Description and Genome-Based Analysis of Vibrio chaetopteri sp. nov., a New Species of the Mediterranei Clade Isolated from a Marine Polychaete
Abstract
:1. Introduction
2. Materials and Methods
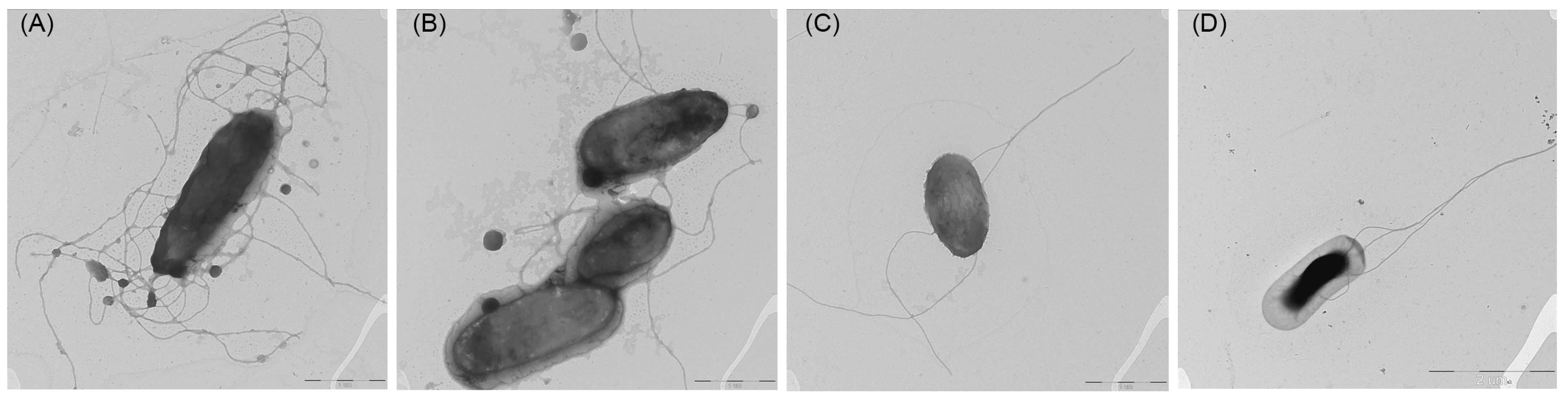
2.1. Isolation and Phenotypic Characterization of Bacteria
2.2. 16S rRNA Gene Sequence Analysis
2.3. Multilocus Sequence Analysis (MLSA)
2.4. Whole-Genome Sequencing and Genome Characterization
3. Results and Discussion
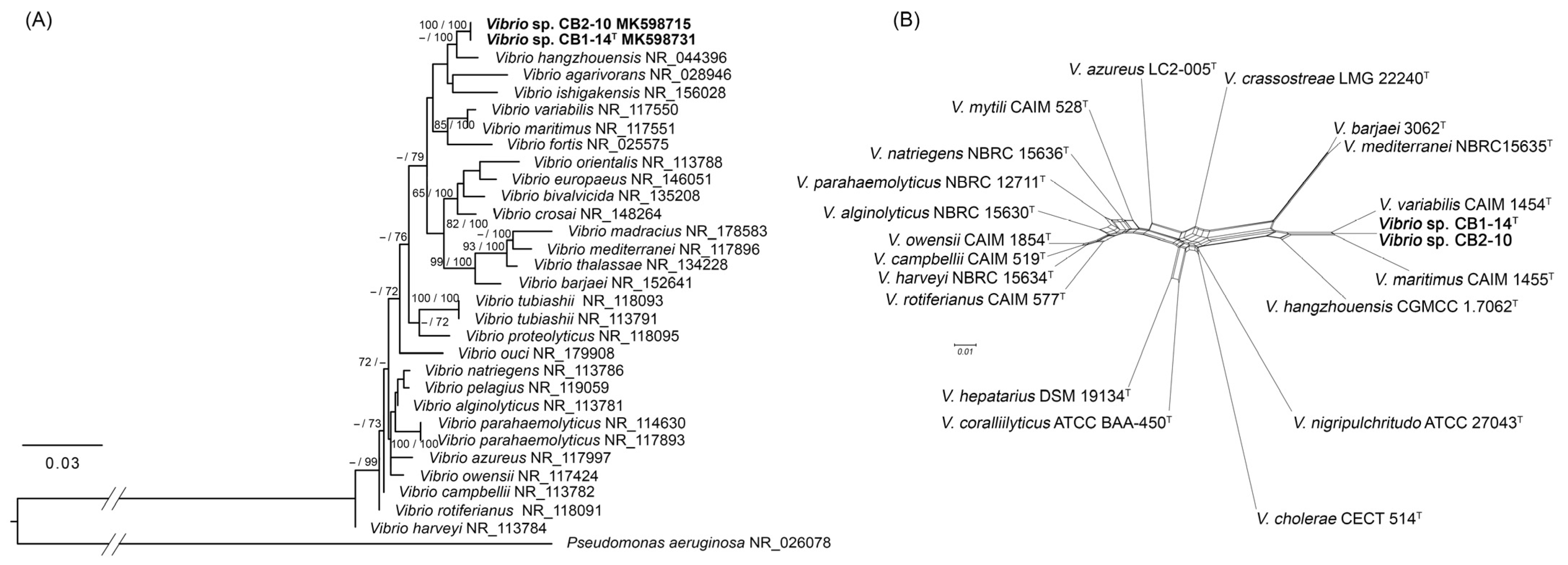
3.1. Phylogenetic Analyses
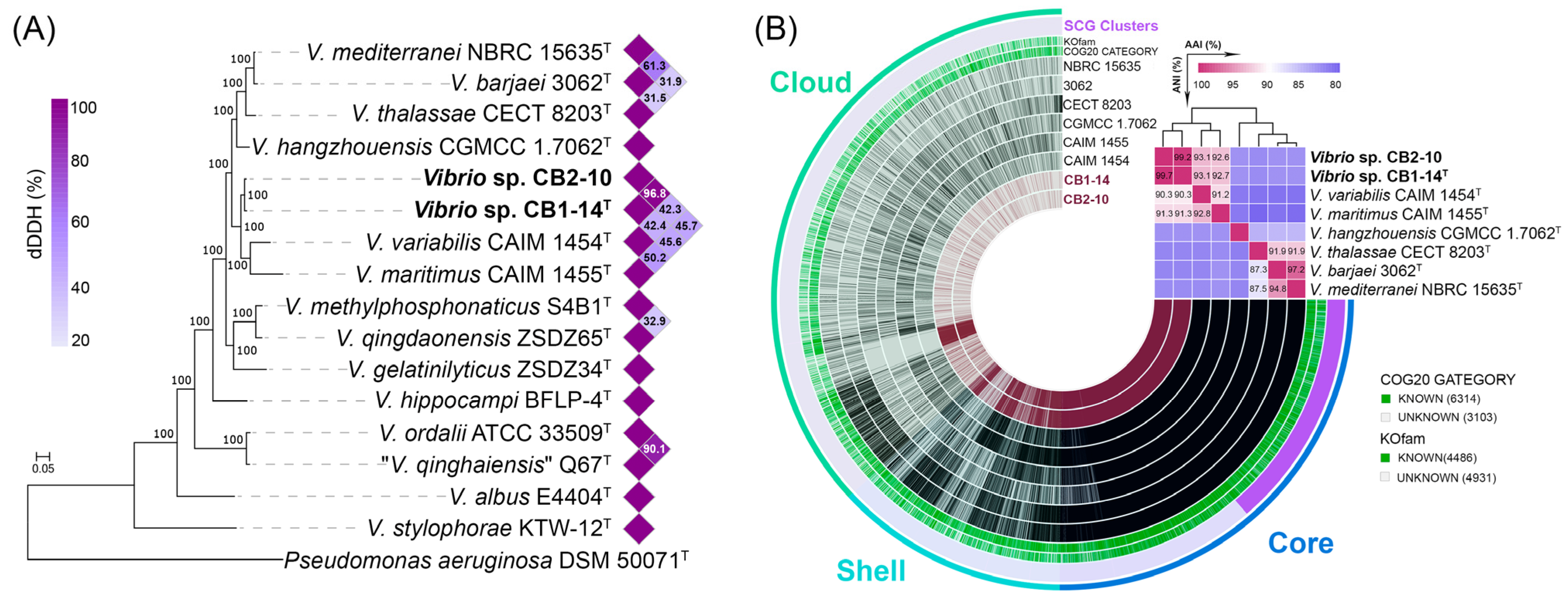
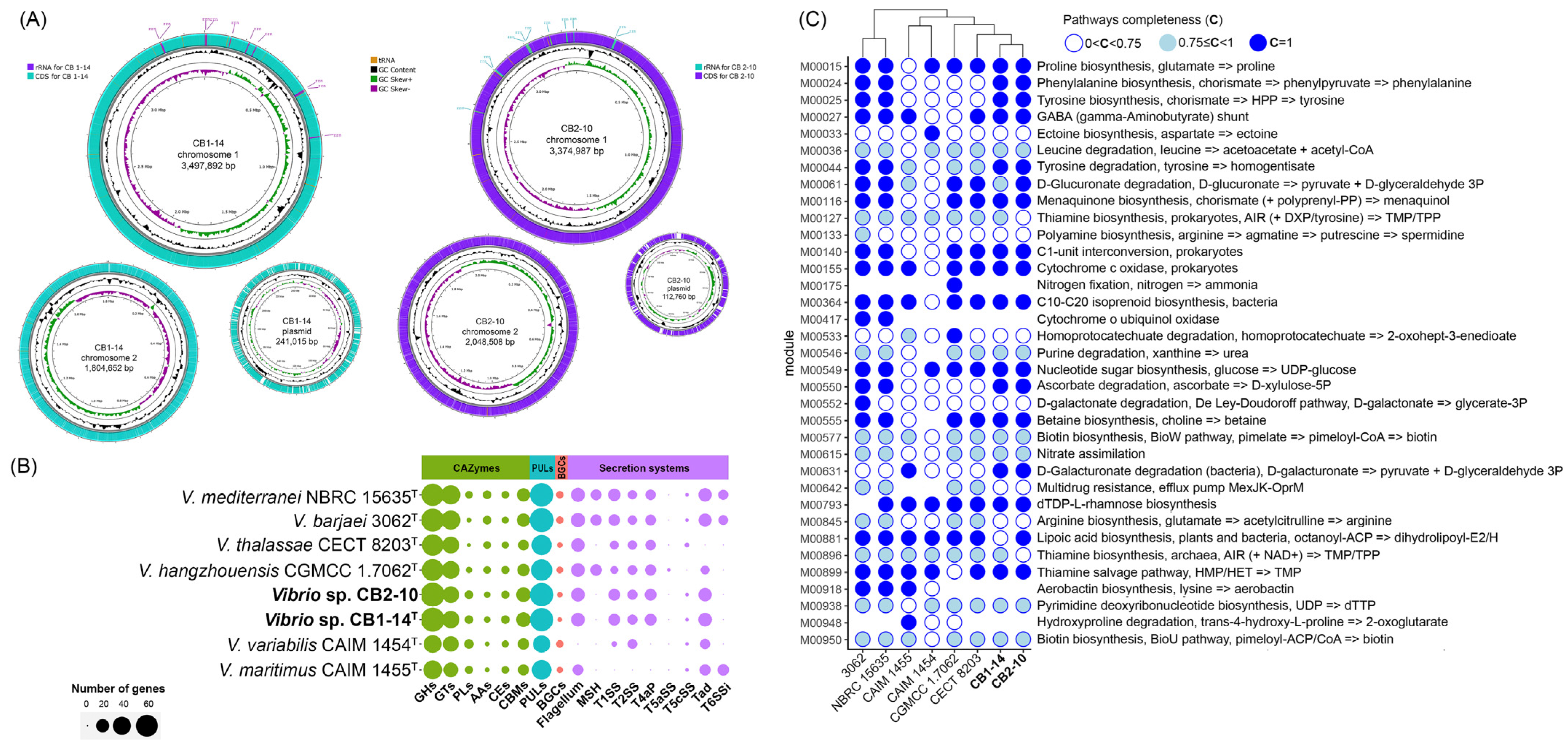
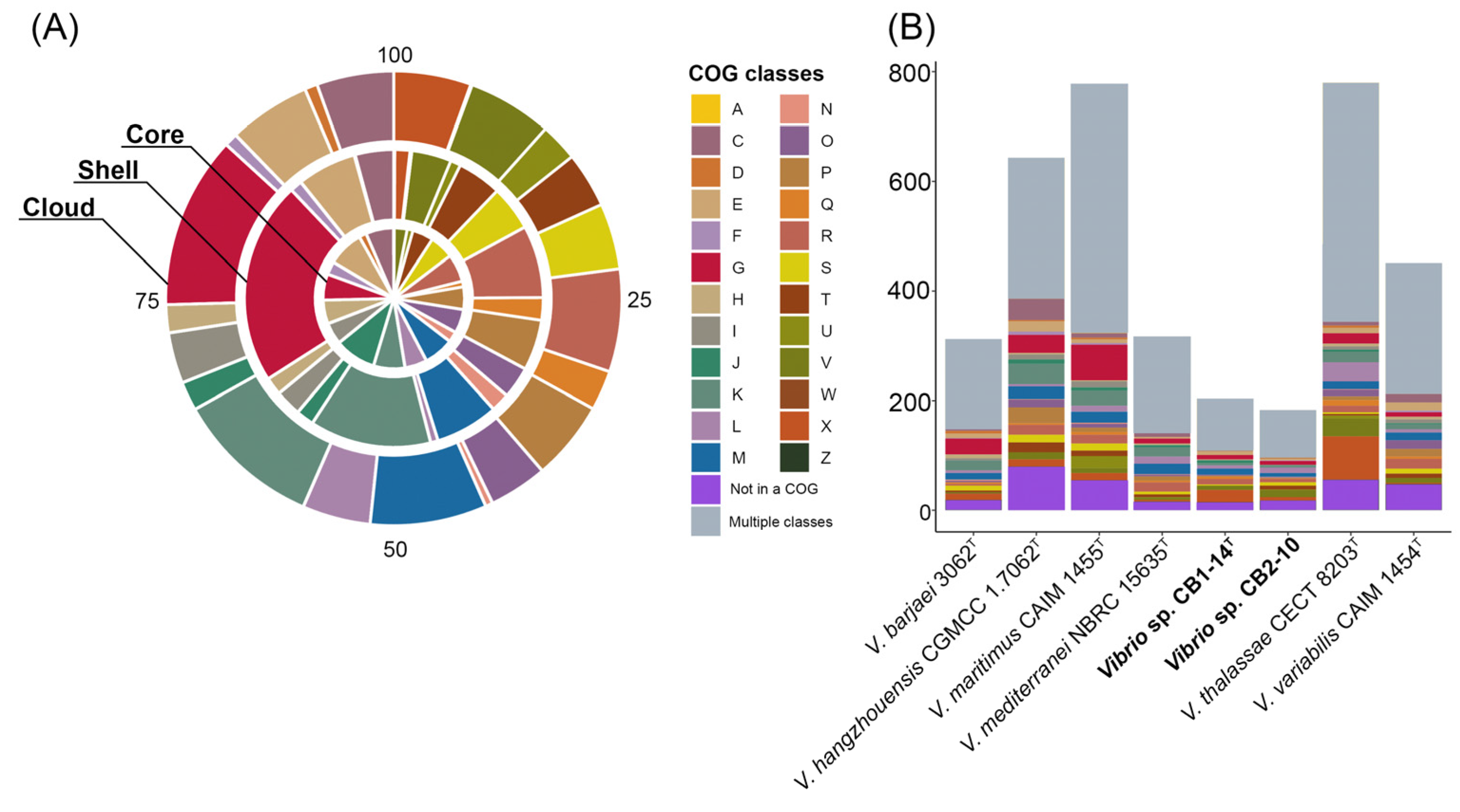
3.2. Genomic Characteristics and Pan-Genome Analysis
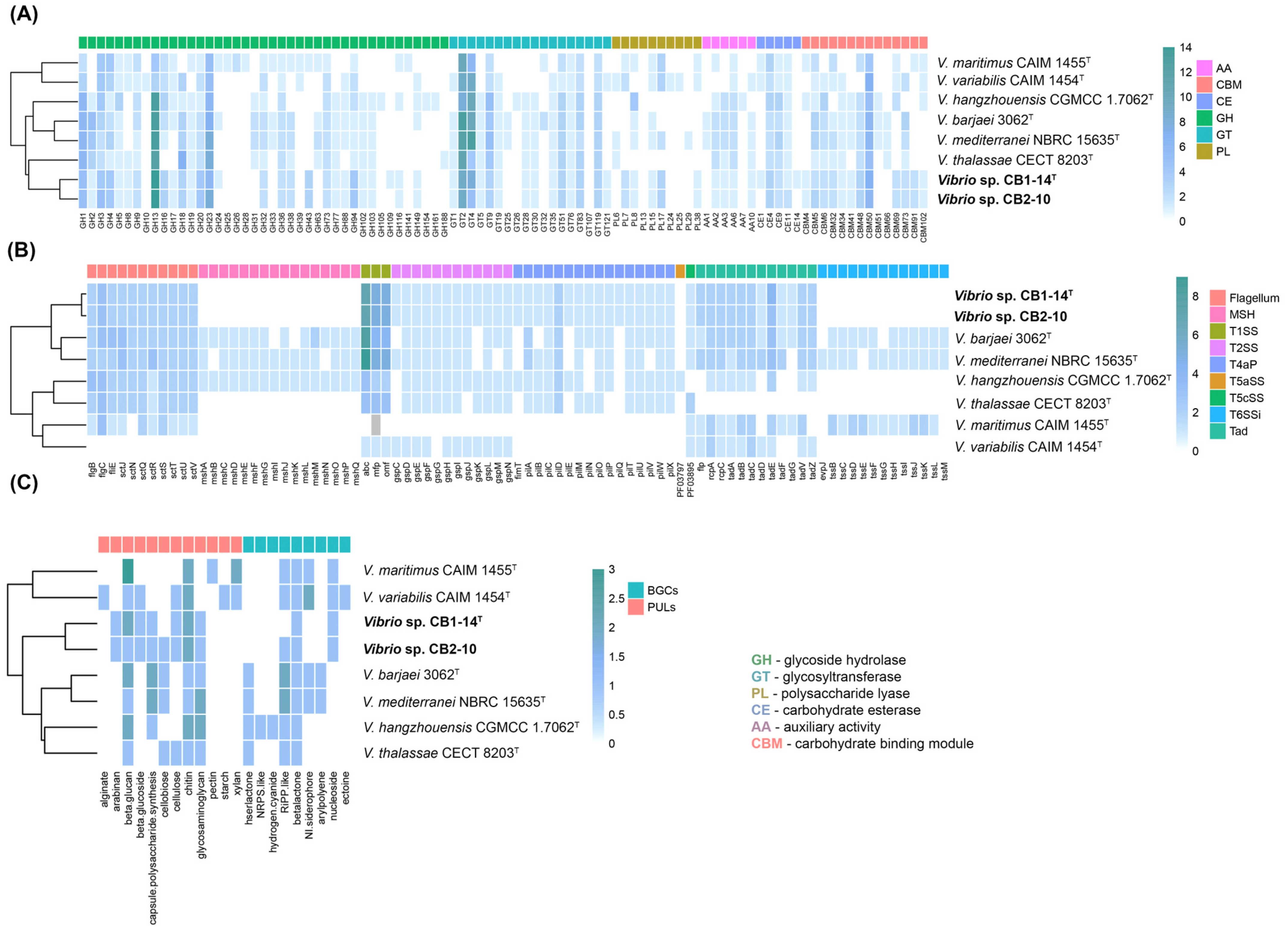
3.3. In Silico Analysis of Hydrolytic and Biosynthetic Potentials
3.4. Phenotypic Characterization and Chemotaxonomy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parte, A.C.; Sarda Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Gil, B.; Thompson, C.C.; Matsumura, Y.; Sawabe, T.; Iida, T.; Christen, R.; Thompson, F.; Sawabe, T. The Family Vibrionaceae. In The Prokaryotes: Gammaproteobacteria; Rosenberg, E., DeLong, E.F., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Farmer, J.J., III; Hickman-Brenner, F.W. The Genera Vibrio and Photobacterium. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 508–563. [Google Scholar]
- Jiang, C.; Tanaka, M.; Nishikawa, S.; Mino, S.; Romalde, J.L.; Thompson, F.L.; Gomez-Gil, B.; Sawabe, T. Vibrio Clade 3.0: New Vibrionaceae evolutionary units using genome-based approach. Curr. Microbiol. 2022, 79, 10. [Google Scholar] [CrossRef] [PubMed]
