Antibiotic Resistance and Virulence Determinants of Pseudomonas aeruginosa Isolates Cultured from Hydrocarbon-Contaminated Environmental Samples
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of P. aeruginosa Strain CHA1
2.2. Whole-Genome Sequencing of P. aeruginosa Strain CHA1
2.3. Detection of Acquired Antibiotic-Resistance Genes
2.4. Searching for Genomic Mutations Causing an Antibiotic-Resistant Phenotype
| Nr. | Strain Code | Country | Sampling Location | Sample Type | NCBI Biosample | Ref. |
|---|---|---|---|---|---|---|
| 1 | CHA1 | Nigeria | Ogoniland, Rivers State | crude oil contaminated soil | SAMN38280573 | this work |
| 2 | PA1-Petro | Brazil | Oilfield in State of Sergipe | oil production water | SAMN20156365 | [40] |
| 3 | CMIP 8.1 | Brazil | Rio de Janeiro | crude oil well | SAMN18912870 | [41] |
| 4 | W-101 | China | Dagang Oil Reservoir | sewage | SAMN09767472 | [42] |
| 5 | 2K-1 | Peru | Talara Oil Refinery | oil-contaminated environment | SAMN29360594 | [43] |
| 6 | 6K-11 | Peru | Talara Oil Refinery | oil-contaminated environment | SAMN29360595 | [43] |
| 7 | ATCC33988 | USA | Ponca City, OK | fuel storage tank | SAMN02767933 | [44] |
| 8 | M8A1 | Colombia | Caño Limon Oilfield | crude oil residual water | SAMN04916455 | [25] |
| 9 | IMP66 | China | Daqing Oilfield | crude oil | SAMN08915456 | [25] |
| 10 | ATCC27853 | USA | Boston Hospital | human blood culture | SAMN29939566 | [45] |
| 11 | M8A4 | Colombia | Caño Limon Oilfield | crude oil residual water | SAMN04916454 | [25] |
| 12 | DQ8 | China | Daqing Oilfield | crude oil polluted soil | SAMN02470947 | [25] |
| 13 | L6-1 | China | Xinjiang Oilfield | oil reservoir production fluid | SAMN04325380 | [46] |
| 14 | 8D | China | Ansai Oilfield | oilfield production water | SAMN27926083 | [47] |
| 15 | PAO1 | Australia | Melbourne | human wound sample | SAMN02603714 | [34,35,36] |
2.5. In Vitro Antibiotic Susceptibility Testing of Strain CHA1
2.6. Detection of P. aeruginosa Virulence Genes
