The Genetic Determinants of Extreme UV Radiation and Desiccation Tolerance in a Bacterium Recovered from the Stratosphere
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. UVCR Survival Assays
2.3. Whole-Genome Sequencing and Comparative Genomics
2.4. ROS Quantification Assays
2.5. Photolyase Activity Assay
2.6. UVCR and Desiccation Exposure for RNAseq Experiments
2.7. RNA Extraction and Sequencing
2.8. Preprocessing, Mapping Sequencing Data, and Differential Expression Analysis
2.9. Gene Set Enrichment Analysis of Differentially Expressed Genes
3. Results
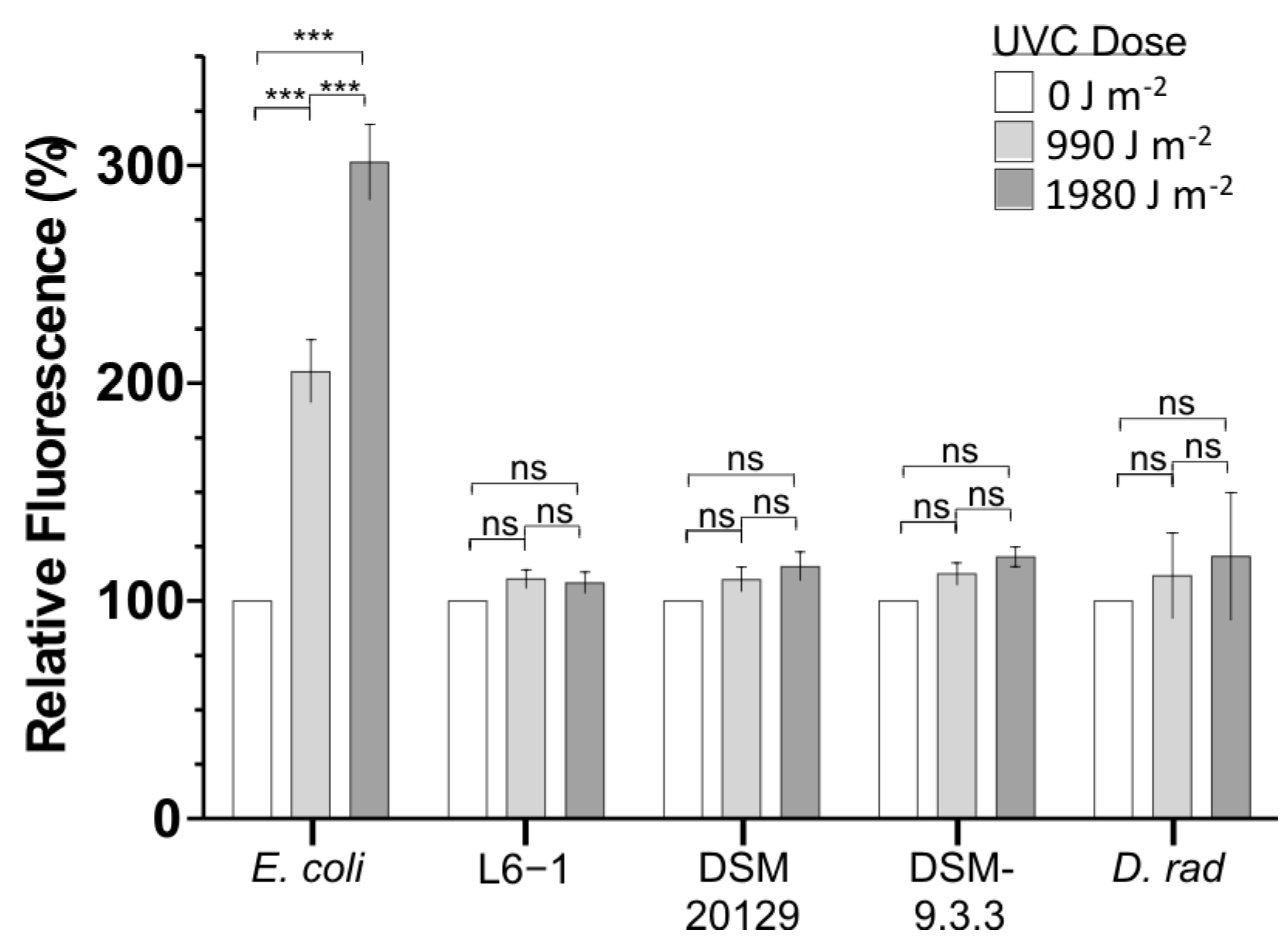
3.1. UVCR Resistance in C. aetherium and Phylogenetically Related Bacteria
3.2. Genetic Differences Between UVCR-Tolerant and -Sensitive Strains
3.3. Directed Evolution of UVCR Tolerance in C. flaccumfaciens DSM 20129
3.4. Intracellular ROS Concentrations After UVCR Exposure
3.5. In Vivo Assessment of Photolyase Activity
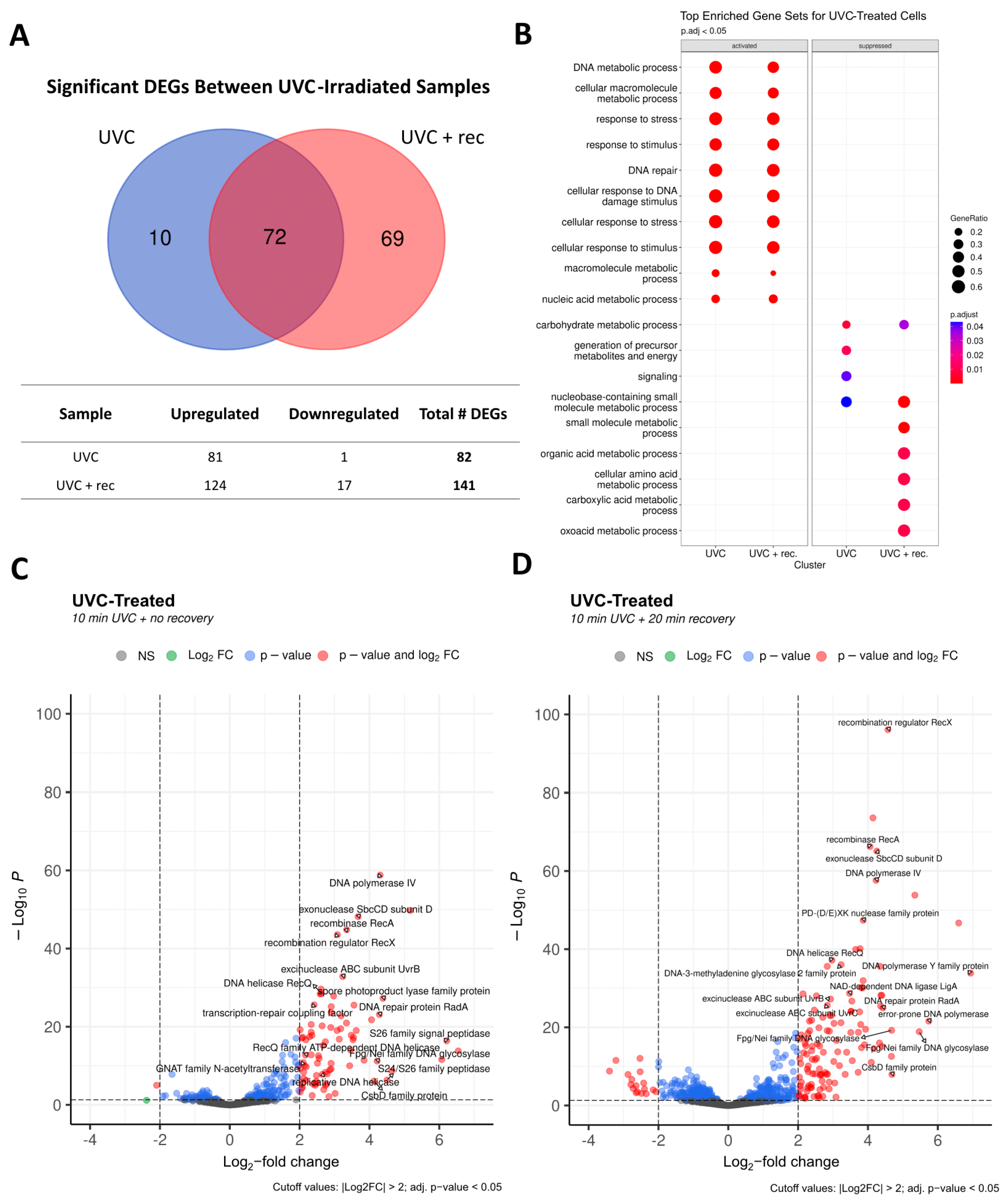
3.6. Differential Expression Analysis
3.7. Transcriptional Changes in Response to UVCR Exposure
3.7.1. DNA Metabolism and Repair in Response to UVCR
3.7.2. Stress Response and Regulation of Transcription and Translation
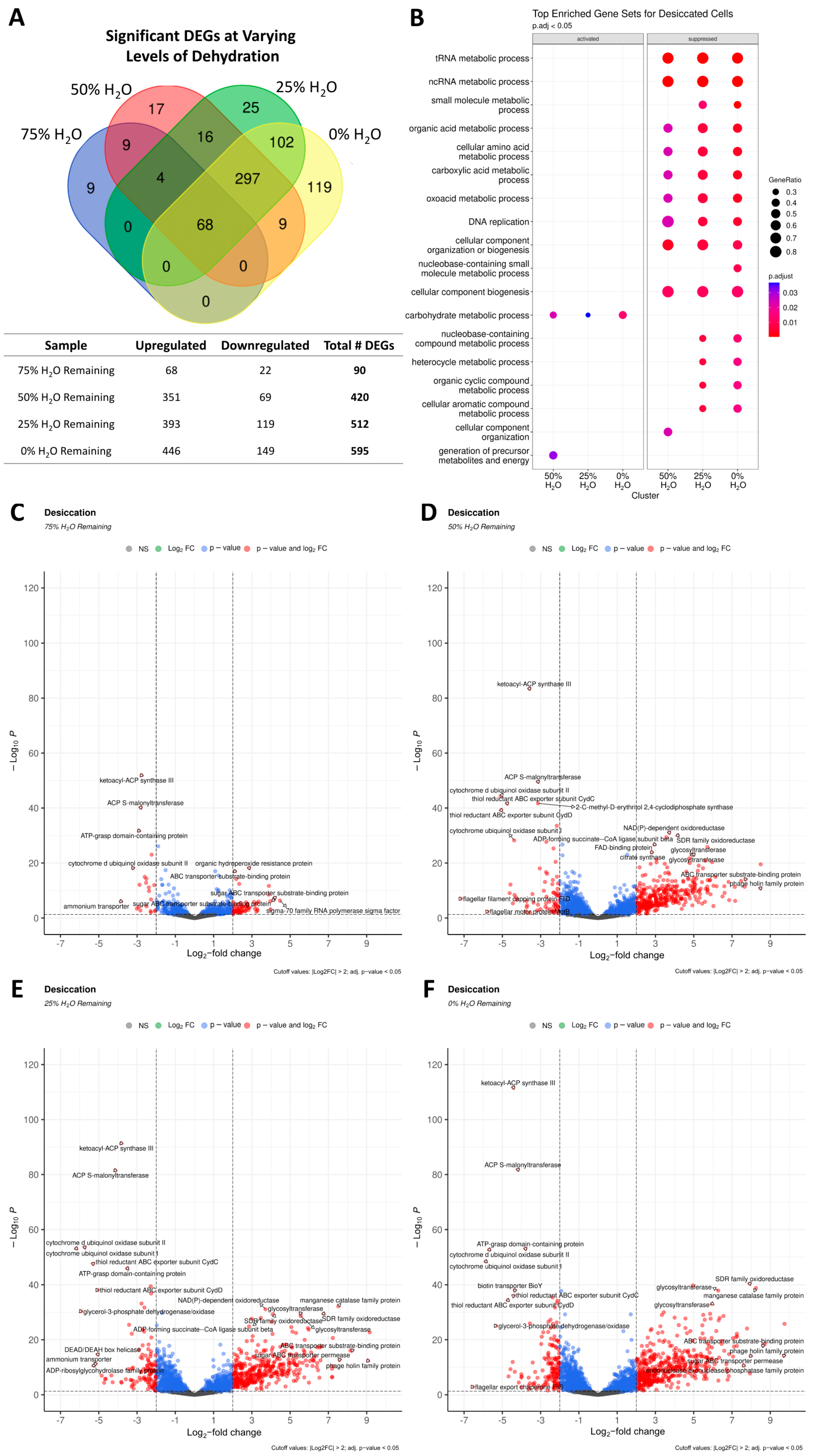
3.8. Transcriptional Changes in Response to Desiccation Stress
3.8.1. DNA Metabolism and Repair in Response to Desiccation
3.8.2. Regulation of Stress Response, Transcription, and Translation
3.8.3. Nutrient Transport and Metabolism
3.8.4. Antioxidants and Detoxification Systems
4. Discussion
4.1. Genetic Basis of UVR Tolerance in Curtobacterium
4.2. Desiccation Response in C. aetherium
4.3. Cross-Tolerance to Environmental Stressors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 6-4PP | pyrimidine (6-4) pyrimidone photoproduct |
| AAI | average amino acid identity |
| ANI | average nucleotide identity |
| ANOVA | Analysis of Variance |
| AP | apurinic/apyrimidinic site |
| ASL | above sea level |
| BER | base excision repair |
| BLAST | Basic Local Alignment Search Tool |
| BRIG | BLAST Ring Image Generator |
| CCP | cytochrome c peroxidase |
| CFU | colony-forming units |
| CPD | cyclobutane pyrimidine dimer |
| DEG | differentially expressed genes |
| DNA | deoxyribonucleic acid |
| DSB | double-strand break |
| dsDNA | double-stranded DNA |
| DSMZ | German Collection of Microorganisms and Cell Cultures |
| GO | Gene Ontology |
| Grx | glutaredoxin |
| GSH | glutathione |
| H2DCFDA | 2′,7′-dichlorodihydrofluoresciendiacetate |
| HR | homologous recombination |
| LB | Luria–Bertani media |
| LD90 | lethal dose reducing the population by 90% |
| MMR | mismatch repair |
| MSH | mycothiol |
| NASA | National Aeronautics and Space Administration |
| NCBI | National Center for Biotechnology Information |
| NER | nucleotide excision repair |
| NHEJ | non-homologous end joining |
| OD600 | optical density (λ = 600 nm) |
| oligos | oligonucleotides |
| PCA | principal component analysis |
| PCR | polymerase chain reaction |
| PGAP | Prokaryotic Genome Annotation Pipeline |
| R2A | Reasoner’s 2A media |
| RAST | Rapid Annotation using Subsystems Technology |
| RH | relative humidity |
| RNA | ribonucleic acid |
| ROS | reactive oxygen species |
| rRNA | ribosomal RNA |
| RT | reverse transcriptase |
| SEM | standard error of the mean |
| SPL | spore photoproduct lyase |
| SSB | single-strand break |
| ssb | single-stranded binding protein |
| ssDNA | single-stranded DNA |
| TM-score | template modeling score |
| tRNA | transfer RNA |
| UVR | ultraviolet radiation |
| UVCR | ultraviolet C radiation |
References
- Douwes, J.; Thorne, P.; Pearce, N.; Heederik, D. Bioaerosol health effects and exposure assessment: Progress and prospects. Ann. Occup. Hyg. 2003, 47, 187–200. [Google Scholar] [CrossRef]