- Sawabe, T.; Kita-Tsukamoto, K.; Thompson, F.L. Inferring the evolutionary history of Vibrios by means of multilocus sequence analysis. J. Bacteriol. 2007, 189, 7932–7936. [Google Scholar] [CrossRef] [PubMed]
- Sawabe, T.; Ogura, Y.; Matsumura, Y.; Feng, G.; Amin, A.R.; Mino, S.; Nakagawa, S.; Sawabe, T.; Kumar, R.; Fukui, Y.; et al. Updating the Vibrio Clades Defined by Multilocus Sequence Phylogeny: Proposal of Eight New Clades, and the Description of Vibrio tritonius sp. Nov. Front. Microbiol. 2013, 4, 414. [Google Scholar] [CrossRef]
- González-Castillo, A.; Enciso-Ibarra, J.; Gomez-Gil, B. Genomic taxonomy of the Mediterranei clade of the genus Vibrio (Gammaproteobacteria). Antonie Van Leeuwenhoek 2020, 113, 851–859. [Google Scholar] [CrossRef]
- Pujalte, M.J.; Garay, E. Proposal of Vibrio mediterranei sp. nov.: A New Marine Member of the Genus Vibrio. Int. J. Syst. Bact. 1986, 36, 278–281. [Google Scholar] [CrossRef]
- Tarazona, E.; Lucena, T.; Arahal, D.R.; Macián, M.C.; Ruvira, M.A.; Pujalte, M.J. Multilocus sequence analysis of putative Vibrio mediterranei strains and description of Vibrio thalassae sp. nov. Syst. Appl. Microbiol. 2014, 37, 320–328. [Google Scholar] [CrossRef]
- Dubert, J.; Balboa, S.; Regueira, M.; González-Castillo, A.; Gómez-Gil, B.; Romalde, J.L. Vibrio barjaei sp. nov., a new species of the Mediterranei clade isolated in a shellfish hatchery. Syst. Appl. Microbiol. 2016, 39, 553–556. [Google Scholar] [CrossRef]
- Xu, X.W.; Wu, Y.H.; Wang, C.S.; Oren, A.; Wu, M. Vibrio hangzhouensis sp. nov., isolated from sediment of the East China Sea. Int. J. Syst. Evol. Microbiol. 2009, 59, 2099–2103. [Google Scholar] [CrossRef]
