Genomic Characterization of Enterococcus casseliflavus Isolated from Beef Cows and Calves
Abstract
1. Introduction
2. Methodology
2.1. Sample Collection and Processing
2.2. Whole Genome Sequencing
2.3. Genomic Analysis of Enterococcus casseliflavus
2.4. Antimicrobial Susceptibility Testing
3. Results
3.1. Recovery of Enterococcus casseliflavus
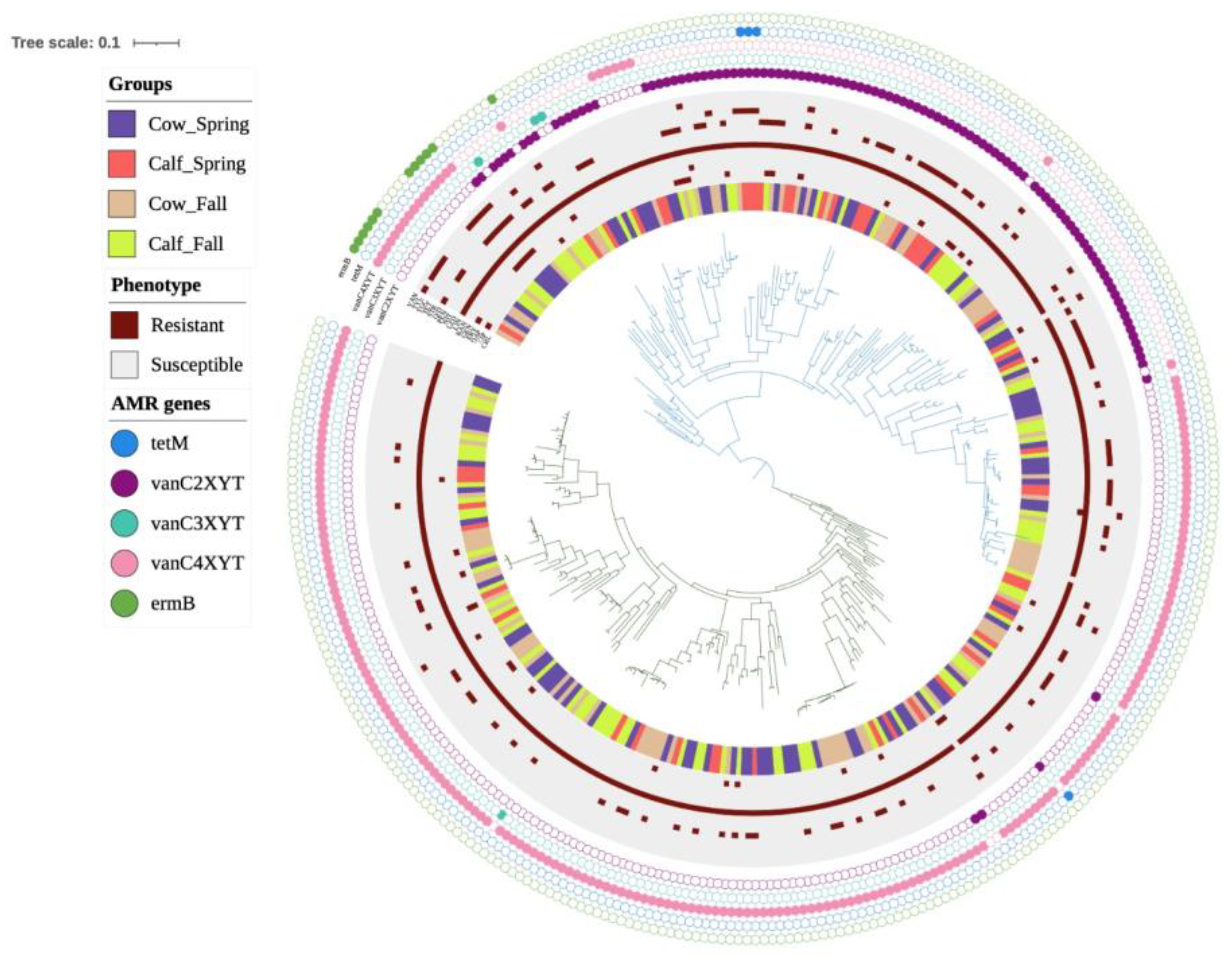
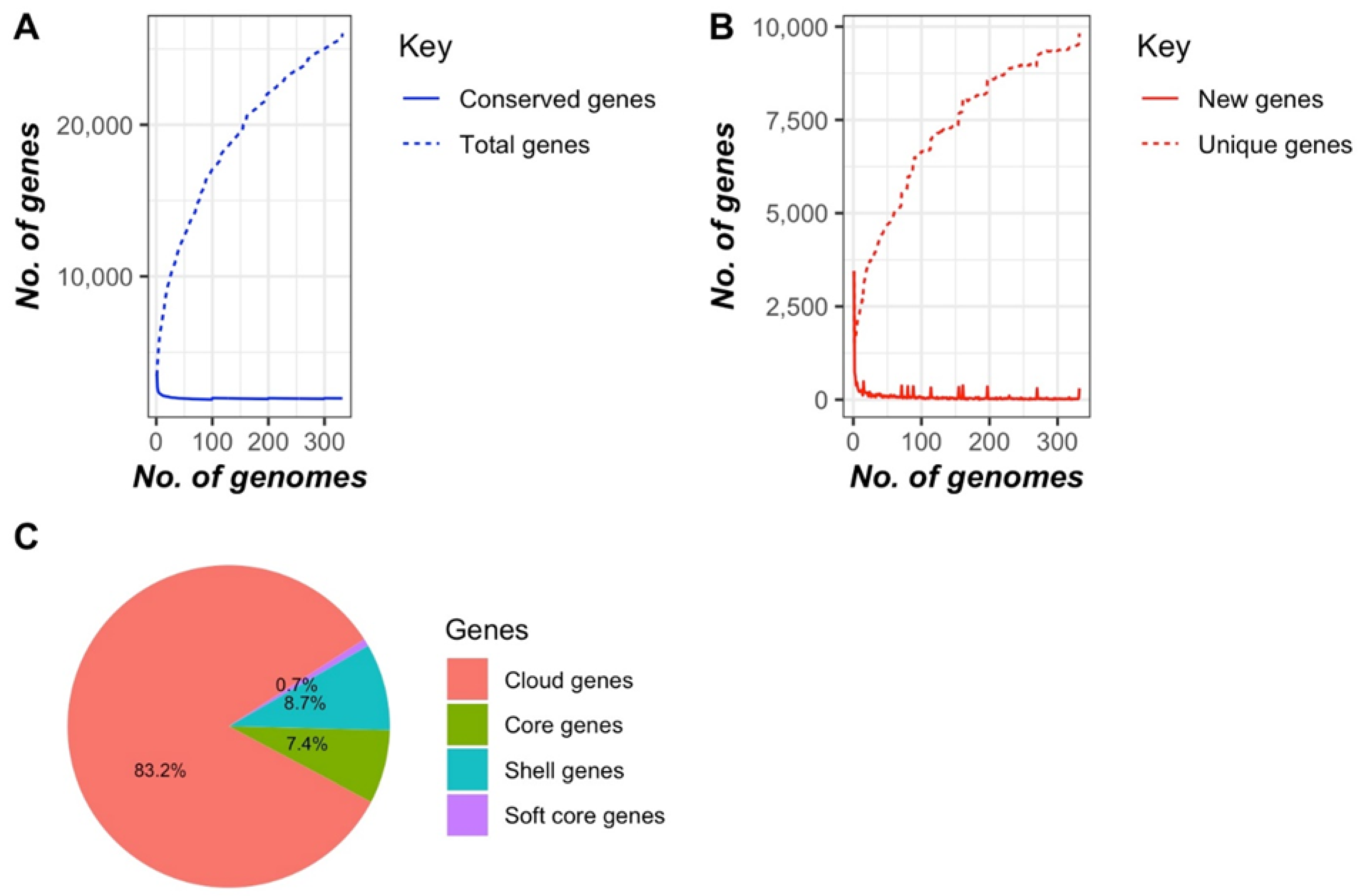
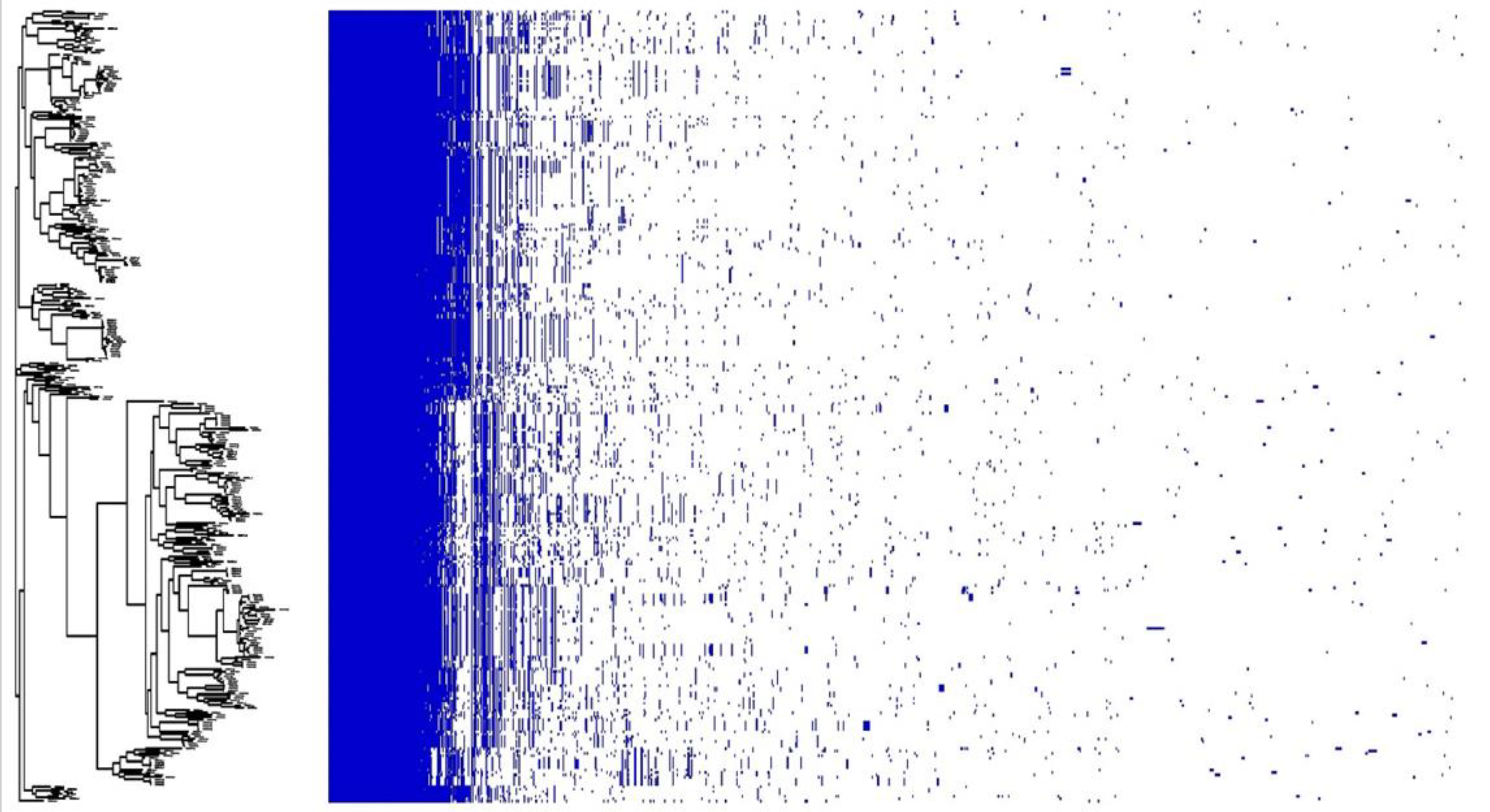
3.2. Genomic Characterization of E. casseliflavus
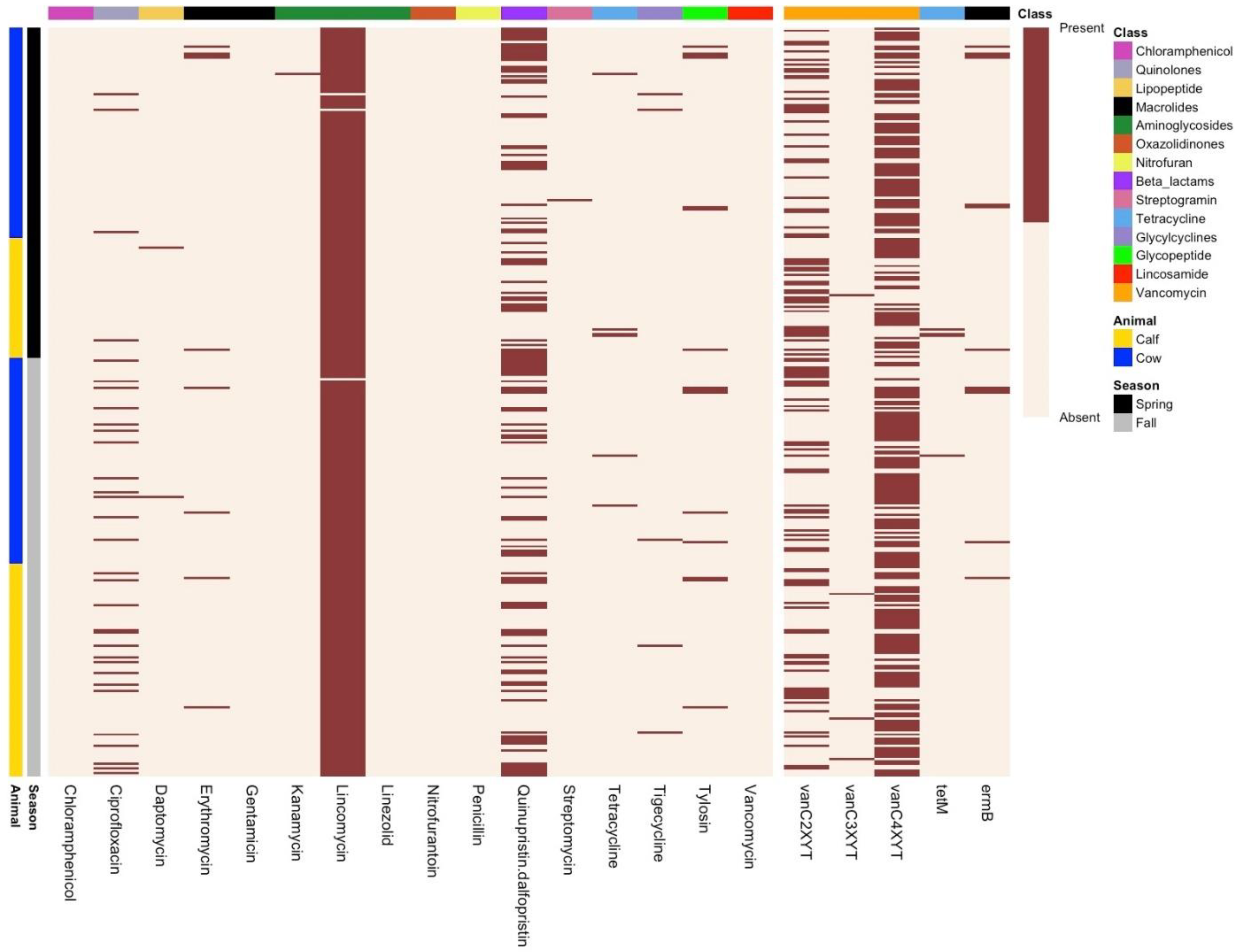
3.3. Phenotypic Resistance Profiling
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Cameron, A.; McAllister, T.A. Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 2016, 7, 68. [Google Scholar] [CrossRef] [PubMed]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 4, 795. [Google Scholar] [CrossRef]
- Gilmore, M.S.; Clewell, D.B.; Ike, Y.; Shankar, N. Enterococci. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Wheeler, A.L.; Hartel, P.G.; Godfrey, D.G.; Hill, J.L.; Segars, W.I. Potential of Enterococcus faecalis as a human fecal indicator for microbial source tracking. J. Environ. Qual. 2002, 31, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 3937. [Google Scholar] [CrossRef]
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef]
- Fiore, E.; Van Tyne, D.; Gilmore, M.S. Pathogenicity of Enterococci. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Yoshino, Y. Enterococcus casseliflavus Infection: A Review of Clinical Features and Treatment. Infect. Drug Resist. 2023, 16, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Lombard, J.E.; Dargatz, D.A.; Fedorka-Cray, P.J. Prevalence, species distribution and antimicrobial resistance of enterococci isolated from US dairy cattle. Lett. Appl. Microbiol. 2010, 52, 41–48. [Google Scholar] [CrossRef]
- Messele, Y.E.; Hasoon, M.F.; Trott, D.J.; Veltman, T.; McMeniman, J.P.; Kidd, S.P.; Low, W.Y.; Petrovski, K.R. Longitudinal Analysis of Antimicrobial Resistance among Enterococcus Species Isolated from Australian Beef Cattle Faeces at Feedlot Entry and Exit. Animals 2022, 12, 2690. [Google Scholar] [CrossRef]
- Fossen, J.D.; Campbell, J.R.; Gow, S.P.; Erickson, N.; Waldner, C.L. Antimicrobial resistance in Enterococcus isolated from western Canadian cow-calf herds. BMC Vet-Res. 2024, 20, 6. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Bharat, A.; Petkau, A.; Avery, B.P.; Chen, J.C.; Folster, J.P.; Carson, C.A.; Kearney, A.; Nadon, C.; Mabon, P.; Thiessen, J.; et al. Correlation between Phenotypic and In Silico Detection of Antimicrobial Resistance in Salmonella enterica in Canada Using Staramr. Microorganisms 2022, 10, 292. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Abricate, Github. Available online: https://github.com/tseemann/abricate (accessed on 25 March 2025).