2.7. Protein Sequence Alignment of AlkB1 and AlkB2 Alkane Hydroxylases
2.8. Construction of Phylogenetic Trees from Whole-Genome Sequence Data
2.9. Assessment of the Contribution of P. aeruginosa Efflux Pumps to Hexane Tolerance
3. Results
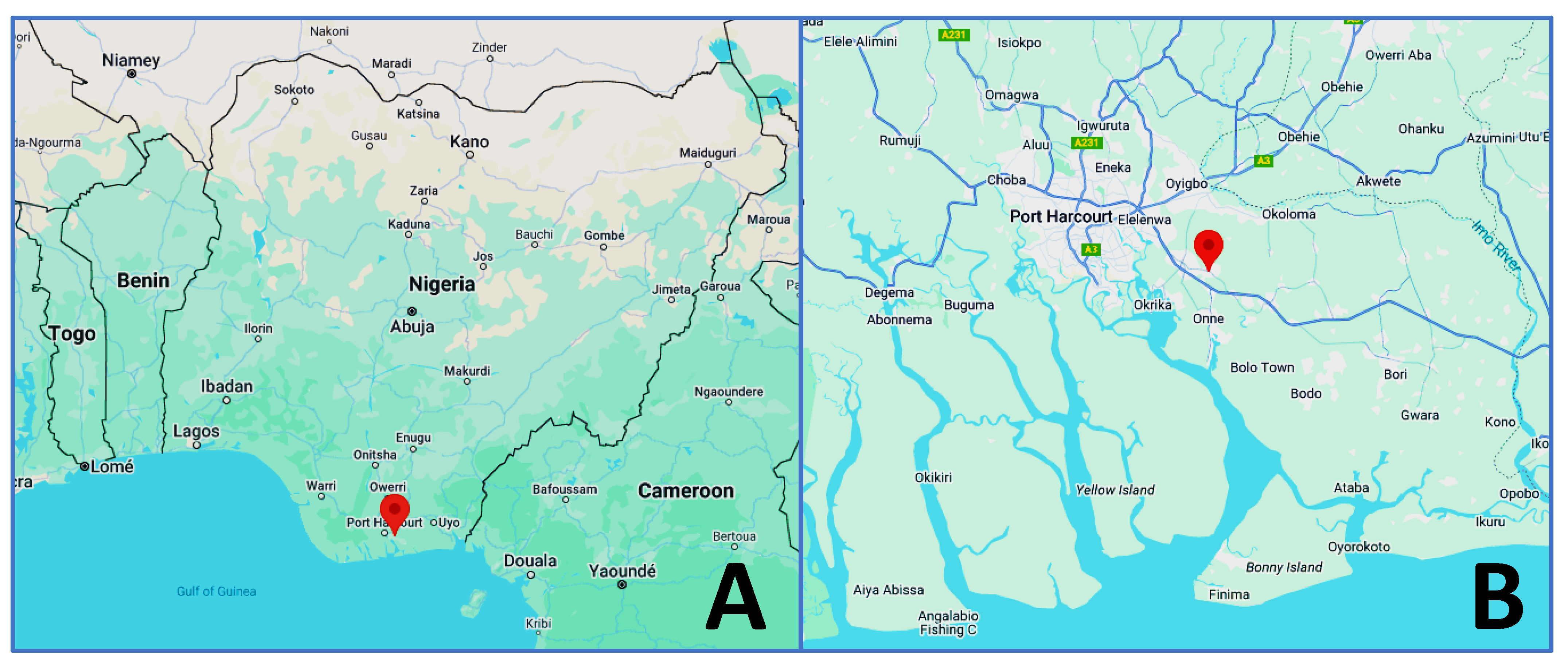
3.1. Characterization of P. aeruginosa Strain CHA1 Cultured from Hydrocarbon-Polluted Soil of the Ejama-Ebubu Community in Nigeria
3.2. In Vitro Antibiotic Susceptibility of P. aeruginosa Strain CHA1
| Testing Method | PIT | CTZ | CEP | IMI | MER | CIP | GEN | TOB | CHL | COL | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Disk diffusion | Inhibitory zone (mm) b | 27 | 20 | 20 | 26 | 25 | 34 | 19 | 23 | 0 | - |
| Interpretation | I | I | R | I | S | I | S | R | |||
| Broth microdilution | MIC (mg/L) | 4 | 2 | 1 | 2 | 1 | 0.25 | 0.5 | 0.5 | 64 | 0.5 |
| Interpretation | I | I | I | I | S | I | WT c | S | R | S | |
| Antibiotic | MIC (mg/L) | ||
|---|---|---|---|
| Ceftazidime | Trimethoprim | Chloramphenicol | |
| −PAβN a | 2 | 128 | 64 |
| +PAβN a | ≤0.25 | ≤8 | ≤2 |
| Fold-change in MIC | ≥8 | ≥16 | ≥32 |
3.3. Assessment of ARGs and Known Genetic Mutations Leading to Antibiotic Resistance in Hydrocarbon-Impacted and Reference P. aeruginosa Strains
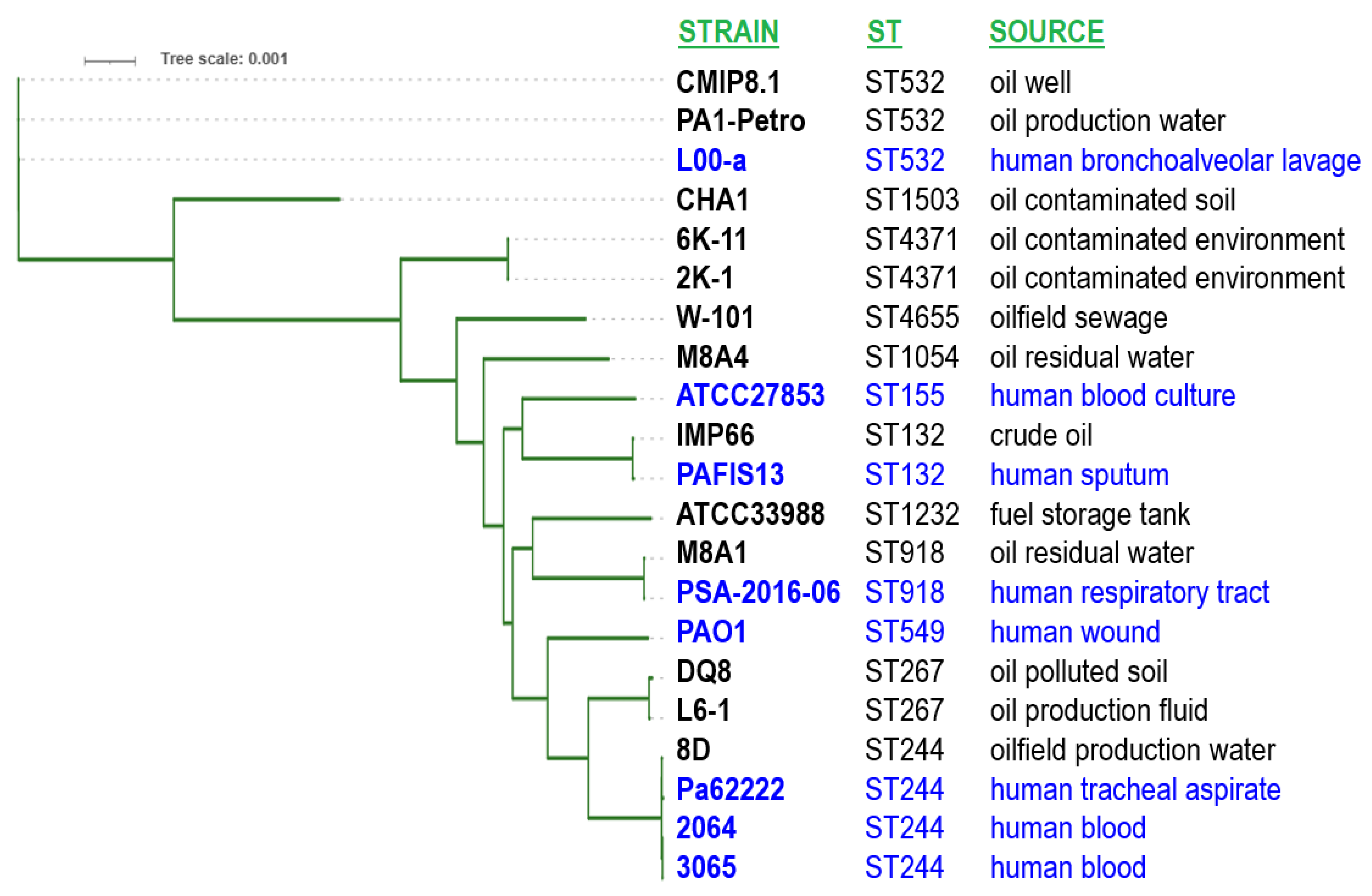
3.4. Phylogenetic Analysis Based on the Whole-Genome Sequence Data
| Strain Code | Country | ST | Sero-Type | OXA-50 Family Variant | catB7 | crpP | aph(3′)-Ia | ampC | ampR | mexR | mexS | mexZ | nalC | nalD | dacB | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PDC-1 | PDC-3 | PDC-5 | PDC-8 | PDC16-L | PDC-59 | PDC-120 | E114A | G283E | M288R | R119C | R244W | FM | V126E | FM | D249N | A75V | E278D | L128M | G71E | S209R | A145V | FM | deletion | A394P | A474T | ||||||||
| PA1-Petro | Brazil | ST532 | O11 | OXA-906 | |||||||||||||||||||||||||||||
| CMIP 8.1 | Brazil | ST532 | O11 | OXA-906 | |||||||||||||||||||||||||||||
| CHA1 | Nigeria | ST1503 | O1 | OXA-1032 | |||||||||||||||||||||||||||||