- Chimetto, L.A.; Cleenwerck, I.; Moreira, A.P.B.; Brocchi, M.; Willems, A.; De Vos, P.; Thompson, F.L. Vibrio variabilis sp. nov. and Vibrio maritimus sp. nov., isolated from Palythoa caribaeorum. Int. J. System. Evol. Microb. 2011, 61, 3009–3015. [Google Scholar] [CrossRef]
- Makarieva, T.; Shubina, L.; Kurilenko, V.; Isaeva, M.; Chernysheva, N.; Popov, R.; Bystritskaya, E.; Dmitrenok, P.; Stonik, V. Marine bacterium Vibrio sp. CB1-14 produces guanidine alkaloid 6-epi-Monanchorin, previously isolated from marine polychaete and sponges. Mar. Drugs 2019, 17, 213. [Google Scholar] [CrossRef] [PubMed]
- Smibert, R.M.; Krieg, N.R. Phenotypic characterization. In Methods for General and Molecular Bacteriology; Gerhardt, P., Murray, R.G.E., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–655. [Google Scholar]
- Romanenko, L.A.; Kurilenko, V.V.; Guzev, K.V.; Svetashev, V.I. Characterization of Labrenzia polysiphoniae sp. nov. isolated from red alga Polysiphonia sp. Arch. Microbiol. 2019, 201, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.D.; Shah, H.N. Fatty acid, menaquinone and polar lipid composition of Rothia dentosacariosa. Arch. Microbiol. 1984, 137, 247–249. [Google Scholar] [CrossRef]
- Hiraishi, A.; Ueda, Y.; Ishihara, J.; Mori, T. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J. Gen. Appl. Microbiol. 1996, 42, 457–459. [Google Scholar] [CrossRef]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. MIDI; Technical Note #101: Newark, DE, USA, 1990. [Google Scholar]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–147. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021, 7, D801–D807. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genomic. Sci. 2014, 8, 10. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Mol. Biol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Kokoulin, M.S.; Savicheva, Y.V.; Otstavnykh, N.Y.; Kurilenko, V.V.; Meleshko, D.A.; Isaeva, M.P. Structure and Biosynthetic Gene Cluster of Sulfated Capsular Polysaccharide from the Marine Bacterium Vibrio sp. KMM 8419. Int. J. Mol. Sci. 2024, 25, 12927. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 15, 2114–2120. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. BioRxiv 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Asnicar, F.; Thomas, A.M.; Beghini, F.; Mengoni, C.; Manara, S.; Manghi, P.; Zhu, Q.; Bolzan, M.; Cumbo, F.; May, U.; et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11, 2500. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Esen, O.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platformfor ‘omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016, 4, e1900v1. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef]
- Ausland, C.; Zheng, J.; Yi, H.; Yang, B.; Li, T.; Feng, X.; Zheng, B.; Yin, Y. dbCAN-PUL: A database of experimentally characterized CAZyme gene clusters and their substrates. Nucleic Acids Res. 2021, 49, D523–D528. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Neron, B.; Denise, R.; Coluzzi, C.; Touchon, M.; Rocha, E.P.; Abby, S.S. MacSyFinder v2: Improved modelling and search engine to identify molecular systems in genomes. Peer Community J. 2023, 3, e28. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Riesco, R.; Trujillo, M.E. Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2024, 74, 006300. [Google Scholar] [CrossRef]
