Fluroxypyr Inhibits Maize Growth by Disturbing the Diversity of the Endophytic Bacterial Communities in Maize Roots
Abstract
1. Introduction
2. Experimental Procedures
2.1. Study Site and Fluroxypyr Treatment
2.2. Soil Sampling
2.3. Analysis of Parameters of Maize
2.4. DNA Extraction, Polymerase Chain Reaction (PCR) Amplification, and Illumina MiSeq Sequencing
2.5. Statistical Analyses
3. Results
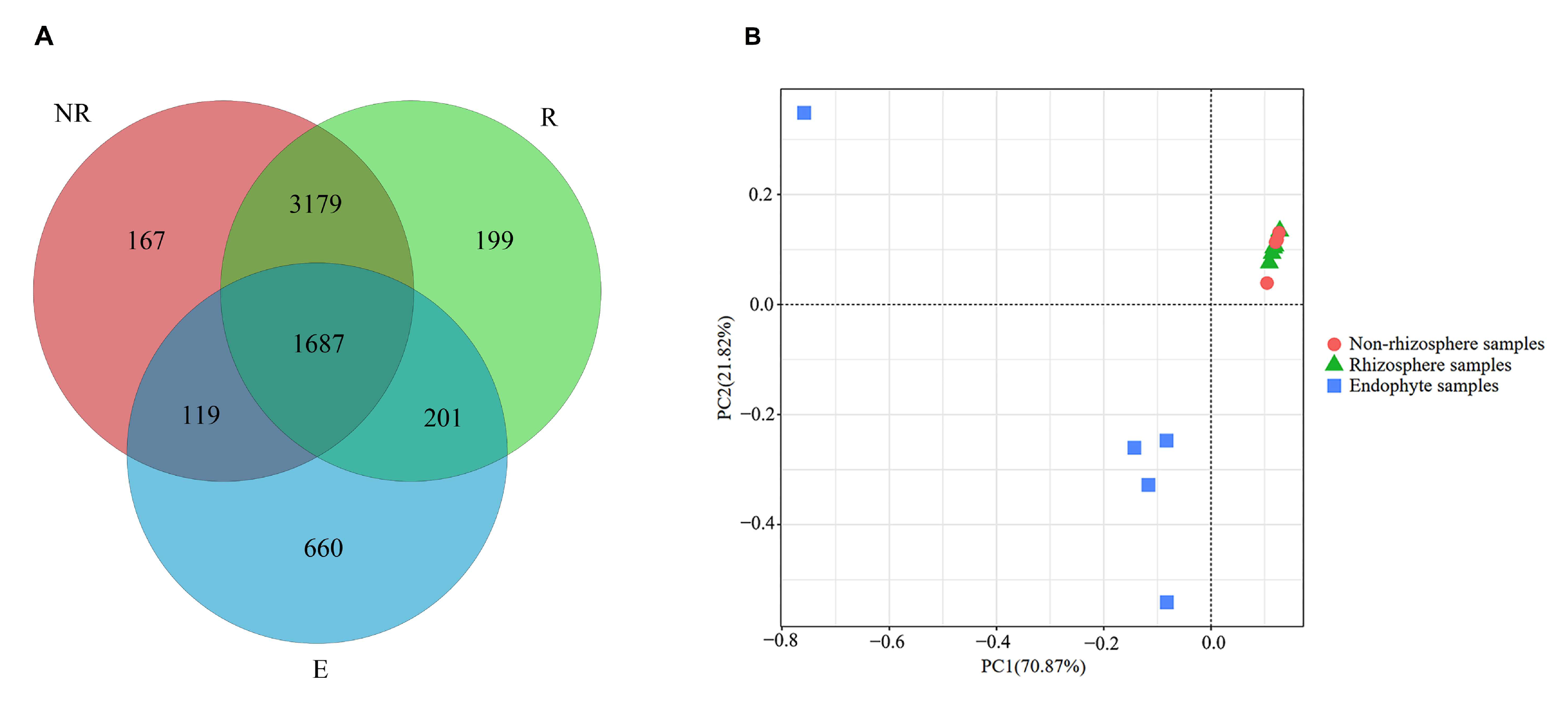
3.1. Similarities in the Bacterial Community in Different Portions of Maize Roots
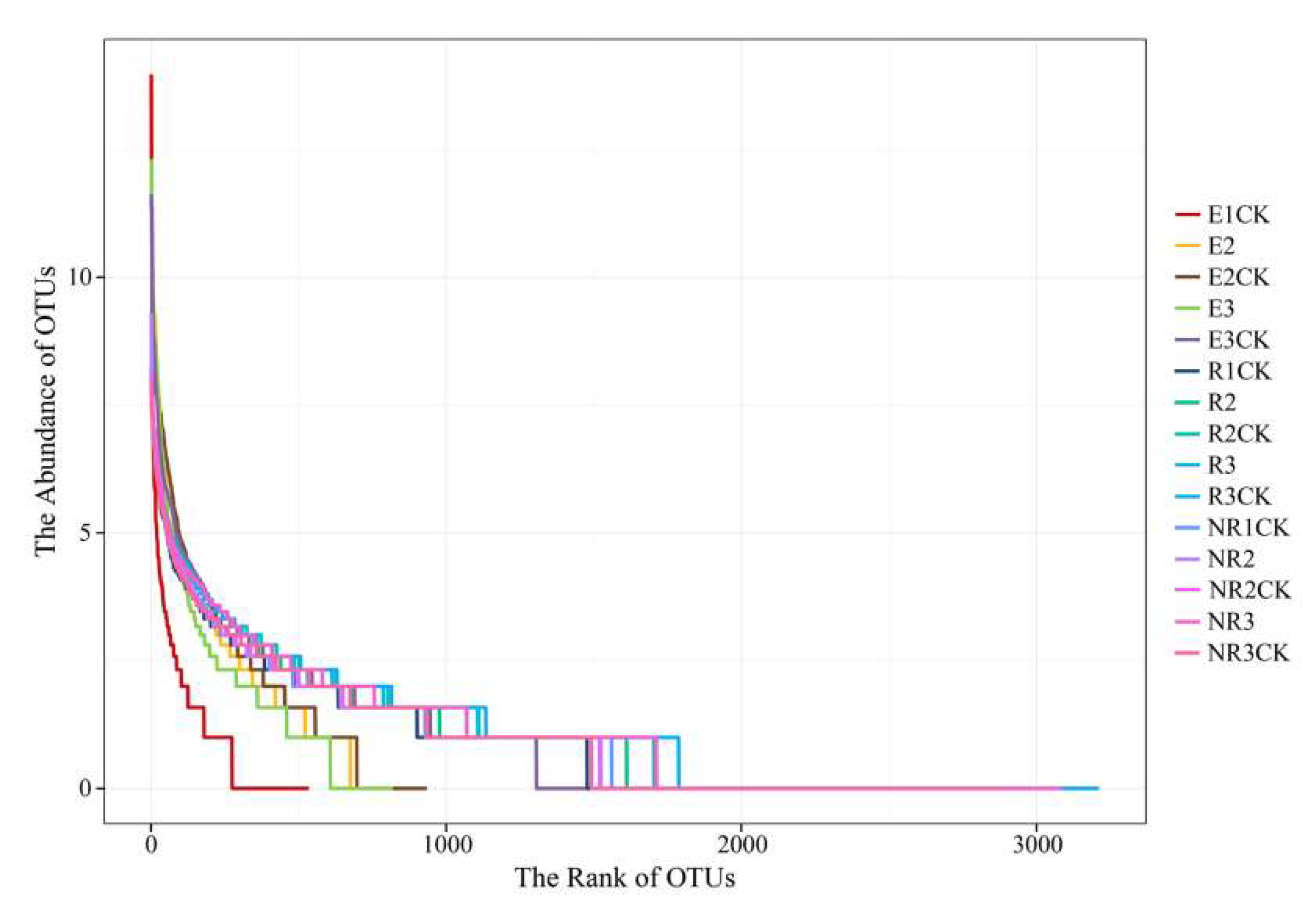
3.2. Effects of Fluroxypyr on the Richness and Diversity of Microbial Communities
3.3. Effects of Fluroxypyr on the Ear Traits and Morphological Observations of the Roots Throughout the Maturity Stage
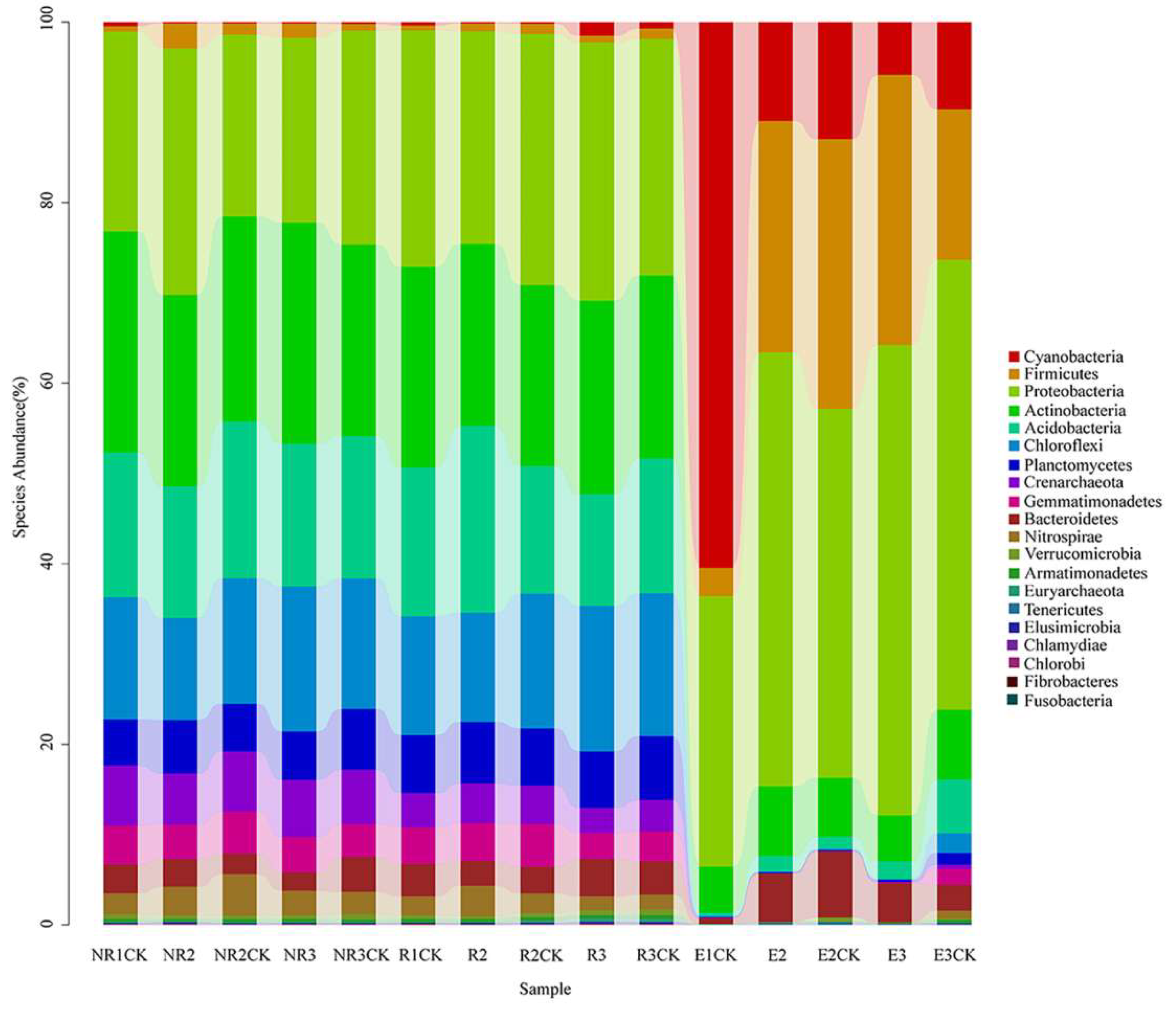
3.4. Effects of Fluroxypyr on the Soil Microbial Community Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chauhan, B.S.; Johnson, D.E. Row spacing and weed control timing affect yield of aerobic rice. Field Crops Res. 2011, 121, 226–231. [Google Scholar] [CrossRef]
- Takim, F. Weed competition in maize (Zea mays L.) as a function of the timing of hand-hoeing weed control in the southern Guinea savanna zone of Nigeria. Acta Agron. Hung. 2012, 60, 257–264. [Google Scholar] [CrossRef]
- Mehmeti, A.; Fetahaj, R.; Demaj, A.; Nishori, F.; Rracaj, V. Evaluation of pre- and post-emergence herbicides for weed control in maize (Zea mays L.). J. Cent. Eur. Agric. 2019, 20, 208–222. [Google Scholar]
- Mesarović, J.; Srdić, J.; Mladenović-Drinić, S.; Dragičević, V.; Simić, M.; Brankov, M.; Milojković-Opsenica, D. Evaluation of the nutritional profile of sweet maize after herbicide and foliar fertilizer application. J. Cereal Sci. 2019, 87, 132–137. [Google Scholar] [CrossRef]
- Altman, J.; Campbell, C.L. Effect of Herbicides on Plant Diseases. Annu. Rev. Phytopathol. 1977, 15, 361–385. [Google Scholar] [CrossRef]
