- Yano, H.; Rogers, L.M.; Knox, M.G.; Heuer, H.; Smalla, K.; Brown, C.J.; Top, E.M. Host range diversification within the IncP-1 plasmid group. Microbiology 2013, 159, 2303–2315. [Google Scholar] [CrossRef]
- Kamachi, K.; Sota, M.; Tamai, Y.; Nagata, N.; Konda, T.; Inoue, T.; Top, E.M.; Arakawa, Y. Plasmid pBP136 from Bordetella pertussis represents an ancestral form of IncP-1β plasmids without accessory mobile elements. Microbiology 2006, 152, 3477–3484. [Google Scholar] [CrossRef]
- Suzuki, H.; Yano, H.; Brown, C.J.; Top, E.M. Predicting plasmid promiscuity based on genomic signature. J. Bacteriol. 2010, 192, 6045–6055. [Google Scholar] [CrossRef]
- Norberg, P.; Bergström, M.; Jethava, V.; Dubhashi, D.; Hermansson, M. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat. Commun. 2011, 2, 268. [Google Scholar] [CrossRef]
- Adamczyk, M.; Jagura-Burdzy, G. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochem. Pol. 2003, 50, 425–453. [Google Scholar] [CrossRef]
- Heinemann, J.A.; Sprague, G.F. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 1989, 340, 205–209. [Google Scholar] [CrossRef]
- Nishikawa, M.; Suzuki, K.; Yoshida, K. Structural and functional stability of IncP plasmids during stepwise transmission by trans-kingdom mating: Promiscuous conjugation of Escherichia coli and Saccharomyces cerevisiae. Jpn. J. Genet. 1990, 65, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Hayman, G.T.; Bolen, P.L. Movement of shuttle plasmids from Escherichia coli into yeasts other than Saccharomyces cerevisiae using trans-kingdom conjugation. Plasmid 1993, 30, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Sawasaki, Y.; Inomata, K.; Yoshida, K. Trans-kingdom conjugation between Agrobacterium tumfaciens and Saccharomyces cerevisiae, a bacterium and a yeast. Plant Cell Physiol. 1996, 37, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.; Cashmore, A.M.; Wilkins, B.M. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae: Involvement of the Tra2 mating system. J. Bacteriol. 1998, 180, 6538–6543. [Google Scholar] [CrossRef]
- Waters, V.L. Conjugation between bacterial and mammalian cells. Nat. Genet. 2001, 29, 375–376. [Google Scholar] [CrossRef]
- Dodsworth, J.A.; Li, L.; Wei, S.; Hedlund, B.P.; Leigh, J.A.; de Figueiredo, P. Interdomain conjugal transfer of DNA from bacteria to archaea. Appl. Environ. Microbiol. 2010, 76, 5644–5647. [Google Scholar] [CrossRef]
- Fink, C.; Beblawy, S.; Enkerlin, A.M.; Mühling, L.; Angenent, L.T.; Molitor, B. A shuttle-vector system allows heterologous gene expression in the thermophilic methanogen Methanothermobacter thermautotrophicus ΔH. mBio 2021, 12, e02766-21. [Google Scholar] [CrossRef]
- Karas, B.J.; Diner, R.E.; Lefebvre, S.C.; McQuaid, J.; Phillips, A.P.R.; Noddings, C.M.; Brunson, J.K.; Valas, R.E.; Deerinck, T.J.; Jablanovic, J.; et al. Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 2015, 6, 6925. [Google Scholar] [CrossRef]
- Awwad, F.; Fantino, E.I.; Héneault, M.; Diaz-Garza, A.M.; Merindol, N.; Custeau, A.; Gélinas, S.E.; Meddeb-Mouelhi, F.; Li, J.; Lemay, J.F.; et al. Bioengineering of the marine diatom Phaeodactylum tricornutum with cannabis genes enables the production of the cannabinoid precursor, olivetolic acid. Int. J. Mol. Sci. 2023, 24, 16624. [Google Scholar] [CrossRef]
- Maeda, Y.; Nakamura, M.; Watanabe, K.; Okamoto, E.; Tanaka, T. Functional analysis of the putative centromere sequences of marine oleaginous diatom Fistulifera solaris. Algal Res. 2023, 74, 103225. [Google Scholar] [CrossRef]
