Genome-Informed Real-Time PCR Assay for Detection of ‘Candidatus Phytoplasma Prunorum,’ Which Is Associated with European Stone Fruit Yellows
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plant Hosts, and DNA Extraction
2.2. Target Identification and Primer Design
2.3. PCR Reaction Conditions
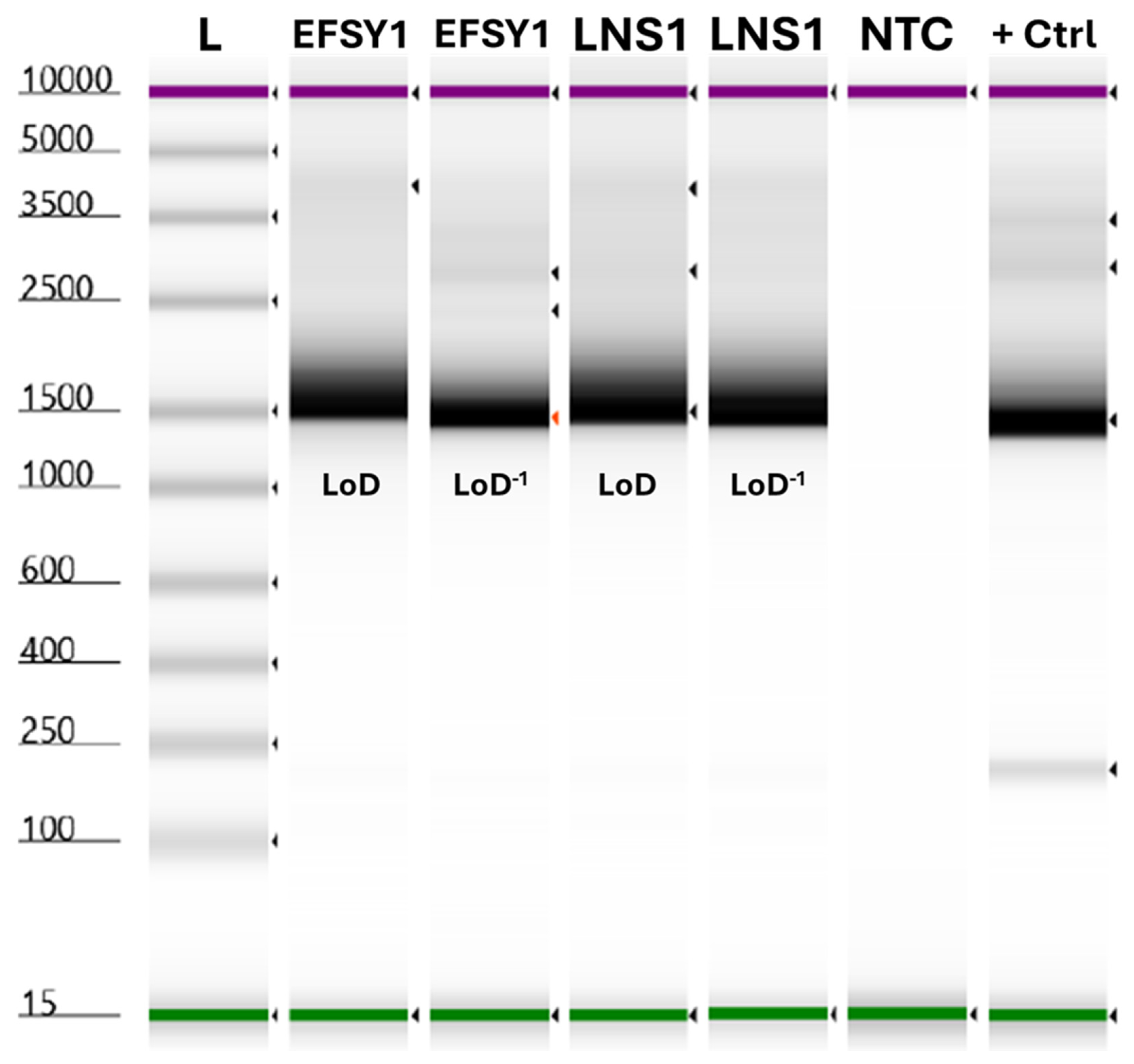
2.4. Gel Electrophoresis and Sequencing
2.5. Synthetic Positive Control
2.6. Assay Validation Testing
3. Results
3.1. Target Identification
3.2. Assay Sensitivity
3.3. Assay Specificity
3.4. Precision and Reproducibility
3.5. Synthetic Control
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
General Disclaimer
Commercial Endorsement Disclaimer
References
- Marcone, C.; Jarausch, B.; Jarausch, W. Candidatus Phytoplasma prunorum, the causal agent of European stone fruit yellows: An overview. J. Plant Pathol. 2010, 92, 19–34. [Google Scholar]
- Seemüller, E.; Schneider, B. ‘Candidatus Phytoplasma mali’, ‘Candidatus Phytoplasma pyri’ and ‘Candidatus Phytoplasma prunorum’, the causal agents of apple proliferation, pear decline and European stone fruit yellows, respectively. Int. J. Syst. Evol. Microbiol. 2004, 54, 1217–1226. [Google Scholar] [CrossRef]
- Carraro, L.; Osler, R. European stone fruit yellows: A destructive disease in the mediterranean basin. In Virus and Virus-Like Diseases of Stone Fruits, with Particular Reference to the Mediterranean Region; Myrta, A., Di, T.B., Savino, V., Eds.; Options Méditerranéennes Série B; CIHEAM-IAMB: Valenzano, Italy, 2003; Volume 45, pp. 113–117. [Google Scholar]
- FAOSTAT. Value of Agricultural Production. Available online: https://www.fao.org/faostat/en/#data/QV (accessed on 16 December 2024).