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal Infection-Treatment and Antibiotic Resistance. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Soltani, M.; Beighton, D.; Philpott-Howard, J.; Woodford, N. Mechanisms of resistance to quinupristin-dalfopristin among isolates of Enterococcus faecium from animals, raw meat, and hospital patients in Western Europe. Antimicrob. Agents Chemother. 2000, 44, 433–436. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcìa-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Himanshu, R.; Prudencio, C.; da Costa, A.C.; Leal, E.; Chang, C.-M.; Pandey, R.P. Systematic Surveillance and Meta-Analysis of Antimicrobial Resistance and Food Sources from China and the USA. Antibiotics 2022, 11, 1471. [Google Scholar] [CrossRef]
- Marques, J.M.; Coelho, M.; Santana, A.R.; Pinto, D.; Semedo-Lemsaddek, T. Dissemination of Enterococcal Genetic Lineages: A One Health Perspective. Antibiotics 2023, 12, 1140. [Google Scholar] [CrossRef]
- Osman, M.; Altier, C.; Cazer, C. Antimicrobial resistance among canine enterococci in the northeastern United States, 2007–2020. Front. Microbiol. 2022, 13, 1025242. [Google Scholar] [CrossRef]
- Prieto, A.M.G.; van Schaik, W.; Rogers, M.R.C.; Coque, T.M.; Baquero, F.; Corander, J.; Willems, R.J.L. Global Emergence and Dissemination of Enterococci as Nosocomial Pathogens: Attack of the Clones? Front. Microbiol. 2016, 7, 788. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; Dapkevicius, M.d.L.E.; Igrejas, G.; Poeta, P. Enterococci, from Harmless Bacteria to a Pathogen. Microorganisms 2020, 8, 1118. [Google Scholar] [CrossRef]
- Mundt, J.O.; Graham, W.F. Streptococcus faecium var. casseliflavus, nov. var. J. Bacteriol. 1968, 95, 2005–2009. [Google Scholar] [CrossRef]
- Badgley, B.D.; Thomas, F.I.M.; Harwood, V.J. The effects of submerged aquatic vegetation on the persistence of environmental populations of Enterococcus spp. Environ. Microbiol. 2010, 12, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Yin, H.; Chen, S.; Peng, H.; Chang, J.; Liu, Z.; Dang, Z. Aerobic degradation of BDE-209 by Enterococcus casseliflavus: Isolation, identification and cell changes during degradation process. J. Hazard. Mater. 2016, 308, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ju, F. Uninheritable but Widespread Bacterial Symbiont Enterococcus casseliflavus Mediates Detoxification of the Insecticide Chlorantraniliprole in the Agricultural Invasive Pest Spodoptera frugiperda. J. Agric. Food Chem. 2024, 72, 18365–18377. [Google Scholar] [CrossRef]
- Safari, R.; Adel, M.; Lazado, C.C.; Caipang, C.M.; Dadar, M. Host-derived probiotics Enterococcus casseliflavus improves resistance against Streptococcus iniae infection in rainbow trout (Oncorhynchus mykiss) via immunomodulation. Fish Shellfish. Immunol. 2016, 52, 198–205. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, K.; Lai, J.; Wu, C.; Shen, J.; Wang, Y. Prevalence and antimicrobial resistance of Enterococcus species of food animal origin from Beijing and Shandong Province, China. J. Appl. Microbiol. 2013, 114, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.-K.; Lai, C.-C.; Wang, J.-Y.; Lin, S.-H.; Liao, C.-H.; Huang, Y.-T.; Wang, C.-Y.; Lin, H.-I.; Hsueh, P.-R. Bacteremia caused by non-faecalis and non-faecium enterococcus species at a Medical center in Taiwan, 2000 to 2008. J. Infect. 2010, 61, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Toc, D.A.; Pandrea, S.L.; Botan, A.; Mihaila, R.M.; Costache, C.A.; Colosi, I.A.; Junie, L.M. Enterococcus raffinosus, Enterococcus durans and Enterococcus avium isolated from a tertiary care hospital in Romania—Retrospective study and brief review. Biology 2022, 11, 598. [Google Scholar] [CrossRef]