- Moriguchi, K.; Edahiro, N.; Yamamoto, S.; Tanaka, K.; Kurata, N.; Suzuki, K. Transkingdom genetic transfer from Escherichia coli to Saccharomyces cerevisiae as a simple gene introduction tool. Appl. Environ. Microbiol. 2013, 79, 4393–4400. [Google Scholar] [CrossRef] [PubMed]
- Brumwell, S.L.; MacLeod, M.R.; Huang, T.; Cochrane, R.R.; Meaney, R.S.; Zamani, M.; Matysiakiewicz, O.; Dan, K.N.; Janakirama, P.; Edgell, D.R.; et al. Designer Sinorhizobium meliloti strains and multi-functional vectors enable direct inter-kingdom DNA transfer. PLoS ONE 2019, 14, e0206781. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, R.R.; Shrestha, A.; Severo De Almeida, M.M.; Agyare-Tabbi, M.; Brumwell, S.L.; Hamadache, S.; Meaney, J.S.; Nucifora, D.P.; Heng Say, H.; Sharma, J.; et al. Superior conjugative plasmids delivered by bacteria to diverse fungi. BioDesign Res. 2022, 2022, 9802168. [Google Scholar] [CrossRef]
- Moriguchi, K.; Yamamoto, S.; Ohmine, Y.; Suzuki, K. A fast and practical yeast transformation method mediated by Escherichia coli based on a trans-kingdom conjugal transfer system: Just mix two cultures and wait one hour. PLoS ONE 2016, 11, e0148989. [Google Scholar] [CrossRef]
- Soltysiak, M.P.M.; Meaney, R.S.; Hamadache, S.; Janakirama, P.; Edgell, D.R.; Karas, B.J. Trans-kingdom conjugation within solid media from Escherichia coli to Saccharomyces cerevisiae. Int. J. Mol. Sci. 2019, 20, 5212. [Google Scholar] [CrossRef]
- Mizuta, M.; Satoh, E.; Katoh, C.; Tanaka, K.; Moriguchi, K.; Suzuki, K. Screening for yeast mutants defective in recipient ability for transkingdom conjugation with Escherichia coli revealed importance of vacuolar ATPase activity in the horizontal DNA transfer phenomenon. Microbiol. Res. 2012, 167, 311–316. [Google Scholar] [CrossRef]
- Moriguchi, K.; Yamamoto, S.; Tanaka, K.; Kurata, N.; Suzuki, K. Trans-kingdom horizontal DNA transfer from bacteria to yeast is highly plastic due to natural polymorphisms in auxiliary nonessential recipient genes. PLoS ONE 2013, 8, e74590. [Google Scholar] [CrossRef]
- Zoolkefli, F.I.R.M.; Moriguchi, K.; Cho, Y.; Kiyokawa, K.; Yamamoto, S.; Suzuki, K. Isolation and analysis of donor chromosomal genes whose deficiency is responsible for accelerating bacterial and trans-kingdom conjugations by IncP1 T4SS machinery. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Moriguchi, K.; Zoolkefli, F.I.R.M.; Abe, M.; Kiyokawa, K.; Yamamoto, S.; Suzuki, K. Targeting antibiotic resistance genes is a better approach to block acquisition of antibiotic resistance than blocking conjugal transfer by recipient cells: A genome-wide screening in Escherichia coli. Front. Microbiol. 2020, 10, 2939. [Google Scholar] [CrossRef]
- Zhang, P.-Y.; Xu, P.-P.; Xia, Z.-J.; Wang, J.; Xiong, J.; Li, Y.-Z. Combined treatment with the antibiotics kanamycin and streptomycin promotes the conjugation of Escherichia coli. FEMS Microbiol. Lett. 2013, 348, 149–156. [Google Scholar] [CrossRef]
- Quail, M.A.; Guest, J.R. Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR–aceEF–lpd Operon of Escherichia coli. Mol. Microbiol. 1995, 15, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Perham, R.N. Swinging arms and swinging domains in multifunctional enzymes: Catalytic machines for multistep reactions. Annu. Rev. Biochem. 2000, 69, 961–1004. [Google Scholar] [CrossRef] [PubMed]