- FAOSTAT. Crops and Livestock Products. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 16 December 2024).
- EPPO. ‘Candidatus Phytoplasma mali’ (PHYPMA). Available online: https://gd.eppo.int/taxon/PHYPMA (accessed on 5 February 2025).
- EPPO. ‘Candidatus Phytoplasma prunorum’ (PHYPPR). Available online: https://gd.eppo.int/taxon/PHYPPR (accessed on 5 February 2025).
- CABI. Phytoplasma Prunorum (Apricot Chlorotic Leafroll). Available online: https://www.cabidigitallibrary.org/doi/full/10.1079/cabicompendium.34065 (accessed on 5 February 2025).
- CABI. Phytoplasma Mali (Apple Proliferation). Available online: https://www.cabidigitallibrary.org/doi/10.1079/cabicompendium.6502 (accessed on 5 February 2025).
- CABI. Phytoplasma Pyri (Pear Decline). Available online: https://www.cabidigitallibrary.org/doi/10.1079/cabicompendium.44021 (accessed on 5 February 2025).
- EPPO. ‘Candidatus Phytoplasma pyri’ (PHYPPY). Available online: https://gd.eppo.int/taxon/PHYPPY (accessed on 5 February 2025).
- Carraro, L.; Loi, N.; Ermacora, P.; Osler, R. High tolerance of European plum varieties to plum leptonecrosis. Eur. J. Plant Pathol. 1998, 104, 141–145. [Google Scholar] [CrossRef]
- Christensen, N.M.; Nicolaisen, M.; Hansen, M.; Schulz, A. Distribution of phytoplasmas in infected plants as revealed by real-time PCR and bioimaging. Mol. Plant-Microbe Interact. 2004, 17, 1175–1184. [Google Scholar] [CrossRef]
- Del Serrone, P.; La Starza, S.; Krystai, L.; Kolber, M.; Barba, M. Occurrence of apple proliferation and pear decline phytoplasmas in diseased pear trees in Hungary. J. Plant Pathol. 1998, 80, 53–58. [Google Scholar]
- Lee, I.-M.; Bertaccini, A.; Vibio, M.; Gundersen, D.E. Detection of multiple phytoplasmas in perennial fruit trees with decline symptoms in Italy. Phytopathology 1995, 85, 728–735. [Google Scholar] [CrossRef]
- Mall, S.; Kirdat, K.; Singh, A.; Tiwarekar, B.; Sathe, S.; Marcone, C.; Yadav, A. Updates on phytoplasma diseases associated with weeds acting as alternate hosts in Asian countries. In Phytoplasma Diseases of Major Crops, Trees, and Weeds; Tiwari, A.K., Caglayan, K., Hoat, T.X., Al Subhi, A., Nejat, N., Reddy, G., Eds.; Academic Press: Cambridge, MA, USA, 2023; Volume 2, pp. 347–365. [Google Scholar]
- Mall, S.; Rao, G.P.; Marcone, C. Phytoplasma diseases of weeds: Detection, taxonomy and diversity. In Recent Trends in Biotechnology and Microbiology; Gaur, R.K., Ed.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2010; pp. 87–108. [Google Scholar]
- Mehle, N.; Brzin, J.; Boben, J.; Hren, M.; Frank, J.; Petrovič, N.; Gruden, K.; Dreo, T.; Žežlina, I.; Seljak, G.; et al. First report of ‘Candidatus Phytoplasma mali’ in Prunus avium, P. armeniaca and P. domestica. Plant Pathol. 2007, 56, 721. [Google Scholar] [CrossRef]
- Hodgetts, J.; Boonham, N.; Mumford, R.; Dickinson, M. Panel of 23S rRNA gene-based real-time PCR assays for improved universal and group-specific detection of phytoplasmas. Appl. Environ. Microbiol. 2009, 75, 2945–2950. [Google Scholar] [CrossRef]
- Jarausch, W.; Fuchs, A.; Jarausch, B. Establishment of a quantitative real-time PCR assay for the specific quantification of Ca. Phytoplasma prunorum in plants and insects. Jul.-Kühn-Arch. 2010, 427, 392. [Google Scholar]
- Martini, M.; Loi, N.; Ermacora, P.; Carraro, L.; Pastore, M. A real-time PCR method for detection and quantification of ‘Candidatus Phytoplasma prunorum’ in its natural hosts. Bull. Insectol. 2007, 60, 251. [Google Scholar]
- Mehle, N.; Nikolić, P.; Gruden, K.; Ravnikar, M.; Dermastia, M. Real-time PCR for specific detection of three phytoplasmas from the apple proliferation group. In Phytoplasma: Methods and Protocols; Dickinson, M., Hodgetts, J., Eds.; Springer: New York, NY, USA, 2013; pp. 269–281. [Google Scholar]
- Nikolić, P.; Mehle, N.; Gruden, K.; Ravnikar, M.; Dermastia, M. A panel of real-time PCR assays for specific detection of three phytoplasmas from the apple proliferation group. Mol. Cell. Probes 2010, 24, 303–309. [Google Scholar] [CrossRef]