- Gangola, S.; Chaube, S.; Bayram, A.; Joshi, S.; Bhandari, G.; Malik, S.; Khan, A.A. Optimizing microbial strain selection for pyrethroid biodegradation in contaminated environments through a TOPSIS-based decision-making system. Sci. Rep. 2024, 14, 14928. [Google Scholar] [CrossRef]
- Benito, N.; Magnoli, K.; Carranza, C.S.; Aluffi, M.E.; Magnoli, C.E.; Barberis, C.L. Influence of a glyphosate-based herbicide on growth parameters and aflatoxin B1 production by Aspergillus section Flavi on maize grains. Rev. Argent. Microbiol. 2021, 53, 162–170. [Google Scholar] [CrossRef]
- Anderson, J.P.E.; Armstrong, R.A.; Smith, S.N. Methods to evaluate pesticide damage to the biomass of the soil microflora. Soil Biol. Biochem. 1981, 13, 149–153. [Google Scholar] [CrossRef]
- Gangola, S.; Bhatt, P.; Kumar, A.J.; Bhandari, G.; Joshi, S.; Punetha, A.; Bhatt, K.; Rene, E.R. Biotechnological tools to elucidate the mechanism of pesticide degradation in the environment. Chemosphere 2022, 296, 133916. [Google Scholar] [CrossRef]
- Ramos, B.; García, J.A.L.; Probanza, A.; Domenech, J.; Mañero, F.J.G. Influence of an indigenous European alder (Alnus glutinosa (L.) Gaertn) rhizobacterium (Bacillus pumilus) on the growth of alder and its rhizosphere microbial community structure in two soils. New For. 2003, 25, 149–159. [Google Scholar] [CrossRef]
- Kuklinsky-Sobral, J.; Araújo, W.L.; Mendes, R.; Geraldi, I.O.; Pizzirani-Kleiner, A.A.; Azevedo, J.L. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ. Microbiol. 2004, 6, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Naik, D.; Smith, E.; Cumming, J.R. Rhizosphere carbon deposition, oxidative stress and nutritional changes in two poplar species exposed to aluminum. Tree Physiol. 2009, 29, 423–436. [Google Scholar] [CrossRef]
- Qiu, M.; Zhang, R.; Xue, C.; Zhang, S.; Li, S.; Zhang, N.; Shen, Q. Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol. Fertil. Soils 2012, 48, 807–816. [Google Scholar] [CrossRef]
- Chen, Z.-J.; Sheng, X.-F.; He, L.-Y.; Huang, Z.; Zhang, W.-H. Effects of root inoculation with bacteria on the growth, Cd uptake and bacterial communities associated with rape grown in Cd-contaminated soil. J. Hazard. Mater. 2013, 244–245, 709–717. [Google Scholar] [CrossRef]