| W-101 | China | ST4655 | O1 | OXA-494-like | |||||||||||||||||||||||||||||
| 2K-1 | Peru | ST4371 | O11 | OXA-494-like | |||||||||||||||||||||||||||||
| 6K-11 | Peru | ST4371 | O11 | OXA-494-like | |||||||||||||||||||||||||||||
| ATCC33988 | USA | ST1232 | O11 | OXA-50 | |||||||||||||||||||||||||||||
| M8A1 | Colombia | ST918 | O6 | OXA-50-like | |||||||||||||||||||||||||||||
| IMP66 | China | ST132 | O6 | OXA-494 | |||||||||||||||||||||||||||||
| ATCC27853 | USA | ST155 | O6 | OXA-396 | |||||||||||||||||||||||||||||
| M8A4 | Colombia | ST1054 | O6 | OXA-396 | |||||||||||||||||||||||||||||
| DQ8 | China | ST267 | O5 | OXA-1026 | |||||||||||||||||||||||||||||
| L6-1 | China | ST267 | O5 | OXA-1026 | |||||||||||||||||||||||||||||
| 8D | China | ST244 | O5 | OXA-847 | |||||||||||||||||||||||||||||
| PAO1 | Australia | ST549 | O5 | OXA-50 | |||||||||||||||||||||||||||||
3.5. Comparing the Prevalence of Virulence and Antibiotic-Resistance Genes Between Clinical and Environmental Isolates
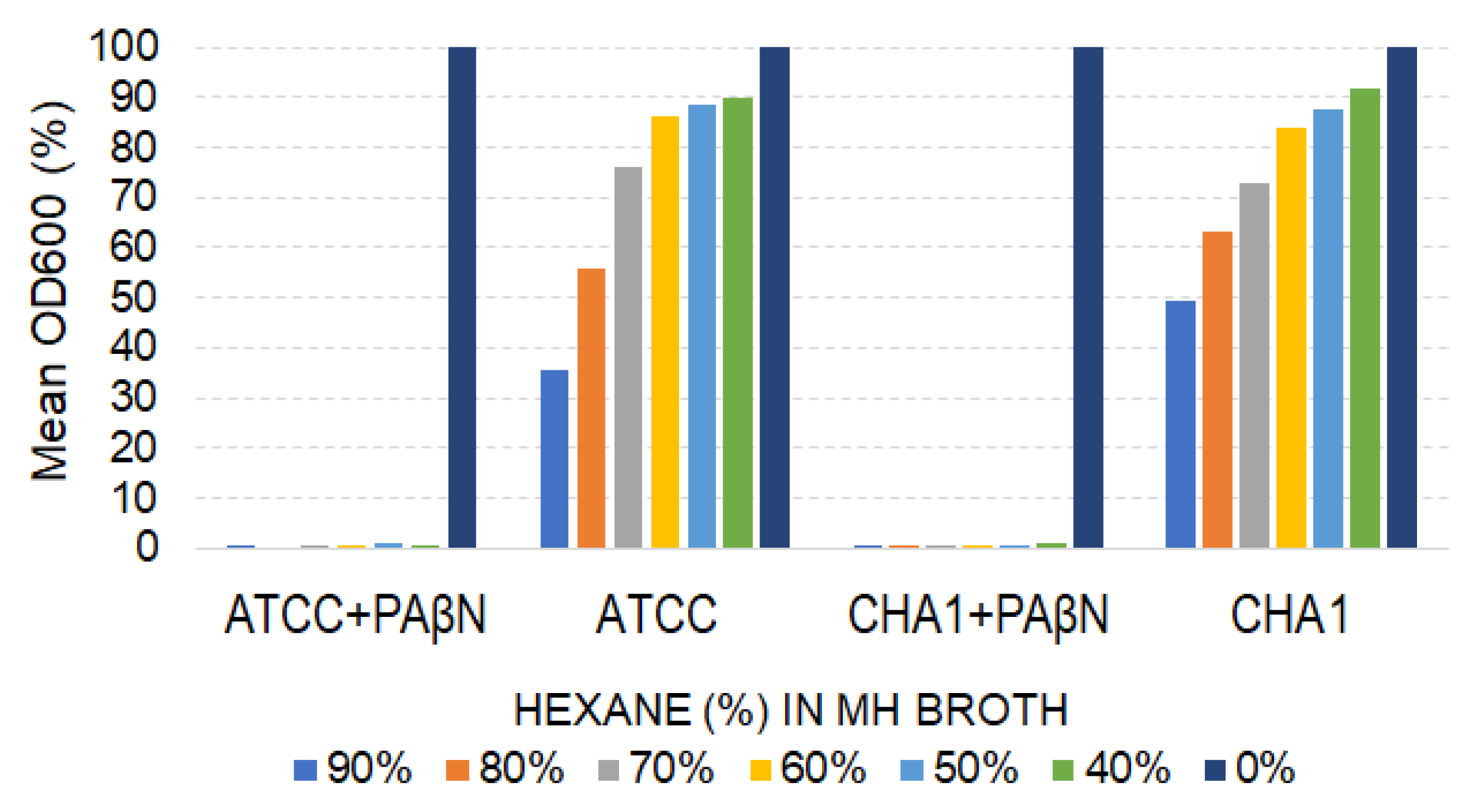
3.6. Contribution of P. aeruginosa Efflux Pumps to Hexane Tolerance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilton, N.; Lyon-Marion, B.A.; Kamath, R.; McVey, K.; Pennell, K.D.; Robbat, A., Jr. Remediation of heavy hydrocarbon impacted soil using biopolymer and polystyrene foam beads. J. Hazard. Mater. 2018, 349, 153–159. [Google Scholar] [CrossRef]
- Chang, S.; Stone, J.; Demes, K.; Piscitelli-Doshkov, M. Consequences of oil spills: A review and framework for informing planning. Ecol. Soc. 2014, 19, 26. [Google Scholar] [CrossRef]
- Michel, J.; Fingas, M. Oil Spills: Causes, consequences, prevention, and countermeasures. In Fossil Fuels: Current Status and Future Directions; World Scientific: Singapore, 2018; pp. 159–201. [Google Scholar]
- Sasu, D.D. Contribution of Oil and Natural Gas Sector to GDP in Nigeria from 4th Quarter of 2018 to the 3rd Quarter of 2022. Available online: https://www.statista.com/topics/6914/oil-industry-in-nigeria/#topicOverview (accessed on 12 March 2025).