- Ran, L.; Wang, X.; He, X.; Guo, R.; Wu, Y.; Zhang, P.; Zhang, X.H. Genomic analysis and chitinase characterization of Vibrio harveyi WXL538: Insight into its adaptation to the marine environment. Front. Microbiol. 2023, 14, 1121720. [Google Scholar] [CrossRef]
- Wang, Z.; Robertson, K.L.; Liu, C.; Liu, J.L.; Johnson, B.J.; Leary, D.H.; Compton, J.R.; Vuddhakul, V.; Legler, P.M.; Vora, G.J. A novel Vibrio beta-glucosidase (LamN) that hydrolyzes the algal storage polysaccharide laminarin. FEMS Microbiol. Ecol. 2015, 91, fiv087. [Google Scholar] [CrossRef]
- Senoura, T.; Ito, S.; Taguchi, H.; Higa, M.; Hamada, S.; Matsui, H.; Ozawa, T.; Jin, S.; Watanabe, J.; Wasaki, J.; et al. New microbial mannan catabolic pathway that involves a novel mannosylglucose phosphorylase. Biochem. Biophys. Res. Commun. 2011, 408, 701–706. [Google Scholar] [CrossRef]
- Hehemann, J.H.; Boraston, A.B.; Czjzek, M. A sweet new wave: Structures and mechanisms of enzymes that digest polysaccharides from marine algae. Curr. Opin. Struc. Biol. 2014, 28, 77–86. [Google Scholar] [CrossRef]
- He, X.; Zhang, Y.; Wang, X.; Zhu, X.; Chen, L.; Liu, W.; Lyu, Q.; Ran, L.; Cheng, H.; Zhang, X.H. Characterization of multiple alginate lyases in a highly efficient alginate-degrading vibrio strain and its degradation strategy. Appl. Environ. Microbiol. 2022, 88, e01389–22. [Google Scholar] [CrossRef]
- Kanonenberg, K.; Spitz, O.; Erenburg, I.N.; Beer, T.; Schmitt, L. Type I secretion system—It takes three and a substrate. FEMS Microbiol. Lett. 2018, 365, fny094. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Lopez, M.; Zorgani, M.A.; Kelley, L.A.; Bailly, X.; Kajava, A.V.; Henderson, I.R.; Polticelli, F.; Pizza, M.; Rosini, R.; Desvaux, M. Identification of the autochaperone domain in the type Va secretion system (T5aSS): Prevalent feature of autotransporters with a β-helical passenger. Front. Microbiol. 2018, 8, 2607. [Google Scholar] [CrossRef]