- Kim, K.H.; Kabir, E.; Jahan, S.A. Airborne bioaerosols and their impact on human health. J. Environ. Sci. 2018, 67, 23. [Google Scholar] [CrossRef]
- Dillon, C.F.; Dillon, M.B. Multiscale Airborne Infectious Disease Transmission. Appl. Environ. Microbiol. 2020, 87, e02314–e02320. [Google Scholar] [CrossRef]
- Yadav, S.; Gettu, N.; Swain, B.; Kumari, K.; Ojha, N.; Gunthe, S.S. Bioaerosol impact on crop health over India due to emerging fungal diseases (EFDs): An important missing link. Environ. Sci. Pollut. Res. Int. 2020, 27, 12802–12829. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.K.M.; Hovmøll, M.S. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 2002, 297, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Phillips, V.T.J.; Andronache, C.; Christner, B.; Morris, C.E.; Sands, D.C.; Bansemer, A.; Lauer, A.; McNaughton, C.; Seman, C. Potential impacts from biological aerosols on ensembles of continental clouds simulated numerically. Biogeosciences 2009, 6, 987–1014. [Google Scholar] [CrossRef]
- Joyce, R.E.; Lavender, H.; Farrar, J.; Werth, J.T.; Weber, C.F.; D’Andrilli, J.; Vaitilingom, M.; Christner, B.C. Biological Ice-Nucleating Particles Deposited Year-Round in Subtropical Precipitation. Appl. Environ. Microbiol. 2019, 85, 1567–1586. [Google Scholar] [CrossRef]
- Fröhlich-Nowoisky, J.; Kampf, C.J.; Weber, B.; Huffman, J.A.; Pöhlker, C.; Andreae, M.O.; Lang-Yona, N.; Burrows, S.M.; Gunthe, S.S.; Elbert, W.; et al. Bioaerosols in the Earth system: Climate, health, and ecosystem interactions. Atmos. Res. 2016, 182, 346–376. [Google Scholar] [CrossRef]
- Smith, D.J.; Griffin, D.W.; McPeters, R.D.; Ward, P.D.; Schuerger, A.C. Microbial survival in the stratosphere and implications for global dispersal. Aerobiologia 2011, 27, 319–332. [Google Scholar] [CrossRef]
- Smith, D.J. Microbes in the upper atmosphere and unique opportunities for astrobiology research. Astrobiology 2013, 13, 981–990. [Google Scholar] [CrossRef]
- Maki, T.; Hara, K.; Kobayashi, F.; Kurosaki, Y.; Kakikawa, M.; Matsuki, A.; Chen, B.; Shi, G.; Hasegawa, H.; Iwasaka, Y. Vertical distribution of airborne bacterial communities in an Asian-dust downwind area, Noto Peninsula. Atmos. Environ. 2015, 119, 282–293. [Google Scholar] [CrossRef]
- Els, N.; Baumann-Stanzer, K.; Larose, C.; Vogel, T.M.; Sattler, B. Beyond the planetary boundary layer: Bacterial and fungal vertical biogeography at Mount Sonnblick, Austria. Geo Geogr. Environ. 2019, 6, e00069. [Google Scholar] [CrossRef]
- Burrows, S.M.; Butler, T.; Jöckel, P.; Tost, H.; Kerkweg, A.; Pöschl, U.; Lawrence, M.G. Bacteria in the global atmosphere—Part 2: Modeling of emissions and transport between different ecosystems. Atmos. Chem. Phys. 2009, 9, 9281–9297. [Google Scholar] [CrossRef]
- Bryan, N.C.; Christner, B.C.; Guzik, T.G.; Granger, D.J.; Stewart, M.F. Abundance and survival of microbial aerosols in the troposphere and stratosphere. ISME J. 2019, 13, 2789–2799. [Google Scholar] [CrossRef]
- Kerr, J.B.; Fioletov, V.E. Surface ultraviolet radiation. Atmos.-Ocean 2008, 46, 159–184. [Google Scholar] [CrossRef]
- Rodriguez-Manfredi, J.A.; de la Torre Juarez, M.; Sanchez-Lavega, A.; Hueso, R.; Martinez, G.; Lemmon, M.T.; Newman, C.E.; Munguira, A.; Hieta, M.; Tamppari, L.K.; et al. The diverse meteorology of Jezero crater over the first 250 sols of Perseverance on Mars. Nat. Geosci. 2023, 16, 19–28. [Google Scholar] [CrossRef]
- Polkko, J.; Hieta, M.; Harri, A.M.; Tamppari, L.; Martínez, G.; Viúdez-Moreiras, D.; Savijärvi, H.; Conrad, P.; Zorzano Mier, M.P.; de la Torre Juarez, M.; et al. Initial Results of the Relative Humidity Observations by MEDA Instrument Onboard the Mars 2020 Perseverance Rover. J. Geophys. Res. Planets 2023, 128, e2022JE007447. [Google Scholar] [CrossRef]