- Cayford, S. The Ogoni Uprising: Oil, Human Rights, and a Democratic Alternative in Nigeria. Afr. Today 1996, 43, 183–197. [Google Scholar]
- Jaja, J.; Obuah, E. The politics of the Ogoni clean-up: Challenges and prospects. Afr. Res. Rev. 2019, 13, 101. [Google Scholar] [CrossRef]
- Sokolo, R.; Atagana, H.; Akani, N. Molecular Characterisation of Culturable Aerobic Hydrocarbon Utilising Bacteria and Fungi in Oil Polluted Soil at Ebubu-Ejama Community, Eleme, Rivers State, Nigeria. J. Adv. Biol. Biotechnol. 2018, 18, 1–7. [Google Scholar] [CrossRef]
- Naku, D. Shell Confirms Fresh Oil Spill in Rivers Community. Available online: https://punchng.com/just-in-shell-confirms-fresh-oil-spill-in-rivers-community (accessed on 23 January 2025).
- United Nations Environment Programme (UNEP). Ogoniland Oil Assessment Reveals Extent of Environmental Contamination and Threats to Human Health. 2017. Available online: https://www.unep.org/news-and-stories/story/unep-ogoniland-oil-assessment-reveals-extent-environmental-contamination-and (accessed on 23 January 2025).
- United Nations Environment Programme (UNEP). Environmental Assessment of Ogoniland. Available online: https://wedocs.unep.org/xmlui/bitstream/handle/20.500.11822/22169/EA_Ogoniland_ES.pdf?sequence=1 (accessed on 23 January 2025).
- Sam, K.; Zabbey, N.; Vincent-Akpu, I.F.; Komi, G.; Onyagbodor, P.O.; Babatunde, B.B. Socio-economic baseline for oil-impacted communities in Ogoniland: Towards a restoration framework in Niger Delta, Nigeria. Environ. Sci. Pollut. Res. 2024, 31, 25671–25687. [Google Scholar] [CrossRef]
- Prince, M.; Igbuku, A. Challenges and Prospect of Environmental Remediation/Restoration in Niger Delta of Nigeria: The Case of Ogoniland. J. Energy Technol. Policy 2015, 5, 1–7. [Google Scholar]
- Akani, N. Physicochemical and Microbiological Assessment of Oil-impacted Freshwater Swamp Vegetation in Ejama-Ebubu in Rivers State -a case study. Curr. Stud. Comp. Educ. Sci. Technol. 2014, 1, 146–155. [Google Scholar]
- Akomah-Abadaike, O.; Abu, G. Isolation and characterization of microorganisms of Ejamah-Ebubu oil spill site. Global J. Pure Appl. Sci. 2018, 24, 99. [Google Scholar] [CrossRef]
- Akomah, O.N.; Abu, G.O. Physicochemical Characteristics and Mycoremediation of Ejamah-Ebubu Oil Spill Site located at Eleme Local Government Area in Rivers State, Nigeria. J. Appl. Sci. Environ. Manag. 2018, 22, 3–6. [Google Scholar] [CrossRef]
- Van Beilen, J.B.; Witholt, B. Alkane degradation by Pseudomonads. In Pseudomonas; Ramos, J.L., Ed.; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2004; Volume 3, pp. 397–423. [Google Scholar]
- Norman, R.S.; Moeller, P.; McDonald, T.J.; Morris, P.J. Effect of pyocyanin on a crude-oil-degrading microbial community. Appl. Environ. Microbiol. 2004, 70, 4004–4011. [Google Scholar] [CrossRef] [PubMed]
- Hamamura, N.; Fukui, M.; Ward, D.M.; Inskeep, W.P. Assessing soil microbial populations responding to crude-oil amendment at different temperatures using phylogenetic, functional gene (alkB) and physiological analyses. Environ. Sci. Technol. 2008, 42, 7580–7586. [Google Scholar] [CrossRef] [PubMed]
- Norman, R.S.; Frontera-Suau, R.; Morris, P.J. Variability in Pseudomonas aeruginosa lipopolysaccharide expression during crude oil degradation. Appl. Environ. Microbiol. 2002, 68, 5096–5103. [Google Scholar] [CrossRef]
- Cai, M.; Nie, Y.; Chi, C.Q.; Tang, Y.Q.; Li, Y.; Wang, X.B.; Liu, Z.S.; Yang, Y.; Zhou, J.; Wu, X.L. Crude oil as a microbial seed bank with unexpected functional potentials. Sci. Rep. 2015, 5, 16057. [Google Scholar] [CrossRef]
- Wolfgang, M.C.; Kulasekara, B.R.; Liang, X.; Boyd, D.; Wu, K.; Yang, Q.; Miyada, C.G.; Lory, S. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2003, 100, 8484–8489. [Google Scholar] [CrossRef] [PubMed]
- Libisch, B.; Balogh, B.; Füzi, M. Identification of two multidrug-resistant Pseudomonas aeruginosa clonal lineages with a countrywide distribution in Hungary. Curr. Microbiol. 2009, 58, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Rojo, F.; Martínez, J.L. Oil Degraders as Pathogens. In Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3293–3303. [Google Scholar]
- Libisch, B. N-Alkane Assimilation by Pseudomonas aeruginosa and Its Interactions with Virulence and Antibiotic Resistance. Antibiotics 2024, 13, 1028. [Google Scholar] [CrossRef]
- Xu, A.; Wang, D.; Ding, Y.; Zheng, Y.; Wang, B.; Wei, Q.; Wang, S.; Yang, L.; Ma, L.Z. Integrated Comparative Genomic Analysis and Phenotypic Profiling of Pseudomonas aeruginosa Isolates from Crude Oil. Front. Microbiol. 2020, 11, 519. [Google Scholar] [CrossRef] [PubMed]
- Keresztény, T.; Libisch, B.; Orbe, S.C.; Nagy, T.; Kerényi, Z.; Kocsis, R.; Posta, K.; Papp, P.P.; Olasz, F. Isolation and characterization of lactic acid bacteria with probiotic attributes from different parts of the gastrointestinal tract of free-living wild boars in Hungary. Probiotics Antimicrob. Proteins 2023, 16, 1221–1239. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Thrane, S.W.; Taylor, V.L.; Lund, O.; Lam, J.S.; Jelsbak, L. Application of Whole-Genome Sequencing Data for O-Specific Antigen Analysis and In Silico Serotyping of Pseudomonas aeruginosa Isolates. J. Clin. Micobiol. 2016, 54, 1782–1788. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.H.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antibiotic Resistance Genes. 2016. Available online: https://github.com/tseemann/abricate (accessed on 1 September 2024).
