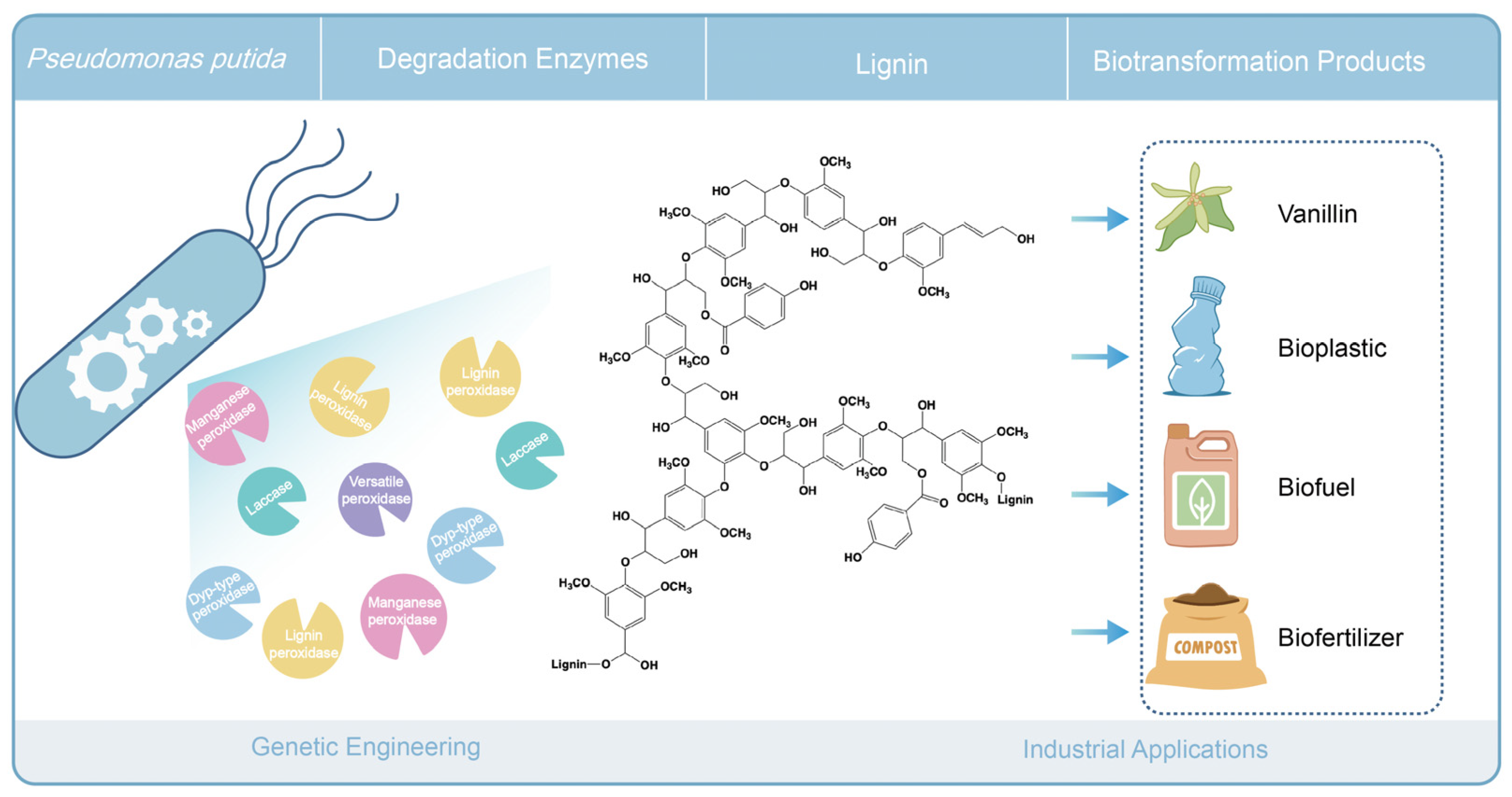
Abstract
Efficient utilization of lignin, a complex polymer in plant cell walls, is one of the key strategies for developing a green and sustainable bioeconomy. However, bioconversion of lignin poses a significant challenge due to its recalcitrant nature. Microorganisms, particularly fungi and bacteria, play a crucial role in lignin biodegradation, using various enzymatic pathways. Among bacteria, Pseudomonas putida is considered a promising host for lignin degradation and valorization, due to its robust and flexible metabolism and its tolerance to many noxious and toxic compounds. This review explores the various mechanisms of lignin breakdown by microorganisms, with a focus on P. putida’s metabolic versatility and genetic engineering potential. By leveraging advanced genetic tools and metabolic pathway optimization, P. putida can be engineered to efficiently convert lignin into valuable bioproducts, offering sustainable solutions for lignin valorization in industrial applications.
1. Introduction
Lignocellulose, composed of intertwined cellulose, hemicellulose, and lignin, is the most abundant renewable material on the Earth [1,2]. Lignin represents a class of complex and rigid organic polymers that form important structural and strengthening support in vascular plants and algae tissues. Lignin is the second most abundant terrestrial polymer on Earth after cellulose. Because of its stability and high recalcitrance, lignin has long been regarded as an industrial byproduct in pulp and paper waste, agricultural residues, and other hydrolytic industries [3,4].