| Feature | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Assembly level | chromosome | chromosome | scaffold | contig | scaffold | contig | contig | contig |
| Genome size (Mb) | 5.5 | 5.5 | 5.1 | 5.6 | 5.5 | 5.1 | 5.8 | 5.1 |
| Number of contigs | 3 | 3 | 207 | 67 | 67 | 60 | 97 | 158 |
| G+C Content (mol%) | 46.1 | 46.1 | 44.5 | 44.0 | 44.0 | 46.5 | 46.0 | 46.5 |
| N50 (Kb) | 3497.9 | 3375.0 | 74.8 | 241.4 | 286.2 | 171.2 | 207.3 | 81.6 |
| L50 | 1 | 1 | 17 | 8 | 7 | 10 | 8 | 16 |
| Coverage | 141.0× | 116.0× | 92.0× | 165× | 100.0× | 246.0× | 31.0× | 17× |
| Total genes | 5015 | 5028 | 4728 | 5217 | 5052 | 4718 | 5523 | 4934 |
| Protein coding genes | 4782 | 4705 | 4635 | 5107 | 4961 | 4631 | 4151 | 3841 |
| rRNAs (5S/16S/23S) | 10/10/10 | 11/11/11 | 6/1/0 | 2/2/2 | 1/1/1 | 2/6/1 | 1/2/1 | 2/4/2 |
| tRNAs | 113 | 110 | 85 | 62 | 39 | 56 | 75 | |
| checkM completeness (%) | 98.62 | 97.61 | 98.51 | 99.62 | 99.61 | 99.55 | 69.68 | 71.75 |
| checkM contamination (%) | 3.15 | 2.78 | 1.82 | 2.04 | 2.60 | 3.79 | 1.55 | 1.48 |
| WGS project/RefSeq | GCF_040412085.2 | GCF_048400125.1 | OANU01 | BCUE01 | LQXO02 | FNVG01 | JYJJ01 | JYJK01 |
| Genome assembly | ASM4041208v1 | ASM4840012v1 | VthalassaeCECT 8203_ Velvet_Prokka | ASM159112v1 | ASM163906v2 | IMG-taxon 2617270906 | ASM326377v1 | ASM326378v1 |
| Feature | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Growth at/in: | ||||||||
| 10 °C | + | + | − | ND | − | − | + | + |
| 28 °C | + | + | + | + | + | + | + | + |