- Libisch, B.; Abdulkadir, S.; Keresztény, T.; Papp, P.P.; Olasz, F.; Fébel, H.; Sándor, Z.J.; Rasschaert, G.; Lambrecht, E.; Heyndrickx, M.; et al. Detection of acquired antibiotic resistance genes in domestic pig (Sus scrofa) and common carp (Cyprinus carpio) intestinal samples by metagenomics analyses in Hungary. Antibiotics 2022, 11, 1441. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; De Cesare, A.; Hilbert, F.; Lindqvist, R.; et al. Statement on how to interpret the QPS qualification on ‘acquired antimicrobial resistance genes’. EFSA J. 2023, 21, e08323. [Google Scholar]
- Holloway, B.W. Genetic recombination in Pseudomonas aeruginosa. Microbiology 1955, 13, 572–581. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Chandler, C.E.; Horspool, A.M.; Hill, P.J.; Wozniak, D.J.; Schertzer, J.W.; Rasko, D.A.; Ernst, R.K. Genomic and phenotypic diversity among ten laboratory isolates of Pseudomonas aeruginosa PAO1. J. Bacteriol. 2019, 201, 10–1128. [Google Scholar] [CrossRef]
- López-Causapé, C.; Cabot, G.; del Barrio-Tofiño, E.; Oliver, A. The versatile mutational resistome of Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 685. [Google Scholar] [CrossRef]
- del Barrio-Tofiño, E.; López-Causapé, C.; Cabot, G.; Rivera, A.; Benito, N.; Segura, C.; Montero, M.M.; Sorlí, L.; Tubau, F.; Gómez-Zorrilla, S.; et al. Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob. Agents Chemother. 2017, 61, 10–1128. [Google Scholar] [CrossRef]
- Cortes-Lara, S.; del Barrio-Tofiño, E.; López-Causapé, C.; Oliver, A.; GEMARA-SEIMC/REIPI Pseudomonas Study Group. Predicting Pseudomonas aeruginosa susceptibility phenotypes from whole genome sequence resistome analysis. Clin. Microbiol. Infect. 2021, 27, 1631–1637. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, H.L.; Dias, G.M.; Neves, B.C. Genome sequence of Pseudomonas aeruginosa PA1-Petro-A role model of environmental adaptation and a potential biotechnological tool. Heliyon 2022, 8, e11566. [Google Scholar] [CrossRef]
- Paixao, C.T.M.; Gomes, M.B.; Hidalgo, K.J.; Valoni-Romao, E.A.; Martins, L.F.; Oliveira, V.M. Genomics and in silico prospecting of biosurfactants from bacteria in the oil industry. In Abstract Book of the 8th International Symposium on Applied Microbiology and Molecular Biology in Oil Systems, Proceedings of the ISMOS TSC, Virtual Symposium, 8–11 June 2021; Caffrey, S., Biwen An, A., Lund Skovhus, T., Whitby, C., Eds.; ISMOS: Nashville, TN, USA, 2021; p. 45. [Google Scholar]
- Lin, S.; Chen, S.; Li, L.; Cao, H.; Li, T.; Hu, M.; Liao, L.; Zhang, L.-H.; Xu, Z. Genome characterization of a uropathogenic Pseudomonas aeruginosa isolate PA_HN002 with cyclic di-GMP-dependent hyper-biofilm production. Front. Cell. Infect. Microbiol. 2022, 12, 956445. [Google Scholar] [CrossRef]
- Palomino, R.A.; Romero, G.; González-Valdez, A.; Soberón-Chávez, G.M.; Gutiérrez, S.; Merino, F.A. Presencia de genes rhlAB, rhlR y rhlC en Pseudomonas aeruginosa nativas sobreproductoras de ramnolípidos. Rev. Peru. Biol. 2017, 24, 293–302. [Google Scholar] [CrossRef]
- Brown, L.M.; Gunasekera, T.S.; Ruiz, O.N. Draft Genome Sequence of Pseudomonas aeruginosa ATCC 33988, a Bacterium Highly Adapted to Fuel-Polluted Environments. Genome Announc. 2014, 2, e01113-14. [Google Scholar] [CrossRef] [PubMed]
- Reller, L.B.; Schoenknecht, F.D.; Kenny, M.A.; Sherris, J.C. Antibiotic susceptibility testing of Pseudomonas aeruginosa: Selection of a control strain and criteria for magnesium and calcium content in media. J. Infect. Dis. 1974, 130, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Wu, Y.; Wang, Q.; Zheng, M.; Cui, Q. Glycerol or crude glycerol as substrates make Pseudomonas aeruginosa achieve anaerobic production of rhamnolipids. Microb. Cell Fact. 2021, 20, 185. [Google Scholar] [CrossRef]
- Xue, S.; Zhao, Y.; Zhou, C.; Zhang, G.; Chen, F.; Wang, S. Improving oil recovery of the heterogeneous low permeability reservoirs by combination of polymer hydrolysis polyacrylamide and two highly biosurfactant-producing bacteria. Sustainability 2021, 14, 423. [Google Scholar] [CrossRef]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing, Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 15.0, Valid from 1 January 2025. 2025. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_15.0_Breakpoint_Tables.pdf (accessed on 6 February 2025).