- Smith, D.J.; Thakrar, P.J.; Bharrat, A.E.; Dokos, A.G.; Kinney, T.L.; James, L.M.; Lane, M.A.; Khodadad, C.L.; Maguire, F.; Maloney, P.R.; et al. A Balloon-Based Payload for Exposing Microorganisms in the Stratosphere (E-MIST). Gravitational Space Res. 2014, 2, 70–80. [Google Scholar]
- Smith, D.J.; Sowa, M.B. Ballooning for Biologists: Mission Essentials for Flying Life Science Experiments to Near Space on NASA Large Scientific Balloons. Gravit. Space Res. 2017, 5, 52. [Google Scholar]
- Pulschen, A.A.; de Araujo, G.G.; de Carvalho, A.C.S.R.; Cerini, M.F.; de Mendonça Fonseca, L.; Galante, D.; Rodrigues, F. Survival of Extremophilic Yeasts in the Stratospheric Environment during Balloon Flights and in Laboratory Simulations. Appl. Environ. Microbiol. 2018, 84, e01942-18. [Google Scholar] [CrossRef]
- Cortesão, M.; Siems, K.; Koch, S.; Beblo-Vranesevic, K.; Rabbow, E.; Berger, T.; Lane, M.; James, L.; Johnson, P.; Waters, S.M.; et al. MARSBOx: Fungal and Bacterial Endurance from a Balloon-Flown Analog Mission in the Stratosphere. Front. Microbiol. 2021, 12, 601713. [Google Scholar] [CrossRef]
- Sinha, R.P.; Häder, D.P. UV-induced DNA damage and repair: A review. Photochem. Photobiol. Sci. 2002, 1, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Pattison, D.I.; Davies, M.J. Actions of ultraviolet light on cellular structures. In Cancer: Cell Structures, Carcinogens and Genomic Instability; Experientia Supplementum (EXS) Series; Springer: Berlin/Heidelberg, Germany, 2006; pp. 131–157. [Google Scholar] [CrossRef]
- Kurth, D.; Belfiore, C.; Gorriti, M.F.; Cortez, N.; Farias, M.E.; Albarracín, V.H. Genomic and proteomic evidences unravel the UV-resistome of the poly-extremophile Acinetobacter sp. Ver3. Front. Microbiol. 2015, 6, 328. [Google Scholar] [CrossRef]
- Takano, H.; Obitsu, S.; Beppu, T.; Ueda, K. Light-Induced Carotenogenesis in Streptomyces coelicolor A3, Identification of an Extracytoplasmic Function Sigma Factor That Directs Photodependent Transcription of the Carotenoid Biosynthesis Gene Cluster. J. Bacteriol. 2005, 187, 1825. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Pérez, M.; Hellingwerf, K.J.; Kort, R. Blue Light Activates the σB-Dependent Stress Response of Bacillus subtilis via YtvA. J. Bacteriol. 2006, 188, 6411. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Garcia-Pichel, F. Microbial ultraviolet sunscreens. Nat. Rev. Microbiol. 2011, 9, 791–802. [Google Scholar] [CrossRef]
- Lemire, J.; Alhasawi, A.; Appanna, V.P.; Tharmalingam, S.; Appanna, V.D. Metabolic defence against oxidative stress: The road less travelled so far. J. Appl. Microbiol. 2017, 123, 798–809. [Google Scholar] [CrossRef]
- Goosen, N.; Moolenaar, G.F. Repair of UV damage in bacteria. DNA Repair 2008, 7, 353–379. [Google Scholar] [CrossRef]
- Tang, M.; Shen, X.; Frank, E.G.; O’Donnell, M.; Woodgate, R.; Goodman, M.F. UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. USA 1999, 96, 8919–8924. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Richa, N.; Kumar, A.; Tyagi, M.B.; Sinha, R.P. Molecular Mechanisms of Ultraviolet Radiation-Induced DNA Damage and Repair. J. Nucleic Acids 2010, 2010, 32. [Google Scholar] [CrossRef]
- Lebre, P.H.; De Maayer, P.; Cowan, D.A. Xerotolerant bacteria: Surviving through a dry spell. Nat. Rev. Microbiol. 2017, 15, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Greffe, V.R.G.; Michiels, J. Desiccation-induced cell damage in bacteria and the relevance for inoculant production. Appl. Microbiol. Biotechnol. 2020, 104, 3757–3770. [Google Scholar] [CrossRef]