- Pignatta, D.; Pollini, C.P.; Giunchedi, L.; Ratti, C.; Reggiani, N.; Forno, F.; Mattedi, L.; Gobber, M.; Miorelli, P.; Ropelato, E. A real-time PCR assay for the detection of European Stone Fruit Yellows Phytoplasma (ESFYP) in plant propagation material. Acta Hortic. 2008, 781, 499–503. [Google Scholar] [CrossRef]
- Torres, E.; Bertolini, E.; Cambra, M.; Montón, C.; Martín, M.P. Real-time PCR for simultaneous and quantitative detection of quarantine phytoplasmas from apple proliferation (16SrX) group. Mol. Cell. Probes 2005, 19, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Yvon, M.; Thébaud, G.; Alary, R.; Labonne, G. Specific detection and quantification of the phytopathogenic agent ‘Candidatus Phytoplasma prunorum’. Mol. Cell. Probes 2009, 23, 227–234. [Google Scholar] [CrossRef]
- Minguzzi, S.; Terlizzi, F.; Lanzoni, C.; Poggi Pollini, C.; Ratti, C. A rapid protocol of crude RNA/DNA extraction for RT-qPCR detection and quantification of ‘Candidatus phytoplasma prunorum’. PLoS ONE 2016, 11, e0146515. [Google Scholar] [CrossRef]
- Aldaghi, M.; Massart, S.; Dutrecq, O.; Bertaccini, A.; Jijakli, M.H.; Lepoivre, P. A simple and rapid protocol of crude DNA extraction from apple trees for PCR and real-time PCR detection of ‘Candidatus Phytoplasma mali’. J. Virol. Methods 2009, 156, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Aldaghi, M.; Massart, S.; Roussel, S.; Jijakli, M.H. Development of a new probe for specific and sensitive detection of ‘Candidatus Phytoplasma mali’ in inoculated apple trees. Ann. Appl. Biol. 2007, 15, 251–258. [Google Scholar] [CrossRef]
- Baric, S. Quantitative real-time PCR analysis of ‘Candidatus Phytoplasma mali’ without external standard curves. Erwerbs-Obstbau 2012, 54, 147–153. [Google Scholar] [CrossRef]
- Baric, S.; Berger, J.; Cainelli, C.; Kerschbamer, C.; Letschka, T.; Dalla Via, J. Seasonal colonisation of apple trees by ‘Candidatus Phytoplasma mali’ revealed by a new quantitative TaqMan real-time PCR approach. Eur. J. Plant Pathol. 2011, 129, 455–467. [Google Scholar] [CrossRef]
- Baric, S.; Dalla-Via, J. A new approach to apple proliferation detection: A highly sensitive real-time PCR assay. J. Microbiol. Methods 2004, 57, 135–145. [Google Scholar] [CrossRef]
- Baric, S.; Kerschbamer, C.; Dalla Via, J. TaqMan real-time PCR versus four conventional PCR assays for detection of apple proliferation phytoplasma. Plant Mol. Biol. Rep. 2006, 24, 169–184. [Google Scholar] [CrossRef]
- Valasevich, N.; Schneider, B. Rapid detection of “Candidatus Phytoplasma mali” by recombinase polymerase amplification assays. J. Phytopathol. 2017, 165, 762–770. [Google Scholar] [CrossRef]
- Yasuhara-Bell, J.; Costanzo, S.; Mavrodieva, V. Validation of a modified quantitative PCR and synthetic DNA gBlocks control for detection of ‘Candidatus Phytoplasma mali.’. Phytopath. Mollicutes 2023, 13, 143–144. [Google Scholar] [CrossRef]
- Fonesco, J.P.; Nunziata, S.; Yasuhara-Bell, J.; Bertaccini, A.; Rivera, Y.; Newberry, E. A draft genome resource for ‘Candidatus Phytoplasma prunorum’, the agent associated with European Stone Fruit Yellows disease. PhytoFrontiers 2024. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, W.; Lee, I.M.; Shao, J.; Suo, X.; Davis, R.E. The iPhyClassifier, an interactive online tool for phytoplasma classification and taxonomic assignment. In Phytoplasma. Methods in Molecular Biology; Dickinson, M., Hodgetts, J., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 938, pp. 329–338. [Google Scholar]
- Bai, X.; Zhang, J.; Ewing, A.; Miller, S.A.; Jancso Radek, A.; Shevchenko, D.V.; Tsukerman, K.; Walunas, T.; Lapidus, A.; Campbell, J.W.; et al. Living with genome instability: The adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 2006, 188, 3682–3696. [Google Scholar] [CrossRef]