- Chen, L.; He, L.; Wang, Q.; Sheng, X. Synergistic effects of plant growth-promoting Neorhizobium huautlense T1-17 and immobilizers on the growth and heavy metal accumulation of edible tissues of hot pepper. J. Hazard. Mater. 2016, 312, 123–131. [Google Scholar] [CrossRef]
- Mahbub, K.R.; Krishnan, K.; Megharaj, M.; Naidu, R. Bioremediation potential of a highly mercury resistant bacterial strain Sphingobium SA2 isolated from contaminated soil. Chemosphere 2016, 144, 330–337. [Google Scholar] [CrossRef]
- Lin, X.; Mou, R.; Cao, Z.; Xu, P.; Wu, X.; Zhu, Z.; Chen, M. Characterization of cadmium-resistant bacteria and their potential for reducing accumulation of cadmium in rice grains. Sci. Total Environ. 2016, 569–570, 97–104. [Google Scholar] [CrossRef]
- Cheng, C.; Nie, Z.-W.; He, L.-Y.; Sheng, X.-F. Rice-derived facultative endophytic Serratia liquefaciens F2 decreases rice grain arsenic accumulation in arsenic-polluted soil. Environ. Pollut. 2020, 259, 113832. [Google Scholar] [CrossRef]
- Ramakrishna, W.; Rathore, P.; Kumari, R.; Yadav, R. Brown gold of marginal soil: Plant growth promoting bacteria to overcome plant abiotic stress for agriculture, biofuels and carbon sequestration. Sci. Total Environ. 2020, 711, 135062. [Google Scholar] [CrossRef]
- Esperschütz, J.; Pritsch, K.; Gattinger, A.; Welzl, G.; Haesler, F.; Buegger, F.; Winkler, J.B.; Munch, J.C.; Schloter, M. Influence of chronic ozone stress on carbon translocation pattern into rhizosphere microbial communities of beech trees (Fagus sylvatica L.) during a growing season. Plant Soil 2009, 323, 85–95. [Google Scholar] [CrossRef]
- Li, X.; Song, Y.; Wang, F.; Bian, Y.; Jiang, X. Combined effects of maize straw biochar and oxalic acid on the dissipation of polycyclic aromatic hydrocarbons and microbial community structures in soil: A mechanistic study. J. Hazard. Mater. 2019, 364, 325–331. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, N.; Guo, X.; Zhang, Y.; Ye, B. Comparative analysis of bacterial community structure in the rhizosphere of maize by high-throughput pyrosequencing. PLoS ONE 2017, 12, e0178425. [Google Scholar] [CrossRef] [PubMed]