- Jeong, S.W.; Jung, J.H.; Kim, M.K.; Seo, H.S.; Lim, H.M.; Lim, S. The three catalases in Deinococcus radiodurans: Only two show catalase activity. Biochem. Biophys. Res. Commun. 2016, 469, 443–448. [Google Scholar] [CrossRef]
- Bagyan, I.; Casillas-Martinez, L.; Setlow, P. The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by σ(F), and katX is essential for hydrogen peroxide resistance of the germinating spore. J. Bacteriol. 1998, 180, 2057–2062. [Google Scholar] [CrossRef] [PubMed]
- Si, M.R.; Zhang, L.; Yang, Z.F.; Xu, Y.X.; Liu, Y.B.; Jiang, C.Y.; Wang, Y.; Shen, X.H.; Liu, S.J. NrdH redoxin enhances resistance to multiple oxidative stresses by acting as a peroxidase cofactor in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2014, 80, 1750–1762. [Google Scholar] [CrossRef]
- Krinsky, N.I. Antioxidant functions of carotenoids. Free Radic. Biol. Med. 1989, 7, 617–635. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Sun, Z.; Shen, S.; Wang, H.; Jiao, J.; Wang, L.; Hu, Y.; Hua, Y. Effects of carotenoids from Deinococcus radiodurans on protein oxidation. Lett. Appl. Microbiol. 2009, 49, 689–694. [Google Scholar] [CrossRef]
- Tuveson, R.W.; Larson, R.A.; Kagan, J. Role of cloned carotenoid genes expressed in Escherichia coli in protecting against inactivation by near-UV light and specific phototoxic molecules. J. Bacteriol. 1988, 170, 4675–4680. [Google Scholar] [CrossRef]
- Reis-Mansur, M.C.P.P.; Cardoso-Rurr, J.S.; Silva, J.V.M.A.; de Souza, G.R.; da Silva Cardoso, V.; Mansoldo, F.R.P.; Pinheiro, Y.; Schultz, J.; Lopez Balottin, L.B.; da Silva, A.J.R.; et al. Carotenoids from UV-resistant Antarctic Microbacterium sp. LEMMJ01. Sci. Rep. 2019, 9, 9554. [Google Scholar] [CrossRef]
- Newton, G.L.; Rawat, M.; La Clair, J.J.; Jothivasan, V.K.; Budiarto, T.; Hamilton, C.J.; Claiborne, A.; Helmann, J.D.; Fahey, R.C. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat. Chem. Biol. 2009, 5, 625–627. [Google Scholar] [CrossRef]
- Perera, V.R.; Newton, G.L.; Pogliano, K. Bacillithiol: A key protective thiol in Staphylococcus aureus. Expert Rev. Anti-Infect. Ther. 2015, 13, 1089. [Google Scholar] [CrossRef] [PubMed]
- Newton, G.L.; Arnold, K.; Price, M.S.; Sherrill, C.; Delcardayre, S.B.; Aharonowitz, Y.; Cohen, G.; Davies, J.; Fahey, R.C.; Davis, C. Distribution of thiols in microorganisms: Mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 1996, 178, 1990–1995. [Google Scholar] [CrossRef]
- Bryan, N.C.; Stewart, M.; Granger, D.; Guzik, T.G.; Christner, B.C. A method for sampling microbial aerosols using high altitude balloons. J. Microbiol. Methods 2014, 107, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Mijatović Scouten, J.; Smith, A.; Ellington, A.J.; Bryan, N.C.; Harveson, R.M.; Kvitko, B.H.; Christner, B.C. Curtobacterium aetherium sp. nov., a Polyextremophilic Plant Pathogen Isolated from the Stratosphere. Microbiol. Spectr. 2025, e01774-24. [Google Scholar] [CrossRef]
- Ellington, A.J.; Bryan, N.C.; Christner, B.C.; Reisch, C.R. Draft Genome Sequences of Actinobacterial and Betaproteobacterial Strains Isolated from the Stratosphere. Microbiol. Resour. Announc. 2021, 10, e1009–e1021. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Konstantinidis, K.T. Bypassing Cultivation to Identify Bacterial Species Culture-independent genomic approaches identify credibly distinct clusters, avoid cultivation bias, and provide true insights into microbial species. Microbe Mag. 2014, 9, 111–118. [Google Scholar]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016, 4, e1900v1. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Madden, T. The BLAST Sequence Analysis Tool. In The NCBI Handbook; NCBI: Bethesda, MD, USA, 2002; pp. 1–15. Available online: https://www.ncbi.nlm.nih.gov/books/NBK21097/ (accessed on 9 February 2022).