- Anzai, T.; Imamura, S.; Ishihama, A.; Shimada, T. Expanded roles of pyruvate-sensing PdhR in transcription regulation of the Escherichia coli K-12 genome: Fatty acid catabolism and cell motility. Microb. Genom. 2020, 6, e000442. [Google Scholar] [CrossRef] [PubMed]
- Masai, H.; Arai, K. DnaA- and PriA-dependent primosomes: Two distinct replication complexes for replication of Escherichia coli chromosome. Front. Biosci. 1996, 1, 48–58. [Google Scholar] [CrossRef]
- Alalam, H.; Graf, F.E.; Palm, M.; Abadikhah, M.; Zackrisson, M.; Boström, J.; Fransson, A.; Hadjineophytou, C.; Persson, L.; Stenberg, S.; et al. A high-throughput method for screening for genes controlling bacterial conjugation of aAntibiotic resistance. mSystems 2020, 5. [Google Scholar] [CrossRef]
- Lee, E.H.; Kornberg, A. Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n’ protein. Proc. Natl. Acad. Sci. USA 1991, 88, 3029–3032. [Google Scholar] [CrossRef]

| Strains | Relevant Characteristics | Source or Reference |
|---|---|---|
| E. coli | ||
| Keio collection | An in-frame single-gene knockout mutant collection derived from BW25113, KanR | NBRP Japan |
| BW25113 | F− Δ(araD-araB)567 ΔlacZ4787(∷rrnB-3) λ-rph-1 Δ(rhaD-rhaB)568 hsdR514 | NBRP Japan |
| SY327 (λpir) | Δ(lac pro) argE(Am) recA56 RifR NalR λpir, RifR NalR | NBRP Japan |
| SY327 (λpir) ΔaceE | Δ(lac pro) argE(Am) recA56 RifR NalR λpir ΔaceE, RifR NalR KanR | This study |
| SY327 (λpir) ΔpriA | Δ(lac pro) argE(Am) recA56 RifR NalR λpir ΔpriA, RifR NalR KanR | This study |
| S17-1 (λpir) | F− RP4-2(KanR∷Tn7 TetR∷Mu-1) pro-82 λpir recA1 endA1 thiE1 hsdR17 creC510 | NBRP Japan |
| S. cerevisiae | ||
| BY4742 | MATα SSD1-V his3∆1 leu2∆0 lys2∆0 ura3∆0 | Invitrogen, Carlsbad, CA |
| Plasmids | Relevant Characteristics | Source or Reference |
|---|---|---|
| pJP5603sacBGmR | Mobilizable plasmid; sacB oriT RP4 GenR Used for the construction of E. coli complementation strains | Zoolkefli et al., 2021 [28] |
| pBBR122∆CmR | Derivative of a commercially provided plasmid vector pBBR122; mobpBBR1′ (non-transmissible) KanR ∆ChlR | Moriguchi et al., 2020 [29] |
| RP4 | IncP1α-type conjugative broad-host-range plasmid; KanR TetR AmpR | Pansegrau et al., 1994 [3] |
| pRH220 | Helper plasmid; traRP4 trbRP4 oriTRP4 ori-pSC101 ChlR | * AB526840 |
| pRS316∷oriTP | Mobilizable plasmid; URA3 CEN6/ARSH4 ori-pMB1 AmpR oriTRP4 | Moriguchi et al., 2013 [21] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moriguchi, K.; Nakamura, K.; Takahashi, Y.; Higo-Moriguchi, K.; Kiyokawa, K.; Suzuki, K. Genome-Wide Survey of Donor Chromosomal Genes Involved in Trans-Kingdom Conjugation via the RP4-T4SS Machinery. Microorganisms 2025, 13, 488. https://doi.org/10.3390/microorganisms13030488
Moriguchi K, Nakamura K, Takahashi Y, Higo-Moriguchi K, Kiyokawa K, Suzuki K. Genome-Wide Survey of Donor Chromosomal Genes Involved in Trans-Kingdom Conjugation via the RP4-T4SS Machinery. Microorganisms. 2025; 13(3):488. https://doi.org/10.3390/microorganisms13030488
Chicago/Turabian StyleMoriguchi, Kazuki, Kazuyuki Nakamura, Yudai Takahashi, Kyoko Higo-Moriguchi, Kazuya Kiyokawa, and Katsunori Suzuki. 2025. "Genome-Wide Survey of Donor Chromosomal Genes Involved in Trans-Kingdom Conjugation via the RP4-T4SS Machinery" Microorganisms 13, no. 3: 488. https://doi.org/10.3390/microorganisms13030488
APA StyleMoriguchi, K., Nakamura, K., Takahashi, Y., Higo-Moriguchi, K., Kiyokawa, K., & Suzuki, K. (2025). Genome-Wide Survey of Donor Chromosomal Genes Involved in Trans-Kingdom Conjugation via the RP4-T4SS Machinery. Microorganisms, 13(3), 488. https://doi.org/10.3390/microorganisms13030488