- El-Zamkan, M.A.; Mohamed, H.M.A. Antimicrobial resistance, virulence genes and biofilm formation in Enterococcus species isolated from milk of sheep and goat with subclinical mastitis. PLoS ONE 2021, 16, e0259584. [Google Scholar] [CrossRef]
- Fossen, J.D.; Campbell, J.R.; Gow, S.P.; Erickson, N.; Waldner, C.L. Antimicrobial Use in Canadian Cow–Calf Herds. Vet. Sci. 2023, 10, 366. [Google Scholar] [CrossRef] [PubMed]
- Brault, S.A.; Hannon, S.J.; Gow, S.P.; Warr, B.N.; Withell, J.; Song, J.; Williams, C.M.; Otto, S.; Booker, C.W.; Morley, P.S. Antimicrobial Use on 36 Beef Feedlots in Western Canada: 2008–2012. Front. Vet. Sci. 2019, 6, 329. [Google Scholar] [CrossRef]
- Zaheer, R.; Cook, S.R.; Klima, C.L.; Stanford, K.; Alexander, T.; Topp, E.; Read, R.R.; McAllister, T.A. Effect of subtherapeutic vs. therapeutic administration of macrolides on antimicrobial resistance in Mannheimia haemolytica and enterococci isolated from beef cattle. Front. Microbiol. 2013, 4, 133. [Google Scholar] [CrossRef]
- Courvalin, P. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 2006, 42 (Suppl. S1), S25–S34. [Google Scholar] [CrossRef]
- Navarro, F.; Courvalin, P. Analysis of genes encoding D-alanine-D-alanine ligase-related enzymes in Enterococcus casseliflavus and Enterococcus flavescens. Antimicrob. Agents Chemother. 1994, 38, 1788–1793. [Google Scholar] [CrossRef]
- Arias, C.A.; Courvalin, P.; Reynolds, P.E. vanC cluster of vancomycin-resistant Enterococcus gallinarum BM4174. Antimicrob. Agents Chemother. 2000, 44, 1660–1666. [Google Scholar] [CrossRef]
- Dutta, I.; Reynolds, P.E. Biochemical and genetic characterization of the vanC-2 vancomycin resistance gene cluster of Enterococcus casseliflavus ATCC 25788. Antimicrob. Agents Chemother. 2002, 46, 3125–3132. [Google Scholar] [CrossRef]
- Clark, N.C.; Teixeira, L.M.; Facklam, R.R.; Tenover, F.C. Detection and differentiation of vanC-1, vanC-2, and vanC-3 glycopeptide resistance genes in enterococci. J. Clin. Microbiol. 1998, 36, 2294–2297. [Google Scholar] [CrossRef] [PubMed]
- Dutta, I.; Reynolds, P.E. The vanC-3 vancomycin resistance gene cluster of Enterococcus flavescens CCM 439. J. Antimicrob. Chemother. 2003, 51, 703–706. [Google Scholar] [CrossRef]
- Flipse, J.; von Wintersdorff, C.J.H.; van Niekerk, J.M.; Jamin, C.; van Tiel, F.H.; Hasman, H.; van Alphen, L.B. Appearance of vanD-positive Enterococcus faecium in a tertiary hospital in the Netherlands: Prevalence of vanC and vanD in hospitalized patients. Sci. Rep. 2019, 9, 6949. [Google Scholar] [CrossRef] [PubMed]
- McGowan, L.L.; Jackson, C.R.; Barrett, J.B.; Hiott, L.M.; Fedorka-Cray, P.J. Prevalence and antimicrobial resistance of enterococci isolated from retail fruits, vegetables, and meats. J. Food Prot. 2006, 69, 2976–2982. [Google Scholar] [CrossRef] [PubMed]
- Bryson, H.M.; Spencer, C.M. Quinupristin-dalfopristin. Drugs 1996, 52, 406–415. [Google Scholar] [CrossRef]
- Menzies-Gow, N.; Young, N. Antibiotic resistance in faecal bacteria isolated from horses receiving virginiamycin for the prevention of pasture-associated laminitis. Vet. Microbiol. 2011, 152, 424–428. [Google Scholar] [CrossRef]