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Pearson, W.R. An Introduction to Sequence Similarity (“Homology”) Searching. Curr. Protoc. Bioinform. 2013, 42, 3.1.1–3.1.8. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics 2011, 12, 402. [Google Scholar] [CrossRef]
- Krueger, F. TrimGalore: A Wrapper Around Cutadapt and FastQC to Consistently Apply Adapter and Quality Trimming to FastQ Files, with Extra Functionality for RRBS Data; Babraham Institute: Cambridge, UK, 2021; Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 3 October 2021).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2021. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 25 September 2022).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Carlson, M.; Pagès, H. AnnotationForge: Tools for Building SQLite-Based Annotation Data Packages. 2022. Available online: https://bioconductor.org/packages/AnnotationForge (accessed on 10 October 2022).
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 2, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef]
- Bol, D.K.; Yasbin, R.E. The isolation, cloning and identification of a vegetative catalase gene from Bacillus subtilis. Gene 1991, 109, 31–37. [Google Scholar] [CrossRef]
- Deatherage, D.E.; Barrick, J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014, 1151, 165–188. [Google Scholar] [CrossRef]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Leapman, R.D.; Lai, B.; Ravel, B.; Li, S.M.W.; Kemner, K.M.; et al. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 2007, 5, 769–779. [Google Scholar] [CrossRef]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Kiang, J.G.; Fukumoto, R.; Lee, D.Y.; Wehr, N.B.; Viteri, G.A.; Berlett, B.S.; Levine, R.L. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS ONE 2010, 5, 10–15. [Google Scholar] [CrossRef]
- Krisko, A.; Radman, M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc. Natl. Acad. Sci. USA 2010, 107, 14373–14377. [Google Scholar] [CrossRef]
- Sancar, A. Structure and function of DNA photolyase. Biochemistry 1994, 33, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Oberpichler, I.; Pierik, A.J.; Wesslowski, J.; Pokorny, R.; Rosen, R.; Vugman, M.; Zhang, F.; Neubauer, O.; Ron, E.Z.; Batschauer, A.; et al. A Photolyase-Like Protein from Agrobacterium tumefaciens with an Iron-Sulfur Cluster. PLoS ONE 2011, 6, e26775. [Google Scholar] [CrossRef]
- Marizcurrena, J.J.; Morel, M.A.; Braña, V.; Morales, D.; Martinez-López, W.; Castro-Sowinski, S. Searching for novel photolyases in UVC-resistant Antarctic bacteria. Extemophiles 2017, 21, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Portero, L.R.; Alonso-Reyes, D.G.; Zannier, F.; Vazquez, M.P.; Farías, M.E.; Gärtner, W.; Albarracín, V.H. Photolyases and Cryptochromes in UV-resistant Bacteria from High-altitude Andean Lakes. Photochem. Photobiol. 2019, 95, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Illumina. Illumina Stranded Total RNA Prep, Ligation with Ribo-Zero Plus Reference Guide; Illumina: San Diego, CA, USA, 2022; Available online: https://www.illumina.com/company/legal.html (accessed on 13 October 2022).
- Haas, B.J.; Chin, M.; Nusbaum, C.; Birren, B.W.; Livny, J. How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics 2012, 13, 734. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, J.; White, K.P. RNA-seq differential expression studies: More sequence or more replication? Bioinformatics 2014, 30, 301. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Chudobova, D.; Cihalova, K.; Jelinkova, P.; Zitka, J.; Nejdl, L.; Guran, R.; Klimanek, M.; Adam, V.; Kizek, R. Effects of Stratospheric Conditions on the Viability, Metabolism and Proteome of Prokaryotic Cells. Atmosphere 2015, 6, 1290–1306. [Google Scholar] [CrossRef]
- Harwood, C.R.; Kikuchi, Y. The ins and outs of Bacillus proteases: Activities, functions, and commercial significance. FEMS Microbiol. Rev. 2022, 46, fuab046. [Google Scholar] [CrossRef]
- Claverys, J.P.; Prudhomme, M.; Martin, B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 2006, 60, 451–475. [Google Scholar] [CrossRef]
- Favrot, L.; Blanchard, J.S.; Vergnolle, O. Bacterial GCN5-Related N-Acetyltransferases: From Resistance to Regulation. Biochemistry 2016, 55, 989. [Google Scholar] [CrossRef]
- Mol, C.D.; Kuo, C.F.; Thayer, M.M.; Cunningham, R.P.; Tainer, J.A. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature 1995, 374, 381–386. [Google Scholar] [CrossRef]
- Ellis, E.M. Microbial aldo-keto reductases. FEMS Microbiol. Lett. 2002, 216, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Liu, X.; Guo, C.; Zhao, J. Methylglyoxal detoxification by an aldo-keto reductase in the cyanobacterium Synechococcus sp. PCC 7002. Microbiology 2006, 152 Pt 7, 2013–2021. [Google Scholar] [CrossRef][Green Version]
- Aguilera, Á.; de Diego-Castilla, G.; Osuna, S.; Bardera, R.; Mendi, S.S.; Blanco, Y.; González-Toril, E. Microbial Ecology in the Atmosphere: The Last Extreme Environment. In Extremophilic Microbes and Metabolites—Diversity, Bioprospecting and Biotechnological Applications; IntechOpen: Rijeka, Croatia, 2018. [Google Scholar] [CrossRef]
- Šantl-Temkiv, T.; Amato, P.; Casamayor, E.O.; Lee, P.K.H.; Pointing, S.B. Microbial ecology of the atmosphere. FEMS Microbiol. Rev. 2022, 46, fuac009. [Google Scholar] [CrossRef]
- Schuerger, A.C.; Mancinelli, R.L.; Kern, R.G.; Rothschild, L.J.; McKay, C.P. Survival of endospores of Bacillus subtilis on spacecraft surfaces under simulated Martian environments: Implications for the forward contamination of Mars. Icarus 2003, 165, 253–276. [Google Scholar] [CrossRef]
- Tauscher, C.; Schuerger, A.C.; Nicholson, W.L. Survival and Germinability of Bacillus subtilis Spores Exposed to Simulated Mars Solar Radiation: Implications for Life Detection and Planetary Protection. Astrobiology 2006, 6, 592–605. [Google Scholar] [CrossRef]
- Schuerger, A.C.; Richards, J.T.; Newcombe, D.A.; Venkateswaran, K. Rapid inactivation of seven Bacillus spp. under simulated Mars UV irradiation. Icarus 2006, 181, 52–62. [Google Scholar] [CrossRef]
- Cockell, C.S.; Schuerger, A.C.; Billi, D.; Friedmann, E.I.; Panitz, C. Effects of a Simulated Martian UV Flux on the Cyanobacterium, Chroococcidiopsis sp. 029. Astrobiology 2005, 5, 127–140. [Google Scholar] [CrossRef]
- Diaz, B.; Schulze-Makuch, D. Microbial Survival Rates of Escherichia coli and Deinococcus radiodurans Under Low Temperature, Low Pressure, and UV-Irradiation Conditions, and Their Relevance to Possible Martian Life. Astrobiology 2006, 6, 332–347. [Google Scholar] [CrossRef]