- Tran-Nguyen, L.T.T.; Kube, M.; Schneider, B.; Reinhardt, R.; Gibb, K.S. Comparative genome analysis of “Candidatus Phytoplasma australiense” (subgroup tuf-Australia I; rp-A) and “Ca. Phytoplasma asteris” strains OY-M and AY-WB. J. Bacteriol. 2008, 190, 3979–3991. [Google Scholar] [CrossRef] [PubMed]
- Kube, M.; Schneider, B.; Kuhl, H.; Dandekar, T.; Heitmann, K.; Migdoll, A.M.; Reinhardt, R.; Seemüller, E. The linear chromosome of the plant-pathogenic mycoplasma ‘Candidatus Phytoplasma mali’. BMC Genom. 2008, 9, 306. [Google Scholar] [CrossRef]
- Mitrović, J.; Siewert, C.; Duduk, B.; Hecht, J.; Mölling, K.; Broecker, F.; Beyerlein, P.; Büttner, C.; Bertaccini, A.; Kube, M. Generation and analysis of draft sequences of ‘Stolbur’ phytoplasma from multiple displacement amplification templates. J. Mol. Microbiol. Biotechnol. 2014, 24, 1–11. [Google Scholar] [CrossRef]
- Oshima, K.; Kakizawa, S.; Nishigawa, H.; Jung, H.Y.; Wei, W.; Suzuki, S.; Arashida, R.; Nakata, D.; Miyata, S.I.; Ugaki, M.; et al. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 2004, 36, 27–29. [Google Scholar] [CrossRef]
- Andersen, M.T.; Liefting, L.W.; Havukkala, I.; Beever, R.E. Comparison of the complete genome sequence of two closely related isolates of ‘Candidatus Phytoplasma australiense’ reveals genome plasticity. BMC Genom. 2014, 14, 529. [Google Scholar] [CrossRef]
- Médigue, C.; Calteau, A.; Cruveiller, S.; Gachet, M.; Gautreau, G.; Josso, A.; Lajus, A.; Langlois, J.; Pereira, H.; Planel, R.; et al. MicroScope—An integrated resource for community expertise of gene functions and comparative analysis of microbial genomic and metabolic data. Brief. Bioinform. 2019, 20, 1071–1084. [Google Scholar] [CrossRef] [PubMed]
- Vallenet, D.; Belda, E.; Calteau, A.; Cruveiller, S.; Engelen, S.; Lajus, A.; Le Fèvre, F.; Longin, C.; Mornico, D.; Roche, D.; et al. MicroScope—An integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013, 41, D636–D647. [Google Scholar] [CrossRef] [PubMed]
- Vallenet, D.; Calteau, A.; Cruveiller, S.; Gachet, M.; Lajus, A.; Josso, A.; Mercier, J.; Renaux, A.; Rollin, J.; Rouy, Z.; et al. MicroScope in 2017: An expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res. 2017, 45, D517–D528. [Google Scholar] [CrossRef]
- Vallenet, D.; Calteau, A.; Dubois, M.; Amours, P.; Bazin, A.; Beuvin, M.; Burlot, L.; Bussell, X.; Fouteau, S.; Gautreau, G.; et al. MicroScope: An integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2020, 48, D579–D589. [Google Scholar] [CrossRef]
- Vallenet, D.; Engelen, S.; Mornico, D.; Cruveiller, S.; Fleury, L.; Lajus, A.; Rouy, Z.; Roche, D.; Salvignol, G.; Scarpelli, C.; et al. MicroScope: A platform for microbial genome annotation and comparative genomics. Database 2009, 2009, bap021. [Google Scholar] [CrossRef] [PubMed]
- Vallenet, D.; Labarre, L.; Rouy, Z.; Barbe, V.; Bocs, S.; Cruveiller, S.; Lajus, A.; Pascal, G.; Scarpelli, C.; Medigue, C. MaGe: A microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006, 34, 53–65. [Google Scholar] [CrossRef]
- Owczarzy, R.; Tataurov, A.V.; Wu, Y.; Manthey, J.A.; McQuisten, K.A.; Almabrazi, H.G.; Pedersen, K.F.; Lin, Y.; Garretson, J.; McEntaggart, N.O.; et al. IDT SciTools: A suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008, 36, W163–W169. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Ew, M.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Deng, S.; Hiruki, C. Amplification of 16S rRNA genes from culturable and nonculturable mollicutes. J. Microbiol. Methods 1991, 14, 53–61. [Google Scholar] [CrossRef]
- Lee, I.-M.; Martini, M.; Marcone, C.; Zhu, S.F. Classification of phytoplasma strains in the elm yellows group (16SrV) and proposal of ‘Candidatus Phytoplasma ulmi’ for the phytoplasma associated with elm yellows. Int. J. Syst. Evol. Microbiol. 2004, 52, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Makarova, O.; Contaldo, N.; Paltrinieri, S.; Kawube, G.; Bertaccini, A.; Nicolaisen, M. DNA barcoding for identification of ‘Candidatus Phytoplasmas’ using a fragment of the elongation factor Tu gene. PLoS ONE 2012, 7, e52092. [Google Scholar] [CrossRef] [PubMed]