- Pu, S.; Yan, C.; Huang, H.; Liu, S.; Deng, D. Toxicity of nano-CuO particles to maize and microbial community largely depends on its bioavailable fractions. Environ. Pollut. 2019, 255, 113248. [Google Scholar] [CrossRef]
- Sanders, G.E.; Pallett, K.E. Comparison of the uptake, movement and metabolism of fluroxypyr in Stellaria media and Viola arvensis. Weed Res. 1987, 27, 159–166. [Google Scholar] [CrossRef]
- Zand, E.; Baghestani, M.A.; Soufizadeh, S.; PourAzar, R.; Veysi, M.; Bagherani, N.; Barjasteh, A.; Khayami, M.M.; Nezamabadi, N. Broadleaved weed control in winter wheat (Triticum aestivum L.) with post-emergence herbicides in Iran. Crop Prot. 2007, 26, 746–752. [Google Scholar] [CrossRef]
- Abouziena, H.F.; El-Karmany, M.F.; Singh, M.; Sharma, S.D. Effect of Nitrogen Rates and Weed Control Treatments on Maize Yield and Associated Weeds in Sandy Soils. Weed Technol. 2007, 21, 1049–1053. [Google Scholar] [CrossRef]
- Grossmann, K. Auxin herbicides: Current status of mechanism and mode of action. Pest Manag. Sci. 2010, 66, 113–120. [Google Scholar] [CrossRef]
- LeClere, S.; Wu, C.; Westra, P.; Sammons, R.D. Cross-resistance to dicamba, 2,4-D, and fluroxypyr in Kochia scoparia is endowed by a mutation in an AUX/IAA gene. Proc. Natl. Acad. Sci. USA 2018, 115, 201712372. [Google Scholar] [CrossRef]
- Breeze, V.G. Growth of tomato plants following exposure to fluroxypyr vapour. Weed Res. 1988, 28, 297–301. [Google Scholar] [CrossRef]
- Guo, M.-J.; Wang, Y.-G.; Yuan, X.-Y.; Dong, S.-Q.; Wen, Y.-Y.; Song, X.-E.; Guo, P.-Y. Responses of the antioxidant system to fluroxypyr in foxtail millet (Setaria italica L.) at the seedling stage. J. Integr. Agric. 2018, 17, 554–565. [Google Scholar] [CrossRef]