- de la Vega, U.P.; Rettberg, P.; Reitz, G. Simulation of the environmental climate conditions on Martian surface and its effect on Deinococcus radiodurans. Adv. Space Res. 2007, 40, 1672–1677. [Google Scholar] [CrossRef]
- Fendrihan, S.; Bérces, A.; Lammer, H.; Musso, M.; Rontó, G.; Polacsek, T.K.; Holzinger, A.; Kolb, C.; Stan-Lotter, H. Investigating the Effects of Simulated Martian Ultraviolet Radiation on Halococcus dombrowskii and Other Extremely Halophilic Archaebacteria. Astrobiology 2009, 9, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Peeters, Z.; Vos, D.; Ten Kate, I.L.; Selch, F.; Van Sluis, C.A.; Sorokin, D.Y.; Muijzer, G.; Stan-Lotter, H.; Van Loosdrecht, M.C.M.; Ehrenfreund, P. Survival and death of the haloarchaeon Natronorubrum strain HG-1 in a simulated Martian environment. Adv. Space Res. 2010, 46, 1149–1155. [Google Scholar] [CrossRef]
- Johnson, A.P.; Pratt, L.M.; Vishnivetskaya, T.; Pfiffner, S.; Bryan, R.A.; Dadachova, E.; Whyte, L.; Radtke, K.; Chan, E.; Tronick, S.; et al. Extended survival of several organisms and amino acids under simulated Martian surface conditions. Icarus 2011, 211, 1162–1178. [Google Scholar] [CrossRef]
- Smith, D.J.; Schuerger, A.C.; Davidson, M.M.; Pacala, S.W.; Bakermans, C.; Onstott, T.C. Survivability of Psychrobacter cryohalolentis K5 Under Simulated Martian Surface Conditions. Astrobiology 2009, 9, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Khodadad, C.L.; Wong, G.M.; James, L.M.; Thakrar, P.J.; Lane, M.A.; Catechis, J.A.; Smith, D.J. Stratosphere Conditions Inactivate Bacterial Endospores from a Mars Spacecraft Assembly Facility. Astrobiology 2017, 17, 337. [Google Scholar] [CrossRef]
- Ye, T.; Wang, B.; Li, C.; Bian, P.; Chen, L.; Wang, G. Exposure of cyanobacterium Nostoc sp. to the Mars-like stratosphere environment. J. Photochem. Photobiol. 2021, 224, 112307. [Google Scholar] [CrossRef]
- Sundin, G.W.; Jacobs, J.L. Ultraviolet Radiation (UVR) Sensitivity Analysis and UVR Survival Strategies of a Bacterial Community from the Phyllosphere of Field-Grown Peanut (Arachis hypogeae L.). Microb. Ecol. 1999, 38, 27–38. [Google Scholar] [CrossRef]
- Kuhlman, K.R.; Allenbach, L.B.; Ball, C.L.; Fusco, W.G.; La Duc, M.T.; Kuhlman, G.M.; Anderson, R.C.; Stuecker, T.; Erickson, I.K.; Benardini, J.; et al. Enumeration, isolation, and characterization of ultraviolet (UV-C) resistant bacteria from rock varnish in the Whipple Mountains, California. Icarus 2005, 174, 585–595. [Google Scholar] [CrossRef]
- Bauermeister, A.; Bentchikou, E.; Moeller, R.; Rettberg, P. Roles of PprA, IrrE, and RecA in the resistance of Deinococcus radiodurans to germicidal and environmentally relevant UV radiation. Arch. Microbiol. 2009, 191, 913–918. [Google Scholar] [CrossRef]
- Jiang, D.; Hatahet, Z.; Melamede, R.J.; Kow, Y.W.; Wallace, S.S. Characterization of Escherichia coli Endonuclease VIII. J. Biol. Chem. 1997, 272, 32230–32239. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M. Substrate specificities and excision kinetics of DNA glycosylases involved in base-excision repair of oxidative DNA damage. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2003, 531, 109–126. [Google Scholar] [CrossRef]
- Kisker, C.; Kuper, J.; Van Houten, B. Prokaryotic Nucleotide Excision Repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012591. [Google Scholar] [CrossRef]
- Amato, P.; Joly, M.; Besaury, L.; Oudart, A.; Taib, N.; Moné, A.I.; Deguillaume, L.; Delort, A.M.; Debroas, D. Active microorganisms thrive among extremely diverse communities in cloud water. PLoS ONE 2017, 12, e0182869. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Scheerer, P.; Oberpichler, I.; Lamparter, T.; Krauß, N. Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore. Proc. Natl. Acad. Sci. USA 2013, 110, 7217–7222. [Google Scholar] [CrossRef] [PubMed]
- Lassalle, F.; Campillo, T.; Vial, L.; Baude, J.; Costechareyre, D.; Chapulliot, D.; Shams, M.; Abrouk, D.; Lavire, C.; Oger-Desfeux, C.; et al. Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens. Genome Biol. Evol. 2011, 3, 762–781. [Google Scholar] [CrossRef]
- Geisselbrecht, Y.; Frühwirth, S.; Schroeder, C.; Pierik, A.J.; Klug, G.; Essen, L.O. CryB from Rhodobacter sphaeroides: A unique class of cryptochromes with new cofactors. EMBO Rep. 2012, 13, 223–229. [Google Scholar] [CrossRef] [PubMed]
- von Zadow, A.; Ignatz, E.; Pokorny, R.; Essen, L.O.; Klug, G. Rhodobacter sphaeroides CryB is a bacterial cryptochrome with (6-4) photolyase activity. FEBS J. 2016, 283, 4291–4309. [Google Scholar] [CrossRef]
- Hördt, A.; López, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Göker, M. Analysis of 1,000+ Type-Strain Genomes Substantially Improves Taxonomic Classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef]
- Dikbas, U.M.; Tardu, M.; Canturk, A.; Gul, S.; Ozcelik, G.; Baris, I.; Ozturk, N.; Kavakli, I.H. Identification and Characterization of a New Class of (6-4) Photolyase from Vibrio cholerae. Biochemistry 2019, 58, 4352–4360. [Google Scholar] [CrossRef]
- Douki, T.; Cadet, J. Individual determination of the yield of the main UV-induced dimeric pyrimidine photoproducts in DNA suggests a high mutagenicity of CC photolesions. Biochemistry 2001, 40, 2495–2501. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Lai, W.; Lyu, C.; Hang, H.; Wang, H. UHPLC-Q-TOF/MS detection of UV-induced TpT dimeric lesions in genomic DNA. J. Chromatogr. 2018, 1096, 135–142. [Google Scholar] [CrossRef]
- Banaś, A.K.; Zgłobicki, P.; Kowalska, E.; Bażant, A.; Dziga, D.; Strzałka, W. All You Need Is Light. Photorepair of UV-Induced Pyrimidine Dimers. Genes 2020, 11, 1304. [Google Scholar] [CrossRef] [PubMed]
- Todo, T.; Takemori, H.; Ryo, H.; Lhara, M.; Matsunaga, T.; Nikaido, O.; Sato, K.; Nomura, T. A new photoreactivating enzyme that specifically repairs ultraviolet light-induced (6-4) photoproducts. Nature 1993, 361, 371–374. [Google Scholar] [CrossRef]
- Vaïtilingom, M.; Amato, P.; Sancelme, M.; Laj, P.; Leriche, M.; Delort, A.M. Contribution of microbial activity to carbon chemistry in clouds. Appl. Environ. Microbiol. 2010, 76, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Slieman, T.A.; Rebeil, R.; Nicholson, W.L. Spore photoproduct (SP) lyase from Bacillus subtilis specifically binds to and cleaves SP (5-thyminyl-5,6-dihydrothymine) but not cyclobutane pyrimidine dimers in UV-irradiated DNA. J. Bacteriol. 2000, 182, 6412–6417. [Google Scholar] [CrossRef]