- Jarausch, W.; Lansac, M.; Saillard, C.; Broquaire, J.M.; Dosba, F. PCR assay for specific detection of European stone fruit yellows phytoplasmas and its use for epidemiological studies in France. Eur. J. Plant Pathol. 1998, 104, 17–27. [Google Scholar] [CrossRef]
- Jarausch, W.; Peccerella, T.; Schwind, N.; Jarausch, B.; Krczal, G. Establishment of a quantitative real-time PCR assay for the quantification of apple proliferation phytoplasmas in plants and insects. Acta Hortic. 2003, 657, 415–420. [Google Scholar] [CrossRef]
- Martini, M.; Ermacora, P.; Falginella, L.; Loi, N.; Carraro, L. Molecular differentiation of ‘Candidatus Phytoplasma mali’ and its spreading in Friuli Venezia Giulia Region (north-east Italy). Acta Hortic. 2008, 781, 395–402. [Google Scholar] [CrossRef]
- Paltrinieri, S.; Martini, M.; Ermacora, P.; Bertaccini, A. Comparison of different detection systems for apple proliferation phytoplasmas in Trentino (North Italy). Acta Hortic. 2008, 781, 453–457. [Google Scholar]

| Organism | RFLP Group | Sample ID | Host a | Host (Common) | Origin | Source b,c |
|---|---|---|---|---|---|---|
| Alternaria alternata | N/A | 22033001-01 (B) | Citrus limon | Lemon | USA, TX | PPCDL |
| Aureobasidium pullulans | N/A | DSM 14940 | Malus domestica | Apple | Germany | Westbridge Ag |
| Aureobasidium pullulans | N/A | DSM 14941 | Malus domestica | Apple | Germany | Westbridge Ag |
| ‘Candidatus Phytoplasma’ | 16SrII-C | CoPh | Cocos nucifera | Coconut | Tanzania | EPPO-Q-bank |
| ‘Candidatus Phytoplasma’ | 16SrIII-B | GY-U | Vitis vinifera var. ‘Chardonnay’ | Chardonnay grape | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma’ | 16SrIX-C | PEY | Pichris echioides | Bristly oxtongue | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma’ | 16SrV-C | ALY | Alnus sp. | Alder | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma’ | 16SrXI-C | BVK | Psammotettix cephalotes | Leafhopper | Germany | EPPO-Q-bank |
| ‘Candidatus Phytoplasma asteris’ | 16SrI-A | 23092601-01 | Vitis spp. | Grape | MN, USA | PPCDL |
| ‘Candidatus Phytoplasma asteris’ | 16SrI-A | 23092601-02 | Vitis spp. | Grape | MN, USA | PPCDL |
| ‘Candidatus Phytoplasma asteris’ | 16SrI-A | 23092601-03 | Vitis spp. | Grape | MN, USA | PPCDL |
| ‘Candidatus Phytoplasma asteris’ | 16SrI-F | AY-A | Prunus armeniaca | Apricot | Spain | EPPO-Q-bank |
| ‘Candidatus Phytoplasma aurantifolia’ | 16SrII-B | WBDL | Citrus spp. | Lime | Oman | EPPO-Q-bank |
| ‘Candidatus Phytoplasma brasilliense’ | 16SrXV-A | SuV | Hibiscus spp. | Hibiscus | France | EPPO-Q-bank |
| ‘Candidatus Phytoplasma mali’ | 16SrX-A | AP-1 | Malus domestica | Apple | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma mali’ | 16SrX-A | AP-15 | Malus domestica | Apple | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma mali’ | 16SrX-A | AP-2 | Malus domestica | Apple | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma mali’ | 16SrX-A | AP-3 | Malus domestica | Apple | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma mali’ | 16SrX-A | APxN | Malus domestica | Apple | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma mali’ | 16SrX-A | C71 | Malus domestica | Apple | Italy | K. Zikeli |
| ‘Candidatus Phytoplasma mali’ | 16SrX-A | P4 | Malus domestica | Apple | Slovenia | N. Mehle |
| ‘Candidatus Phytoplasma mali’ | 16SrX-A | P6 | Pooled Malus domestica/Cacopsylla picta | Apple/psyllid (jumping plant lice) | Slovenia | N. Mehle |
| ‘Candidatus Phytoplasma palmae’ | 16SrIV-D | 19101702-01 | Phoenix sylvestris | Wild date palm | MS, USA | PPCDL |
| ‘Candidatus Phytoplasma palmae’ | 16SrIV-D | 19110501-02 | Phoenix sylvestris | Wild date palm | MS, USA | PPCDL |