| 32 °C | + | + | + | ND | + | + | + | + |
| 0.5% NaCl | + | + | + | ND | − | + | + | + |
| 3% NaCl | + | + | + | + | + | + | + | + |
| 6% NaCl | + | + | − | + | + | + | + | + |
| pH | 5–11 | 5–11 | 5–10.5 | ND | ND | 6–10 | 5–12 | 6–12 |
| Tyrosine hydrolysis | + | + | + | ND | ND | + | ND | ND |
| Starch hydrolysis | + | + | + | + | + | + | ND | + |
| Gelatin hydrolysis | + | + | − | − | + | + | ND | + |
| Tween 80 hydrolysis | + | + | − | + | + | − | + | + |
| Tween 40 hydrolysis | + | + | ND | ND | ND | ND | ND | ND |
| Tween 20 hydrolysis | + | + | ND | ND | ND | + | ND | ND |
| DNA hydrolysis | + | + | + | + | ND | − | ND | ND |
| H2S production | + | + | + | ND | − | + | − | − |
| Nitrate reduction | + | + | + | ND | ND | − | + | + |
| API 20E tests: | ||||||||
| ONPG | + | + | + | ND | + | ND | + | + |
| Citrate | − | − | − | ND | − | + | − | − |
| Urease | − | − | − | ND | − | − | − | − |
| Glucose | + | + | + | ND | + | ND | + | + |
| Mannitol | + | + | + | ND | + | ND | + | + |
| Inositol | − | w | − | + | ND | ND | + | − |
| Sorbitol | − | − | − | ND | + | ND | − | − |
| L-rhamnose | + | + | − | ND | − | ND | − | + |
| D-sucrose | + | + | − | + | + | + | + | + |
| D-melibiose | − | w | − | ND | − | ND | − | − |
| Amygdalin | + | + | + | ND | + | ND | + | + |
| L-arabinose | − | − | − | ND | − | ND | − | ND |
| Fatty Acid | 1 | 2 | 3 |
|---|---|---|---|
| C12:0 | 3.65 | 3.60 | 2.54 |
| C12:0 3-OH | 2.22 | 1.53 | 1.65 |
| iso-C14:0 | 2.58 | 1.76 | 1.50 |
| C14:0 | 5.67 | 5.50 | 5.56 |
| iso-C15:0 | 0.73 | 1.54 | 0.41 |
| anteiso-C15:0 | 0.51 | 0.27 | 1.28 |
| C15:0 | 0.47 | 0.63 | 4.33 |
| C14:0 3-OH | 2.27 | 1.57 | 2.67 |
| iso-C16:0 | 5.80 | 4.61 | 2.35 |