- Rebeil, R.; Sun, Y.; Chooback, L.; Pedraza-Reyes, M.; Kinsland, C.; Begley, T.P.; Nicholson, W.L. Spore photoproduct lyase from Bacillus subtilis spores is a novel iron-sulfur DNA repair enzyme which shares features with proteins such as class III anaerobic ribonucleotide reductases and pyruvate-formate lyases. J. Bacteriol. 1998, 180, 4879–4885. [Google Scholar] [CrossRef]
- Mohr, S.C.; Sokolov, N.V.H.A.; Chaomei, H.; Setlow, P. Binding of small acid-soluble spore proteins from Bacillus subtilis changes the conformation of DNA from B to A. Proc. Natl. Acad. Sci. USA 1991, 88, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, W.L.; Setlow, B.; Setlow, P. Ultraviolet irradiation of DNA complexed with alpha/beta-type small, acid-soluble proteins from spores of Bacillus or Clostridium species makes spore photoproduct but not thymine dimers. Proc. Natl. Acad. Sci. USA 1991, 88, 8288–8292. [Google Scholar] [CrossRef]
- Whelan, D.R.; Hiscox, T.J.; Rood, J.I.; Bambery, K.R.; McNaughton, D.; Wood, B.R. Detection of an en masse and reversible B- to A-DNA conformational transition in prokaryotes in response to desiccation. J. R. Soc. Interface 2014, 11, 20140454. [Google Scholar] [CrossRef]
- Douki, T.; Cadet, J. Formation of the spore photoproduct and other dimeric lesions between adjacent pyrimidines in UVC-irradiated dry DNA. Photochem. Photobiol. Sci. 2003, 2, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Pedraza-Reyes, M.; Gutierrez-Corona, F.; Nicholson, W.L. Temporal regulation and forespore-specific expression of the spore photoproduct lyase gene by sigma-G RNA polymerase during Bacillus subtilis sporulation. J. Bacteriol. 1994, 176, 3983–3991. [Google Scholar] [CrossRef]
- Manzanera, M. Dealing with water stress and microbial preservation. Environ. Microbiol. 2020, 23, 3351–3359. [Google Scholar] [CrossRef]
- Potts, M. Desiccation tolerance of prokaryotes. Microbiol. Rev. 1994, 58, 755. [Google Scholar] [CrossRef]
- Billi, D.; Potts, M. Life and death of dried prokaryotes. Res. Microbiol. 2002, 153, 7–12. [Google Scholar] [CrossRef]
- Li, H.; Bhaskara, A.; Megalis, C.; Tortorello, M.L. Transcriptomic analysis of Salmonella desiccation resistance. Foodborne Pathog. Dis. 2012, 9, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Finn, S.; Condell, O.; McClure, P.; Amézquita, A.; Fanning, S. Mechanisms of survival, responses, and sources of Salmonella in low-moisture environments. Front. Microbiol. 2013, 4, 331. [Google Scholar] [CrossRef]
- Shuman, S.; Glickman, M.S. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007, 5, 852–861. [Google Scholar] [CrossRef]
- Bhattarai, H.; Gupta, R.; Glickman, M.S. DNA Ligase C1 Mediates the LigD-Independent Nonhomologous End-Joining Pathway of Mycobacterium smegmatis. J. Bacteriol. 2014, 196, 3366. [Google Scholar] [CrossRef]
- Pitcher, R.S.; Green, A.J.; Brzostek, A.; Korycka-Machala, M.; Dziadek, J.; Doherty, A.J. NHEJ protects mycobacteria in stationary phase against the harmful effects of desiccation. DNA Repair 2007, 6, 1271–1276. [Google Scholar] [CrossRef][Green Version]
- Moeller, R.; Setlow, P.; Reitz, G.; Nicholson, W.L. Roles of small, acid-soluble spore proteins and core water content in survival of Bacillus subtilis spores exposed to environmental solar UV radiation. Appl. Environ. Microbiol. 2009, 75, 5202–5208. [Google Scholar] [CrossRef] [PubMed]
- Singh, H. Desiccation and radiation stress tolerance in cyanobacteria. J. Basic Microbiol. 2018, 58, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Romano, I.; Camerlingo, C.; Vaccari, L.; Birarda, G.; Poli, A.; Fujimori, A.; Lepore, M.; Moeller, R.; Di Donato, P. Effects of Ionizing Radiation and Long-Term Storage on Hydrated vs. Dried Cell Samples of Extremophilic Microorganisms. Microorganisms 2022, 10, 190. [Google Scholar] [CrossRef]
- Rainey, F.A.; Ray, K.; Ferreira, M.; Gatz, B.Z.; Nobre, M.F.; Bagaley, D.; Rash, B.A.; Park, M.J.; Earl, A.M.; Shank, N.C.; et al. Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran Desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl. Environ. Microbiol. 2005, 71, 5225–5235. [Google Scholar] [CrossRef]
- Slade, D.; Radman, M. Oxidative Stress Resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 2011, 75, 133–191. [Google Scholar] [CrossRef]
- Kottemann, M.; Kish, A.; Iloanusi, C.; Bjork, S.; DiRuggiero, J. Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles 2005, 9, 219–227. [Google Scholar] [CrossRef]
- Shukla, M.; Chaturvedi, R.; Tamhane, D.; Vyas, P.; Archana, G.; Apte, S.; Bandekar, J.; Desai, A. Multiple-stress tolerance of ionizing radiation-resistant bacterial isolates obtained from various habitats: Correlation between stresses. Curr. Microbiol. 2007, 54, 142–148. [Google Scholar] [CrossRef]
- Romanovskaya, V.A.; Rokitko, P.V.; Mikheev, A.N.; Gushcha, N.I.; Malashenko, Y.R.; Chernaya, N.A. The Effect of γ-Radiation and Desiccation on the Viability of the Soil Bacteria Isolated from the Alienated Zone around the Chernobyl Nuclear Power Plant. Microbiology 2002, 71, 608–613. [Google Scholar] [CrossRef]
- Musilova, M.; Wright, G.; Ward, J.M.; Dartnell, L.R. Isolation of Radiation-Resistant Bacteria from Mars Analog Antarctic Dry Valleys by Preselection, and the Correlation between Radiation and Desiccation Resistance. Astrobiology 2015, 15, 1076. [Google Scholar] [CrossRef]
- Mattimore, V.; Battista, J.R. Radioresistance of Deinococcus radiodurans: Functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 1996, 178, 633. [Google Scholar] [CrossRef]
- Santos, A.L.; Oliveira, V.; Baptista, I.; Henriques, I.; Gomes, N.C.M.; Almeida, A.; Correia, A.; Cunha, Â. Wavelength dependence of biological damage induced by UV radiation on bacteria. Arch. Microbiol. 2013, 195, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Fredrickson, J.K.; Li, S.W.; Gaidamakova, E.K.; Matrosova, V.Y.; Zhai, M.; Sulloway, H.M.; Scholten, J.C.; Brown, M.G.; Balkwill, D.L.; Daly, M.J. Protein oxidation: Key to bacterial desiccation resistance? ISME J. 2008, 2, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Song, M.; Wei, X.; Zhang, H.; Bai, Z.; Zhuang, X. Responses of Phyllosphere Microbiome to Ozone Stress: Abundance, Community Compositions and Functions. Microorganisms 2022, 10, 680. [Google Scholar] [CrossRef] [PubMed]
- NASA Ozone Watch: Latest Status of Ozone. 2023. Available online: https://ozonewatch.gsfc.nasa.gov/SH.html (accessed on 19 March 2023).