- Winkler, M.L.; Papp-Wallace, K.M.; Hujer, A.M.; Domitrovic, T.N.; Hujer, K.M.; Hurless, K.N.; Tuohy, M.; Hall, G.; Bonomo, R.A. Unexpected challenges in treating multidrug-resistant Gram-negative bacteria: Resistance to ceftazidime-avibactam in archived isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 59, 1020–1029. [Google Scholar] [CrossRef]
- Li, X.; Zhou, L.; Lei, T.; Zhang, X.; Yao, J.; He, J.; Liu, H.; Cai, H.; Ji, J.; Zhu, Y.; et al. Genomic epidemiology and ceftazidime-avibactam high-level resistance mechanisms of Pseudomonas aeruginosa in China from 2010 to 2022. Emerg. Microbes Infect. 2024, 13, 2324068. [Google Scholar] [CrossRef]
- Du, S.J.; Kuo, H.C.; Cheng, C.H.; Fei, A.C.Y.; Wei, H.W.; Chang, S.K. Molecular mechanisms of ceftazidime resistance in Pseudomonas aeruginosa isolates from canine and human infections. Vet. Med. 2010, 55, 172–182. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33 (Suppl. 1), D325–D328. [Google Scholar] [CrossRef]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; Van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Mao, G.; Wang, Y.; Bartlam, M. Structural insights into diversity and n-alkane biodegradation mechanisms of alkane hydroxylases. Front. Microbiol. 2013, 4, 58. [Google Scholar] [CrossRef]
- Smits, T.H.; Witholt, B.; van Beilen, J.B. Functional characterization of genes involved in alkane oxidation by Pseudomonas aeruginosa. Antonie Van Leeuwenhoek 2003, 84, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Libisch, B.; Poirel, L.; Lepsanovic, Z.; Mirovic, V.; Balogh, B.; Pászti, J.; Hunyadi, Z.; Dobák, A.; Füzi, M.; Nordmann, P. Identification of PER-1 extended-spectrum β-lactamase producing Pseudomonas aeruginosa clinical isolates of the international clonal complex CC11 from Hungary and Serbia. FEMS Immunol. Med. Microbiol. 2008, 54, 330–338. [Google Scholar] [CrossRef]
- White, P.A.; Stokes, H.W.; Bunny, K.L.; Hall, R.M. Characterisation of a chloramphenicol acetyltransferase determinant found in the chromosome of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 1999, 175, 27–35. [Google Scholar] [CrossRef]
- Zubyk, H.L.; Wright, G.D. CrpP is not a fluoroquinolone-inactivating enzyme. Antimicrob. Agents Chemother. 2021, 65, 10–1128. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.M.; Poirel, L.; Nordmann, P. Extended-spectrum cephalosporinases in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009, 53, 1766–1771. [Google Scholar] [CrossRef]
- Girlich, D.; Naas, T.; Nordmann, P. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2004, 48, 2043–2048. [Google Scholar] [CrossRef]
- Tauch, A.; Krieft, S.; Kalinowski, J.; Pühler, A. The 51,409-bp R-plasmid pTP10 from the multiresistant clinical isolate Corynebacterium striatum M82B is composed of DNA segments initially identified in soil bacteria and in plant, animal, and human pathogens. Mol. Gen. Genet. 2000, 263, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, O.N.; Brown, L.M.; Striebich, R.C.; Smart, C.E.; Bowen, L.L.; Lee, J.S.; Little, B.; Mueller, S.; Gunasekera, T. Effect of conventional and alternative fuels on a marine bacterial community and the significance to bioremediation. Energy Fuels 2016, 30, 434–444. [Google Scholar] [CrossRef]
- Gunasekera, T.S.; Bowen, L.L.; Zhou, C.E.; Howard-Byerly, S.C.; Foley, W.S.; Striebich, R.C.; Dugan, L.C.; Ruiz, O.N. Transcriptomic analyses elucidate adaptive differences of closely related strains of Pseudomonas aeruginosa in fuel. Appl. Environ. Microbiol. 2017, 83, e03249-16. [Google Scholar] [CrossRef]
- Li, X.Z.; Poole, K. Organic solvent-tolerant mutants of Pseudomonas aeruginosa display multiple antibiotic resistance. Can. J. Microbiol. 1999, 45, 18–22. [Google Scholar] [CrossRef]
- Fraud, S.; Campigotto, A.J.; Chen, Z.; Poole, K. MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa: Involvement in chlorhexidine resistance and induction by membrane-damaging agents dependent upon the AlgU stress response sigma factor. Antimicrob Agents Chemother 2008, 52, 4478–4482. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Rojo-Molinero, E.; Arca-Suarez, J.; Beşli, Y.; Bogaerts, P.; Cantón, R.; Cimen, C.; Croughs, P.D.; Denis, O.; Giske, C.G.; et al. Pseudomonas aeruginosa antimicrobial susceptibility profiles, resistance mechanisms and international clonal lineages: Update from ESGARS-ESCMID/ISARPAE Group. Clin. Microbiol. Infect. 2024, 30, 469–480. [Google Scholar] [CrossRef]
- Masuda, N.; Sakagawa, E.; Ohya, S.; Gotoh, N.; Tsujimoto, H.; Nishino, T. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000, 44, 3322–3327. [Google Scholar] [CrossRef]
- Esquisabel, A.C.; Rodríguez, M.C.; Campo-Sosa, A.O.; Rodriguez, C.; Martínez-Martínez, L. Mechanisms of resistance in clinical isolates of Pseudomonas aeruginosa less susceptible to cefepime than to ceftazidime. Clin. Microbiol. Infect. 2011, 17, 1817–1822. [Google Scholar] [CrossRef]