- Schwarz, S.; Shen, J.; Kadlec, K.; Wang, Y.; Michael, G.B.; Feßler, A.T.; Vester, B. Lincosamides, Streptogramins, Phenicols, and Pleuromutilins: Mode of Action and Mechanisms of Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a027037. [Google Scholar] [CrossRef]
- Simjee, S.; White, D.G.; Wagner, D.D.; Meng, J.; Qaiyumi, S.; Zhao, S.; McDermott, P.F. Identification of vat (E) in Enterococcus faecalis isolates from retail poultry and its transferability to Enterococcus faecium. Antimicrob. Agents Chemother. 2002, 46, 3823–3828. [Google Scholar] [CrossRef]
- Petersen, A.; Jensen, L.B. Analysis of gyrA and parC mutations in enterococci from environmental samples with reduced susceptibility to ciprofloxacin. FEMS Microbiol. Lett. 2004, 231, 73–76. [Google Scholar] [CrossRef]
- Agius, J.E.; Phalen, D.N.; Rose, K.; Eden, J.-S. Genomic Insights Into the Pathogenicity of a Novel Biofilm-Forming Enterococcus sp. Bacteria (Enterococcus lacertideformus) Identified in Reptiles. Front. Microbiol. 2021, 12, 635208. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Pang, S.; Abraham, S.; Coombs, G.W. Molecular characterization and evolution of the first outbreak of vancomycin-resistant Enterococcus faecium in Western Australia. Int. J. Antimicrob. Agents 2019, 53, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Zhang, W.; Song, Y.; Liu, W.; Xu, H.; Xi, X.; Menghe, B.; Zhang, H.; Sun, Z. Comparative genomic analysis of the genus Enterococcus. Microbiol. Res. 2017, 196, 95–105. [Google Scholar] [CrossRef]
- Palmer, K.L.; Godfrey, P.; Griggs, A.; Kos, V.N.; Zucker, J.; Desjardins, C.; Cerqueira, G.; Gevers, D.; Walker, S.; Wortman, J.; et al. Comparative Genomics of Enterococci: Variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 2012, 3, e00318-11. [Google Scholar] [CrossRef] [PubMed]

| Assembly Stats | Average Value |
|---|---|
| N50 contig length (bp) | 406,085 |
| Number of contigs | 35 (range: 8–114) |
| Number of contigs >= 1 kb | 28 |
| Number of contigs in N50 | 4 |
| Genome size (bp) | 3,674,039 |
| GC content of contigs | 42.4% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaidi, S.-e.-Z.; Zaheer, R.; Zovoilis, A.; Fossen, J.; Van Domselaar, G.; Waldner, C.; McAllister, T.A. Genomic Characterization of Enterococcus casseliflavus Isolated from Beef Cows and Calves. Microorganisms 2025, 13, 907. https://doi.org/10.3390/microorganisms13040907
Zaidi S-e-Z, Zaheer R, Zovoilis A, Fossen J, Van Domselaar G, Waldner C, McAllister TA. Genomic Characterization of Enterococcus casseliflavus Isolated from Beef Cows and Calves. Microorganisms. 2025; 13(4):907. https://doi.org/10.3390/microorganisms13040907
Chicago/Turabian StyleZaidi, Sani-e-Zehra, Rahat Zaheer, Athanasios Zovoilis, Jayce Fossen, Gary Van Domselaar, Cheryl Waldner, and Tim A. McAllister. 2025. "Genomic Characterization of Enterococcus casseliflavus Isolated from Beef Cows and Calves" Microorganisms 13, no. 4: 907. https://doi.org/10.3390/microorganisms13040907
APA StyleZaidi, S.-e.-Z., Zaheer, R., Zovoilis, A., Fossen, J., Van Domselaar, G., Waldner, C., & McAllister, T. A. (2025). Genomic Characterization of Enterococcus casseliflavus Isolated from Beef Cows and Calves. Microorganisms, 13(4), 907. https://doi.org/10.3390/microorganisms13040907