| ‘Candidatus Phytoplasma pini’-related | 16SrXXI-B | MDPP | Pinus pungens | Mountain pine | MD, USA | S. Costanzo |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 19122001-22 | Prunus spp. | - | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 19122001-41 | Prunus spp. | - | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 20012301-04 | Prunus spp. | Cherry | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 20012301-05 | Prunus spp. | Cherry | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 20012301-06 | Prunus spp. | Cherry | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 20012301-07 | Prunus spp. | Cherry | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 20012301-08 | Prunus spp. | Cherry | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 20012301-09 | Prunus spp. | Cherry | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 20012301-10 | Prunus spp. | Cherry | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 20012301-11 | Prunus spp. | Cherry | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 21021201-02 | Prunus spp. | - | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pruni’ rrnA | 16SrIII-A rrn | 21021201-03 | Prunus spp. | - | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma prunorum’ | 16SrX-F | ESFY1 | Prunus salicina | Japanese/Chinese plum | Italy | K. Zikeli |
| ‘Candidatus Phytoplasma prunorum’ | 16SrX-F | ESFY-1A | Prunus armeniaca | Apricot | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma prunorum’ | 16SrX-F | ESFY-1P | Prunus salicina | Japanese plum | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma prunorum’ | 16SrX-F | ESFY-1PE | Prunus persica | Peach | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma prunorum’ | 16SrX-F | ESFY-2A | Prunus armeniaca | Apricot | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma prunorum’ | 16SrX-F | ESFY-2P | Prunus salicina | Japanese plum | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma prunorum’ | 16SrX-F | ESFY-2PE | Prunus persica | Peach | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma prunorum’ | 16SrX-F | LNp | Prunus salicina | Japanese plum | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma prunorum’ | 16SrX-F | LNS1 | Prunus salicina | Japanese plum | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma prunorum’ | 16SrX-F | LNS2 | Prunus salicina | Japanese plum | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma prunorum’ | 16SrX-F | P1 | Prunus americana and P. persica | American plum and peach | Slovenia | N. Mehle |
| ‘Candidatus Phytoplasma pyri’ | 16SrX-C | 20012301-03 | Pyrus spp. | Pear | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pyri’ | 16SrX-C | 21021201-01 | Pyrus spp. | Pear | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pyri’ | 16SrX-C | 23111702-01 | Pyrus spp. | Pear | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma pyri’ | 16SrX-C | P3 | Pyrus communis | European/common pear | Slovenia | N. Mehle |
| ‘Candidatus Phytoplasma pyri’ | 16SrX-C | PD | Pyrus communis | European/common pear | Germany | EPPO-Q-bank |
| ‘Candidatus Phytoplasma rubi’ | 16SrV-E | RuS | Rubus spp. | Brambles | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma solani’ | 16SrXII-A | CH-1 | Vitis vinifera var. ’Chardonnay’ | Chardonnay grape | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma solani’ | 16SrXII-A | LNIV | Prunus salicina | Japanese plum | Italy | EPPO-Q-bank |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 19041105-01 | Alnus spp. | Alder | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 19081401-01 | Alnus spp. | Alder | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 19122001-01 | Alnus spp. | Alder | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 19122001-26 | Alnus spp. | Alder | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 19122001-32 | Alnus spp. | Alder | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 19122001-40 | Prunus spp. | - | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 19122001-42 | Alnus spp. | Alder | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 21012601-01 | Alnus spp. | Alder | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 21012601-02 | Alnus spp. | Alder | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 21031201-01 | Malus spp. | Apple | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 21031201-02 | Malus spp. | Apple | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 21042602-01 | Malus spp. | Apple | WA, USA | PPCDL |