- Cao, H.; Xia, T.; Li, Y.; Xu, Z.; Bougouffa, S.; Lo, Y.K.; Bajic, V.B.; Luo, H.; Woo, P.C.Y.; Yan, A. Uncoupled quorum sensing modulates the interplay of virulence and resistance in a multidrug-resistant clinical Pseudomonas aeruginosa isolate belonging to the MLST550 clonal complex. Antimicrob. Agents Chemother. 2019, 63, 10–1128. [Google Scholar] [CrossRef]
- Horna, G.; López, M.; Guerra, H.; Saénz, Y.; Ruiz, J. Interplay between MexAB-OprM and MexEF-OprN in clinical isolates of Pseudomonas aeruginosa. Sci. Rep. 2018, 8, 16463. [Google Scholar] [CrossRef]
- Ai, L.; Mei, F.; Peng, R. An Overview of the Role of Membrane Proteins in Microbial Solvents Tolerance. Protein Pept. Lett. 2023, 30, 137–145. [Google Scholar] [PubMed]
- Suresh, M.; Nithya, N.; Jayasree, P.R.; Vimal, K.P.; Manish Kumar, P.R. Mutational analyses of regulatory genes, mexR, nalC, nalD and mexZ of mexAB-oprM and mexXY operons, in efflux pump hyperexpressing multidrug-resistant clinical isolates of Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 2018, 34, 83. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.P.; Xu, Y.H.; Wang, Z.X.; Fang, Y.P.; Shen, J.L. Overexpression of MexAB-OprM efflux pump in carbapenem-resistant Pseudomonas aeruginosa. Arch. Microbiol. 2016, 198, 565–571. [Google Scholar] [CrossRef]
- Hocquet, D.; Nordmann, P.; El Garch, F.; Cabanne, L.; Plésiat, P. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2006, 50, 1347–1351. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Zhang, L.; Poole, K. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J. Bacteriol. 1998, 180, 2987–2991. [Google Scholar] [CrossRef]
- del Barrio-Tofiño, E.; López-Causapé, C.; Oliver, A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob. Agent. 2020, 56, 106196. [Google Scholar] [CrossRef]
- Campa, M.F.; Wolfe, A.K.; Techtmann, S.M.; Harik, A.M.; Hazen, T.C. Unconventional oil and gas energy systems: An unidentified hotspot of antimicrobial resistance? Front. Microbiol. 2019, 10, 2392. [Google Scholar] [CrossRef]
- Mack, A.R.; Hujer, A.M.; Mojica, M.F.; Taracila, M.A.; Feldgarden, M.; Haft, D.H.; Klimke, W.; Prasad, A.B.; Bonomo, R.A. β-Lactamase diversity in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2025, 69, e00785-24. [Google Scholar] [CrossRef]
- Partridge, S.R.; Brown, H.J.; Stokes, H.W.; Hall, R.M. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 2001, 45, 1263–1270. [Google Scholar] [CrossRef]
- Maurya, A.P.; Rajkumari, J.; Pandey, P. Enrichment of antibiotic resistance genes (ARGs) in polyaromatic hydrocarbon–contaminated soils: A major challenge for environmental health. Environ. Sci. Pollut. Res. 2021, 28, 12178–12189. [Google Scholar] [CrossRef]
- Wong, M.H.; Minkina, T.; Vasilchenko, N.; Sushkova, S.; Delegan, Y.; Ranjan, A.; Saxena, P.; Tarigholizadeh, S.; Dudnikova, T.; Barbashev, A.; et al. Assessment of antibiotic resistance genes in soils polluted by chemical and technogenic ways with poly-aromatic hydrocarbons and heavy metals. Environ. Res. 2024, 252, 118949. [Google Scholar] [CrossRef] [PubMed]
- Furneri, P.M.; Garozzo, A.; Musumarra, M.P.; Scuderi, A.C.; Russo, A.; Bonfiglio, G. Effects on adhesiveness and hydrophobicity of sub-inhibitory concentrations of netilmicin. Int. J. Antimicrob. Agents 2003, 22, 164–167. [Google Scholar] [CrossRef]
- Fonseca, A.; Sousa, J. Effect of antibiotic-induced morphological changes on surface properties, motility and adhesion of nosocomial Pseudomonas aeruginosa strains under different physiological states. J. Appl. Microbiol. 2007, 103, 1828–1837. [Google Scholar] [CrossRef]
- Cunningham, C.J.; Kuyukina, M.S.; Ivshina, I.B.; Konev, A.I.; Peshkur, T.A.; Knapp, C.W. Potential risks of antibiotic resistant bacteria and genes in bioremediation of petroleum hydrocarbon contaminated soils. Environ. Sci. Process. Impacts 2020, 22, 1110–1124. [Google Scholar] [CrossRef]
- Streling, A.P.; Cayô, R.; Nodari, C.S.; Almeida, L.G.P.; Bronze, F.; Siqueira, A.V.; Matos, A.P.; Oliveira, V.; Vasconcelos, A.T.R.; Marcondes, M.F.M.; et al. Kinetics Analysis of β-Lactams Hydrolysis by OXA-50 Variants of Pseudomonas aeruginosa. Microb. Drug Resist. 2022, 28, 849–852. [Google Scholar] [CrossRef]
- Cabrera, R.; Fernández-Barat, L.; Vázquez, N.; Alcaraz-Serrano, V.; Bueno-Freire, L.; Amaro, R.; López-Aladid, R.; Oscanoa, P.; Muñoz, L.; Vila, J.; et al. Resistance mechanisms and molecular epidemiology of Pseudomonas aeruginosa strains from patients with bronchiectasis. J. Antimicrob. Chemother. 2022, 77, 1600–1610. [Google Scholar] [CrossRef] [PubMed]