- Sun, H.; Xu, G.; Zhan, H.; Chen, H.; Sun, Z.; Tian, B.; Hua, Y. Identification and evaluation of the role of the manganese efflux protein in Deinococcus radiodurans. BMC Microbiol. 2010, 10, 1–8. [Google Scholar] [CrossRef]
- Cox, M.M.; Battista, J.R. Deinococcus radiodurans—The consummate survivor. Nat. Rev. Microbiol. 2005, 3, 882–892. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar]
- Ye, Y.; Godzik, A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 2003, 19, ii246–ii255. [Google Scholar]
- Li, Z.; Jaroszewski, L.; Iyer, M.; Sedova, M.; Godzik, A. FATCAT 2.0: Towards a better understanding of the structural diversity of proteins. Nucleic Acids Res. 2020, 48, W60–W64. [Google Scholar]
- Schuster, L.A.; Reisch, C.R. Plasmids for Controlled and Tunable High-Level Expression in E. coli. Appl. Environ. Microbiol. 2022, 88, e00939-22. [Google Scholar] [CrossRef]

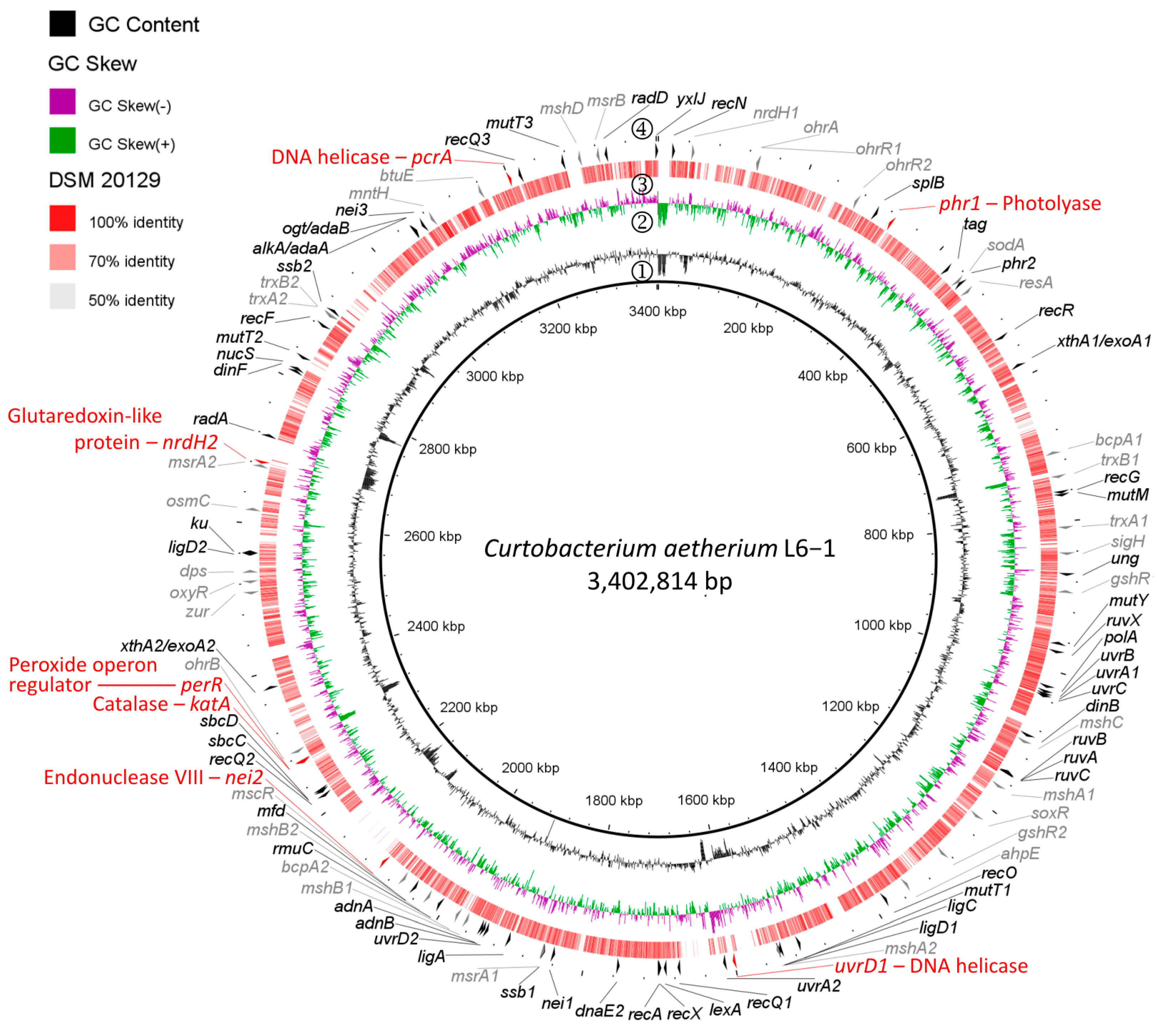

| Strain | Chr | Plasmids | Size (Mbp) | GC % | Genes | rRNA | tRNA | Proteins | ANI (±SD) % | AAI (±SD) % |
|---|---|---|---|---|---|---|---|---|---|---|
| Curtobacterium aetherium L6-1 | 1 | 0 | 3.4 | 72.0 | 3158 | 12 | 46 | 3072 | 83.64 ± 4.97 | 79.97 ± 13.57 |
| Curtobacterium flaccumfaciens DSM 20129 | 1 | 1 | 3.8 | 70.9 | 3616 | 9 | 47 | 3517 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ellington, A.J.; Schult, T.J.; Reisch, C.R.; Christner, B.C. The Genetic Determinants of Extreme UV Radiation and Desiccation Tolerance in a Bacterium Recovered from the Stratosphere. Microorganisms 2025, 13, 756. https://doi.org/10.3390/microorganisms13040756
Ellington AJ, Schult TJ, Reisch CR, Christner BC. The Genetic Determinants of Extreme UV Radiation and Desiccation Tolerance in a Bacterium Recovered from the Stratosphere. Microorganisms. 2025; 13(4):756. https://doi.org/10.3390/microorganisms13040756
Chicago/Turabian StyleEllington, Adam J., Tyler J. Schult, Christopher R. Reisch, and Brent C. Christner. 2025. "The Genetic Determinants of Extreme UV Radiation and Desiccation Tolerance in a Bacterium Recovered from the Stratosphere" Microorganisms 13, no. 4: 756. https://doi.org/10.3390/microorganisms13040756
APA StyleEllington, A. J., Schult, T. J., Reisch, C. R., & Christner, B. C. (2025). The Genetic Determinants of Extreme UV Radiation and Desiccation Tolerance in a Bacterium Recovered from the Stratosphere. Microorganisms, 13(4), 756. https://doi.org/10.3390/microorganisms13040756