| ‘Candidatus Phytoplasma vitis’-related | 16SrV-C | 21042602-02 | Malus spp. | Apple | WA, USA | PPCDL |
| Colletotrichum gloeospiriodes | N/A | 22033001-01 (C) | Citrus limon | Lemon | USA, TX | PPCDL |
| Colletotrichum gloeospiriodes | N/A | 22033001-01 (D1) | Citrus limon | Lemon | USA, TX | PPCDL |
| Colletotrichum queenslandicum | N/A | 17030501 (C-11) | Citrus limon | Lemon | USA, TX | PPCDL |
| Colletotrichum queenslandicum | N/A | 17030501 (C-5) | Citrus limon | Lemon | USA, TX | PPCDL |
| Erwinia amylovora | N/A | Ea110 | Malus domestica | Apple | USA, OR | V. Stockwell |
| Erwinia amylovora | N/A | Ea153 | Malus domestica | Apple | USA, OR | V. Stockwell |
| Erwinia amylovora | N/A | Fire blight | Malus domestica | Apple | USA, MD | J. Yasuhara-Bell |
| Erwinia amylovora | N/A | Fire blight | Malus domestica | Apple | USA, MD | PGQP |
| Erwinia amylovora | N/A | Parkdale | Malus domestica | Apple | USA, MI | V. Stockwell |
| Erwinia aphidicola | N/A | 17110702-04 | Cucumis melo var. ’mlada’ | Mlada melon | USA, CA | PPCDL |
| Erwinia billingae | N/A | Eh24 | Pyrus spp. | Pear | Turkey | V. Stockwell |
| Erwinia billingae | N/A | NCPPB 661T | Pyrus communis | European/common pear | UK | NCPPB |
| Erwinia persicina | N/A | 19122706-01 | Cucumis melo | Melon | USA, CA | PPCDL |
| Erwinia persicina | N/A | LA611 | Rubus spp. | Raspberry | USA, WA | V. Stockwell |
| Erwinia persicina | N/A | LA659 | Rubus spp. | Raspberry | USA, WA | V. Stockwell |
| Erwinia piriflorinigrans | N/A | CFBP 5888T | Pyrus communis | European/common pear | Spain | CFPB |
| Erwinia pyrifoliae | N/A | 23-02275 | Fragaria × ananassa | Strawberry | USA, OH | PPCDL |
| Erwinia pyrifoliae | N/A | Ejp 556 | Pyrus pyrifolia | Asian Pear | Japan | A. Svircev |
| Erwinia pyrifoliae | N/A | Ejp 617 | Pyrus pyrifolia | Asian Pear | Japan | A. Svircev |
| Erwinia pyrifoliae | N/A | Ep 28/96 | Pyrus pyrifolia | Asian Pear | Korea | A. Svircev |
| Erwinia pyrifoliae | N/A | Ep 4/97 | Pyrus pyrifolia | Asian Pear | Korea | A. Svircev |
| Erwinia rhapontici | N/A | 19122702-01 | Delphinium spp. | Larkspur | USA, CA | PPCDL |
| Erwinia tasmaniensis | N/A | Et1/99T | Malus domestica | Apple | Australia | V. Stockwell |
| Erwinia uzenensis | N/A | NCPPB 4475T | Pyrus communis | European/common pear | Japan | NCPPB |
| Monilinia fructicola | N/A | 19090401-01A | Prunus persica | Peach | USA, WV | K. Zeller |
| Monilinia fructicola | N/A | 19090401-02A | Malus domestica | Apple | USA, MN | PPCDL |
| Monilinia fructicola | N/A | Mf35 | Malus domestica | Apple | USA, MN | PPCDL |
| Monilinia fructigena | N/A | Mfg2-GE-A E | Malus domestica | Apple | Hungary | K. Zeller |
| Monilinia laxa | N/A | PSG1 | Prunus persica | Peach | Italy | K. Zeller |
| Monilinia polystroma | N/A | SP61 | Prunus spp. | Plum | Poland | K. Zeller |
| Neofabraea alba | N/A | PD-1696 | Malus domestica | Apple | USA, WA | T. Wilson |
| Neofabraea sp. | N/A | PD-1597 | Malus domestica | Apple | USA, WA | T. Wilson |
| Neofabraea sp. | N/A | PD-1617 | Malus domestica | Apple | USA, WA | T. Wilson |
| Neofabraea sp. | N/A | PD-1655B | Malus domestica | Apple | USA, WA | T. Wilson |
| Pantoea agglomerans | N/A | 23091801-01 | Triticum aestivum | Wheat | USA, CO | PPCDL |
| Pantoea agglomerans | N/A | 23091801-03 | Triticum aestivum | Wheat | USA, CO | PPCDL |
| Pantoea agglomerans | N/A | 23091801-04 | Triticum aestivum | Wheat | USA, CO | PPCDL |
| Pantoea agglomerans | N/A | E325 | Malus domestica | Apple | USA, WA | V. Stockwell |
| Pantoea allii | N/A | 23091801-02 | Triticum aestivum | Wheat | USA, CO | PPCDL |