- Wu, G.L.; Cui, J.; Tao, L.; Yang, H. Fluroxypyr triggers oxidative damage by producing superoxide and hydrogen peroxide in rice (Oryza sativa). Ecotoxicology 2009, 19, 124. [Google Scholar] [CrossRef]
- Hong, C.; Si, Y.; Xing, Y.; Li, Y. Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ. Sci. Pollut. Res. 2015, 22, 10788–10799. [Google Scholar] [CrossRef]
- Gangola, S.; Bhandari, G.; Joshi, S.; Sharma, A.; Simsek, H.; Bhatt, P. Esterase and ALDH dehydrogenase-based pesticide degradation by Bacillus brevis 1B from a contaminated environment. Environ. Res. 2023, 232, 116332. [Google Scholar] [CrossRef]
- Gangola, S.; Joshi, S.; Kumar, S.; Sharma, B.; Sharma, A. Differential proteomic analysis under pesticides stress normal conditions in Bacillus cereus 2D. PLoS ONE 2021, 16, e0253106. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; van Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef]
- Lamb, T.G.; Tonkyn, D.W.; Kluepfel, D.A. Movement of Pseudomonas aureofaciens from the rhizosphere to aerial plant tissue. Can. J. Microbiol. 1996, 42, 1112–1120. [Google Scholar] [CrossRef]
- Handelsman, J.; Stabb, E. Biocontrol of Soilborne Plant Pathogens. Plant Cell 1996, 8, 1855–1869. [Google Scholar]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Llirós, M.; Trias, R.; Borrego, C.; Bañeras, L. Specific Archaeal Communities are Selected on the Root Surfaces of Ruppia spp. and Phragmites australis. Wetlands 2014, 34, 403–411. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an Ecological Classification of Soil Bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Strap, J.L. Actinobacteria—Plant Interactions: A Boon to Agriculture. In Bacteria in Agrobiology: Plant Growth Responses; Springer: Berlin/Heidelberg, Germany, 2011; pp. 285–307. [Google Scholar] [CrossRef]
- Fang, H.; Yu, Y.; Chu, X.; Wang, X.; Yang, X.; Yu, J. Degradation of chlorpyrifos in laboratory soil and its impact on soil microbial functional diversity. J. Environ. Sci. 2009, 21, 380–386. [Google Scholar] [CrossRef]