- Libisch, B. Molecular Typing Methods for the Genus Pseudomonas. In Molecular Typing in Bacterial Infections. Infectious Disease; De Filippis, I., McKee, M., Eds.; Humana Press: Totowa, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Morales-Espinosa, R.; Delgado, G.; Espinosa, L.F.; Isselo, D.; Mendez, J.L.; Rodriguez, C.; Miranda, G.; Cravioto, A. Fingerprint analysis and identification of strains ST309 as a potential high-risk clone in a Pseudomonas aeruginosa population isolated from children with bacteremia in Mexico City. Front. Microbiol. 2017, 8, 313. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.; Aujoulat, F.; Benaoudia, M.; Jumas-Bilak, E.; Chiron, R.; Marchandin, H. Highly diverse dynamics of Pseudomonas aeruginosa colonization from initial detection in cystic fibrosis patients: A 7-year longitudinal genetic diversity study. Infect. Genet. Evol. 2023, 115, 105513. [Google Scholar] [CrossRef]
- Wendel, A.F.; Malecki, M.; Mattner, F.; Xanthopoulou, K.; Wille, J.; Seifert, H.; Higgins, P.G. Genomic-based transmission analysis of carbapenem-resistant Pseudomonas aeruginosa at a tertiary care centre in Cologne (Germany) from 2015 to 2020. JAC-Antimicrob. Resist. 2022, 4, dlac057. [Google Scholar] [CrossRef]
- Guzvinec, M.; Izdebski, R.; Butic, I.; Jelic, M.; Abram, M.; Koscak, I.; Baraniak, A.; Hryniewicz, W.; Gniadkowski, M.; Tambic Andrasevic, A. Sequence types 235, 111, and 132 predominate among multidrug-resistant Pseudomonas aeruginosa clinical isolates in Croatia. Antimicrob. Agents Chemother. 2014, 58, 6277–6283. [Google Scholar] [CrossRef]
- Nemec, A.; Krizova, L.; Maixnerova, M.; Musilek, M. Multidrug-resistant epidemic clones among bloodstream isolates of Pseudomonas aeruginosa in the Czech Republic. Res. Microbiol. 2010, 161, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Fethi, M.; Rojo-Bezares, B.; Arfaoui, A.; Dziri, R.; Chichón, G.; Barguellil, F.; López, M.; El Asli, M.S.; Toledano, P.; Ouzari, H.-I.; et al. High Prevalence of GES-5 Variant and Co-Expression of VIM-2 and GES-45 among Clinical Pseudomonas aeruginosa Strains in Tunisia. Antibiotics 2023, 12, 1394. [Google Scholar] [CrossRef]
- Wei, L.; Wu, Q.; Zhang, J.; Guo, W.; Gu, Q.; Wu, H.; Wang, J.; Lei, T.; Xue, L.; Zhang, Y.; et al. Prevalence, virulence, antimicrobial resistance, and molecular characterization of Pseudomonas aeruginosa isolates from drinking water in China. Front. Microbiol. 2020, 11, 544653. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Vázquez, M.; Sola-Campoy, P.J.; Zurita, Á.M.; Ávila, A.; Gómez-Bertomeu, F.; SolÍs, S.; López-Urrutia, L.; Gónzalez-BarberÁ, E.M.; Cercenado, E.; Bautista, V.; et al. Carbapenemase-producing Pseudomonas aeruginosa in Spain: Interregional dissemination of the high-risk clones ST175 and ST244 carrying blaVIM-2, blaVIM-1, blaIMP-8, blaVIM-20 and blaKPC-2. Int. J. Antimicrob. Agents 2020, 56, 106026. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xie, L.; Wang, C.; Zhou, Q.; Jelsbak, L. Comparative whole-genome analysis of China and global epidemic Pseudomonas aeruginosa high-risk clones. J. Glob. Antimicrob. Resist. 2023, 35, 149–158. [Google Scholar] [CrossRef]
- Fang, Y.; Baloch, Z.; Zhang, W.; Hu, Y.; Zheng, R.; Song, Y.; Tai, W.; Xia, X. Emergence of carbapenem-resistant ST244, ST292, and ST2446 Pseudomonas aeruginosa clones in burn patients in Yunnan province. Infect. Drug Resist. 2022, 15, 1103–1114. [Google Scholar] [CrossRef]
- Hu, F.; Wang, P.; Li, Y.; Ling, J.; Ruan, Y.; Yu, J.; Zhang, L. Bioremediation of environmental organic pollutants by Pseudomonas aeruginosa: Mechanisms, methods and challenges. Environ. Res. 2023, 239, 117211. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ozoaduche, C.L.; Libisch, B.; Itoro, D.; Idemudia, I.B.; Posta, K.; Olasz, F. Antibiotic Resistance and Virulence Determinants of Pseudomonas aeruginosa Isolates Cultured from Hydrocarbon-Contaminated Environmental Samples. Microorganisms 2025, 13, 688. https://doi.org/10.3390/microorganisms13030688
Ozoaduche CL, Libisch B, Itoro D, Idemudia IB, Posta K, Olasz F. Antibiotic Resistance and Virulence Determinants of Pseudomonas aeruginosa Isolates Cultured from Hydrocarbon-Contaminated Environmental Samples. Microorganisms. 2025; 13(3):688. https://doi.org/10.3390/microorganisms13030688
Chicago/Turabian StyleOzoaduche, Chioma Lilian, Balázs Libisch, Daniel Itoro, Iyore Blessing Idemudia, Katalin Posta, and Ferenc Olasz. 2025. "Antibiotic Resistance and Virulence Determinants of Pseudomonas aeruginosa Isolates Cultured from Hydrocarbon-Contaminated Environmental Samples" Microorganisms 13, no. 3: 688. https://doi.org/10.3390/microorganisms13030688
APA StyleOzoaduche, C. L., Libisch, B., Itoro, D., Idemudia, I. B., Posta, K., & Olasz, F. (2025). Antibiotic Resistance and Virulence Determinants of Pseudomonas aeruginosa Isolates Cultured from Hydrocarbon-Contaminated Environmental Samples. Microorganisms, 13(3), 688. https://doi.org/10.3390/microorganisms13030688