| Pantoea ananatis | N/A | CES-14 (SM-272) | - | - | Philippines | S. Miller |
| Pantoea ananatis | N/A | CES-5 (JM-55) | - | Soil | Philippines | S. Miller |
| Pantoea ananatis | N/A | E | Oryza sativa | Rice | USA, AR | J. Leach; E. Peachey |
| Pantoea stewartii | N/A | DCop3-07 | Zea mays | Corn | - | S. Miller; D. Coplin |
| Pantoea stewartii | N/A | PP685 | - | - | S. Miller | |
| Pantoea vagans | N/A | C9-1 | Malus domestica | Apple | USA, MI | V. Stockwell |
| Phacidiopycnis wasingtonensis | N/A | PD-1597 | Malus domestica | Apple | USA, WA | T. Wilson |
| Phacidiopycnis wasingtonensis | N/A | PD-1655B | Malus domestica | Apple | USA, WA | T. Wilson |
| Pseudomonas fluorescens | N/A | A506 | Pyrus spp. | Pear | USA, CA | V. Stockwell |
| Pseudomonas syringae | N/A | JL2583 | Vaccinium spp. | Blueberry | USA, OR | V. Stockwell |
| Sphaeropsis pyriputrescens | N/A | PD-1655B | Malus domestica | Apple | USA, WA | T. Wilson |
| Venturia inaequalis | N/A | Apple Scab | Malus domestica | Apple | USA, MD | PGQP |
| Xanthomonas arboricola pv. corylina | N/A | JL2611 | Corylus avellana | Hazelnut | USA, OR | V. Stockwell |
| Xyllela fastidiosa | N/A | XFS 253 | Salix spp. | Willow | USA, WA | T. Wilson |
| Xyllela fastidiosa | N/A | XFS 254 | Lonicera spp. | Honeysuckle | USA, WA | T. Wilson |
| Xyllela fastidiosa | N/A | XFS 946 | Rosa spp. | Rose | USA, WA | T. Wilson |
| Xyllela fastidiosa subsp. multiplex | N/A | Peach Leaves Georgia | Prunus persica | Peach | USA, GA | Y.K. Jo |
| Xyllela fastidiosa subsp. multiplex | N/A | Peach Texas A&M (1) | Prunus persica | Peach | USA, TX | Y.K. Jo |
| Target Organism | Primer | Primer Sequence (5′->3′) | Final Concentration (nM) | Gene Target | Amplicon (bp) | Ref. |
|---|---|---|---|---|---|---|
| ‘Candidatus Phytoplasma prunorum’ | ESFY-PE639-F | ACAGGCCGCGAATTTATTACT | 200 | Putative effector (639 nt) | 139 | This study |
| ESFY-PE639-R | GGACCGATGCTTTCACTGTT | 200 | ||||
| ESFY-PE639-P | FAM-AGATCAACGCGTACAAGCAACAGA-QSY | 100 | ||||
| ‘Candidatus Phytoplasma mali’ | fimpAP | GGTTCAGTTGTTGGTGCTT | 100 | imp | 88 | [34,35] |
| rimpAP | TTTSTTGTTTACTTTKTGATGAAA | 500 | ||||
| qimpAP | FAM-AGGCCAGAAACTAATAGACCAAGCT-QSY | 100 | ||||
| ‘Candidatus Phytoplasma’ | JH-F 1 | GGTCTCCGAATGGGAAAACC | 240 | 23S | 142–149 | [19] |
| JH-F all | ATTTCCGAATGGGGCAACC | 240 | ||||
| JH-R | CTCGTCACTACTACCRGAATCGTTATTAC | 240 | ||||
| JH-P uni | FAM-AACTGAAATATCTAAGTAAC-MGB-NFQ | 120 | ||||
| Plant host | F | GACTACGTCCCTGCCCTTTG | 48 | 18S | 68 | [13] |
| R-mod a | AACAYTTCACCGGAYCATTCA | 48 | ||||
| Probe b | ABY-ACACACCGCCCGTCGCTCC-QSY | 24 | ||||
| ‘Candidatus Phytoplasma’ | P1 | AAGAGTTTGATCCTGGCTCAGGATT | 400 | 16S | 1538 c | [53,54] |
| P1A | AACGCTGGCGGCGCGCCTAATAC | 400 | ||||
| 16S-SR | GGTCTGTCAAAACTGAAGATG | 400 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yasuhara-Bell, J.; Rivera, Y. Genome-Informed Real-Time PCR Assay for Detection of ‘Candidatus Phytoplasma Prunorum,’ Which Is Associated with European Stone Fruit Yellows. Microorganisms 2025, 13, 929. https://doi.org/10.3390/microorganisms13040929
Yasuhara-Bell J, Rivera Y. Genome-Informed Real-Time PCR Assay for Detection of ‘Candidatus Phytoplasma Prunorum,’ Which Is Associated with European Stone Fruit Yellows. Microorganisms. 2025; 13(4):929. https://doi.org/10.3390/microorganisms13040929
Chicago/Turabian StyleYasuhara-Bell, Jarred, and Yazmín Rivera. 2025. "Genome-Informed Real-Time PCR Assay for Detection of ‘Candidatus Phytoplasma Prunorum,’ Which Is Associated with European Stone Fruit Yellows" Microorganisms 13, no. 4: 929. https://doi.org/10.3390/microorganisms13040929
APA StyleYasuhara-Bell, J., & Rivera, Y. (2025). Genome-Informed Real-Time PCR Assay for Detection of ‘Candidatus Phytoplasma Prunorum,’ Which Is Associated with European Stone Fruit Yellows. Microorganisms, 13(4), 929. https://doi.org/10.3390/microorganisms13040929