- Bhandari, G.; Gangola, S.; Bhatt, P.; Rafatullah, M. Editorial: Potential of the plant rhizomicrobiome for bioremediation of contaminants in agroecosystems. Front. Plant Sci. 2024, 15, 1397360. [Google Scholar] [CrossRef]
- Inzé, D.; Follin, A.; Van Lijsebettens, M.; Simoens, C.; Genetello, C.; Van Montagu, M.; Schell, J. Genetic analysis of the individual T-DNA genes of Agrobacterium tumefaciens; further evidence that two genes are involved in indole-3-acetic acid synthesis. Mol. Genet. Genom. 1984, 194, 265–274. [Google Scholar] [CrossRef]
- Barazani, O.; Friedman, J. Is IAA the Major Root Growth Factor Secreted from Plant-Growth-Mediating Bacteria? J. Chem. Ecol. 1999, 25, 2397–2406. [Google Scholar] [CrossRef]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Cherdyntseva, T.A.; Botina, S.G.; Netrusov, A.I. Bacteria associated with orchid roots and microbial production of auxin. Microbiol. Res. 2007, 162, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Gangola, S.; Sharma, A.; Bhatt, P.; Khati, P.; Chaudhary, P. Presence of esterase and laccase in Bacillus subtilis facilitates biodegradation and detoxification of cypermethrin. Sci. Rep. 2018, 8, 12755. [Google Scholar] [CrossRef]
- Sachdev, D.; Nema, P.; Dhakephalkar, P.; Zinjarde, S.; Chopade, B. Assessment of 16S rRNA gene-based phylogenetic diversity and promising plant growth-promoting traits of Acinetobacter community from the rhizosphere of wheat. Microbiol. Res. 2010, 165, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Purohit, A.; Rai, S.K.; Chownk, M.; Sangwan, R.S.; Yadav, S.K. Xylanase from Acinetobacter pittii MASK 25 and developed magnetic cross-linked xylanase aggregate produce predominantly xylopentose and xylohexose from agro biomass. Bioresour. Technol. 2017, 244, 793–799. [Google Scholar] [CrossRef]
- Higa, T.; Kinjo, S. Effect of Lactic Acid Fermentation Bacteria on Plant Growth and Soil Humus Formation. In Proceedings of the 1th International Conference on Kyusei Nature Farming, Khon Kaen, Thailand, 17–21 October 1989. [Google Scholar]