| C16:1ω9c | 20.34 | 20.95 | 24.89 |
| C16:1ω7c | 17.96 | 18.89 | 5.67 |
| C16:0 | 11.79 | 12.56 | 12.40 |
| C17:1ω8c | 0.34 | 0.46 | 3.62 |
| C18:1ω9c | 9.56 | 8.83 | 16.70 |
| C18:1ω7c | 7.67 | 7.51 | 0.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurilenko, V.; Bystritskaya, E.; Otstavnykh, N.; Velansky, P.; Lichmanuk, D.; Savicheva, Y.; Romanenko, L.; Isaeva, M. Description and Genome-Based Analysis of Vibrio chaetopteri sp. nov., a New Species of the Mediterranei Clade Isolated from a Marine Polychaete. Microorganisms 2025, 13, 638. https://doi.org/10.3390/microorganisms13030638
Kurilenko V, Bystritskaya E, Otstavnykh N, Velansky P, Lichmanuk D, Savicheva Y, Romanenko L, Isaeva M. Description and Genome-Based Analysis of Vibrio chaetopteri sp. nov., a New Species of the Mediterranei Clade Isolated from a Marine Polychaete. Microorganisms. 2025; 13(3):638. https://doi.org/10.3390/microorganisms13030638
Chicago/Turabian StyleKurilenko, Valeriya, Evgenia Bystritskaya, Nadezhda Otstavnykh, Peter Velansky, Darina Lichmanuk, Yulia Savicheva, Lyudmila Romanenko, and Marina Isaeva. 2025. "Description and Genome-Based Analysis of Vibrio chaetopteri sp. nov., a New Species of the Mediterranei Clade Isolated from a Marine Polychaete" Microorganisms 13, no. 3: 638. https://doi.org/10.3390/microorganisms13030638
APA StyleKurilenko, V., Bystritskaya, E., Otstavnykh, N., Velansky, P., Lichmanuk, D., Savicheva, Y., Romanenko, L., & Isaeva, M. (2025). Description and Genome-Based Analysis of Vibrio chaetopteri sp. nov., a New Species of the Mediterranei Clade Isolated from a Marine Polychaete. Microorganisms, 13(3), 638. https://doi.org/10.3390/microorganisms13030638