- Li, B. Isolation and Functional Identification of Acinetobacter zJU-1 and Its Effect on Vegetable Growth. Master’s Thesis, Zhejiang University, Hangzhou, China, 2020. [Google Scholar]
- Das, J.; Sarkar, P. Remediation of arsenic in mung bean (Vigna radiata) with growth enhancement by unique arsenic-resistant bacterium Acinetobacter lwoffii. Sci. Total Environ. 2018, 624, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by random forest. R News 2002, 2, 18–22. [Google Scholar]
- Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef]
- McArdle, B.H.; Anderson, M.J. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 2001, 82, 290–297. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and aualitative β diversity measures lead to different Insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Warton, D.I.; Wright, S.T.; Wang, Y. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol. Evol. 2012, 3, 89–101. [Google Scholar] [CrossRef]
- Chen, H.; Jiang, W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front. Microbiol. 2014, 5, 508. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, N.; Liu, Y.-X.; Zhang, X.; Hu, B.; Qin, Y.; Xu, H.; Wang, H.; Guo, X.; Qian, J.; et al. Root microbiota shift in rice correlates with resident time in the field and developmental stage. Sci. China Life Sci. 2018, 61, 613–621. [Google Scholar] [CrossRef]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut Microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef]
- Zhou, W.; Li, Y.; Liu, X.; He, S.; Huang, J.C. Comparison of microbial communities in different sulfur-based autotrophic denitrification reactors. Appl. Microbiol. Biotechnol. 2017, 101, 447–453. [Google Scholar] [CrossRef]


| Sample | Chao1 | ACE | Simpson | Shannon | |
|---|---|---|---|---|---|
| Non-rhizosphere | NR1CK | 4634 | 5043 | 0.9957 | 9.83 |
| NR2 | 4398 | 4694 | 0.9947 | 9.70 | |
| NR2CK | 4395 | 4717 | 0.9966 | 9.86 | |
| NR3 | 4548 | 4835 | 0.9967 | 9.93 | |
| NR3CK | 4570 | 4908 | 0.9971 | 10.01 | |
| Rhizosphere | R1CK | 4395 | 4731 | 0.9967 | 9.92 |
| R2 | 4550 | 4917 | 0.9967 | 9.88 | |
| R2CK | 5061 | 5269 | 0.9971 | 10.04 | |
| R3 | 4641 | 4854 | 0.9970 | 10.12 | |
| R3CK | 4760 | 5041 | 0.9972 | 10.13 | |
| Endophyte | E1CK | 888 | 935 | 0.6033 | 2.47 |
| E2 | 1055 | 1069 | 0.9576 | 6.11 | |
| E2CK | 1134 | 1128 | 0.9645 | 6.66 | |
| E3 | 969 | 972 | 0.9292 | 5.57 | |
| E3CK | 2576 | 2593 | 0.9620 | 7.10 | |
| Treatment | Ear Weight (kg) | Ear Length (cm) | Bald Tip Length (cm) | 100-Grain Weight (g) |
|---|---|---|---|---|
| F | 0.71 ± 0.02 | 19.63 ± 0.63 | 1.87 ± 0.31 | 39.44 ± 0.37 |
| NF | 0.75 ± 0.01 | 20.15 ± 0.32 | 1.42 ± 0.48 | 41.02 ± 0.30 |
| Source of variation Treatment | 0.035 * | 0.275 | 0.248 | 0.005 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; Liu, N.; Shi, S.; Li, J.; Geng, R.; Fang, L.; Wang, Y.; Lin, M.; Chen, J.; Si, Y.; et al. Fluroxypyr Inhibits Maize Growth by Disturbing the Diversity of the Endophytic Bacterial Communities in Maize Roots. Microorganisms 2025, 13, 728. https://doi.org/10.3390/microorganisms13040728
Zhang G, Liu N, Shi S, Li J, Geng R, Fang L, Wang Y, Lin M, Chen J, Si Y, et al. Fluroxypyr Inhibits Maize Growth by Disturbing the Diversity of the Endophytic Bacterial Communities in Maize Roots. Microorganisms. 2025; 13(4):728. https://doi.org/10.3390/microorganisms13040728
Chicago/Turabian StyleZhang, Gangrui, Nan Liu, Shengbo Shi, Jinghua Li, Rui Geng, Longyu Fang, Yuanyuan Wang, Mingchun Lin, Junfeng Chen, Yanru Si, and et al. 2025. "Fluroxypyr Inhibits Maize Growth by Disturbing the Diversity of the Endophytic Bacterial Communities in Maize Roots" Microorganisms 13, no. 4: 728. https://doi.org/10.3390/microorganisms13040728
APA StyleZhang, G., Liu, N., Shi, S., Li, J., Geng, R., Fang, L., Wang, Y., Lin, M., Chen, J., Si, Y., Shan, K., Zhou, Z., Men, M., Qiao, X., & Hao, L. (2025). Fluroxypyr Inhibits Maize Growth by Disturbing the Diversity of the Endophytic Bacterial Communities in Maize Roots. Microorganisms, 13(4), 728. https://doi.org/10.3390/microorganisms13040728