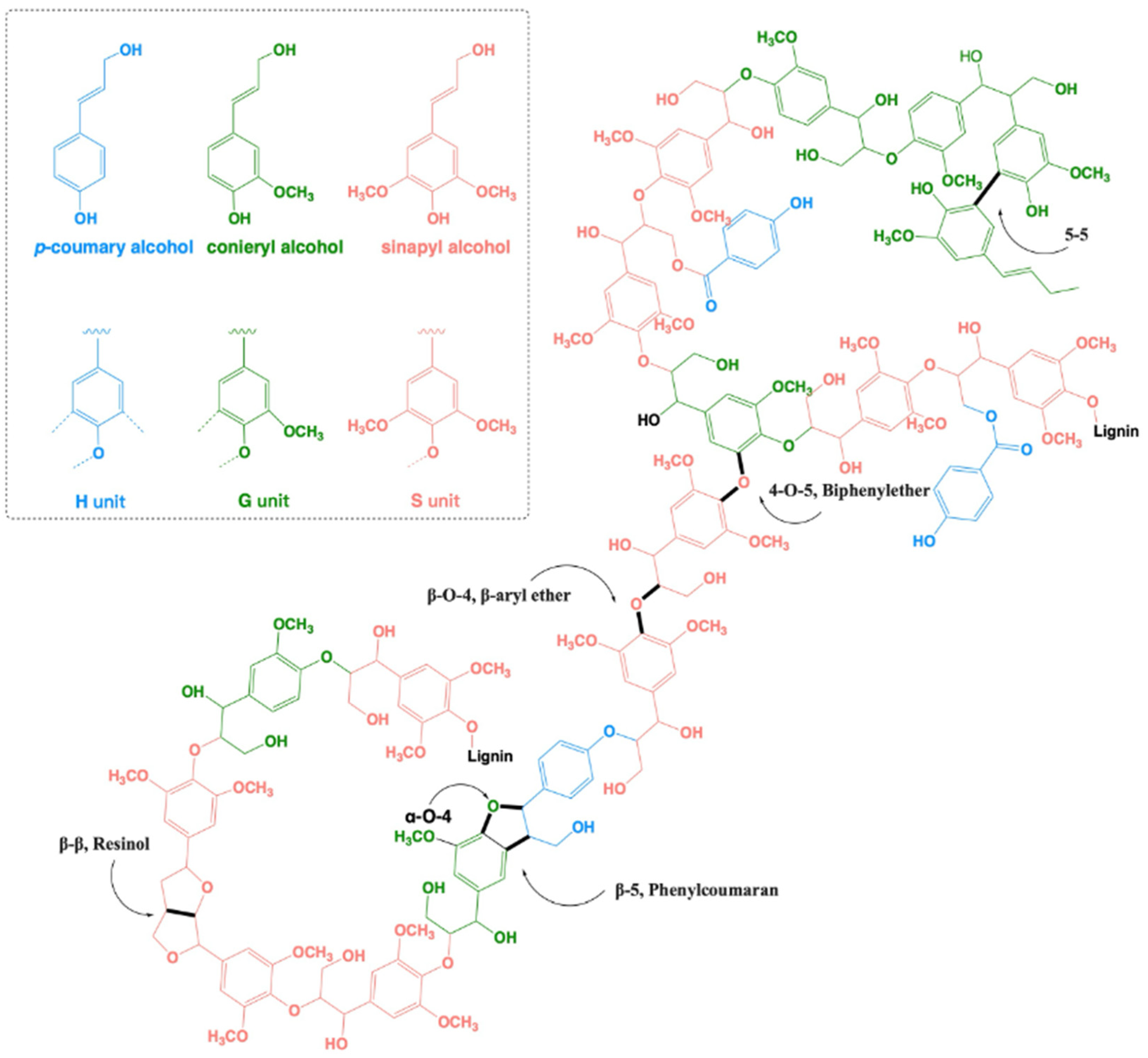
The complex aromatic polymer is synthesized in plants mainly from three basic building blocks: p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol. Consequently, the complex heterogenic polymeric lignin network mainly consists of three recognizable basic units: p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) moieties [5]. Gymnosperms (softwood, i.e., conifer, pine, cedar, spruce, etc.) mostly contain guaiacyl-based lignin (G), whereas dicotyledonous plants (legumes, beans, sunflower, tomato, etc.) mainly contain guaiacyl-syringyl-based lilac lignin (G-S), and monocotyledonous plants (maize, wheat, rice, cane, etc.) mainly contain guaiacyl-syringyl-hydroxyphenyl-based lilac lignin (G-S-H) [6] (Figure 1). Lignin polymerization in plants occurs via the formation of oxidative radicals of these structural units, followed by combinatorial radical coupling [7]. Among the variety of linkages, the β-O-4 bond is the most prominent one (40–50%), followed by β-β, β-5, 5-5, 5-O-4, and α-O-4 linkages [8,9,10,11] (for details, see Figure 1).
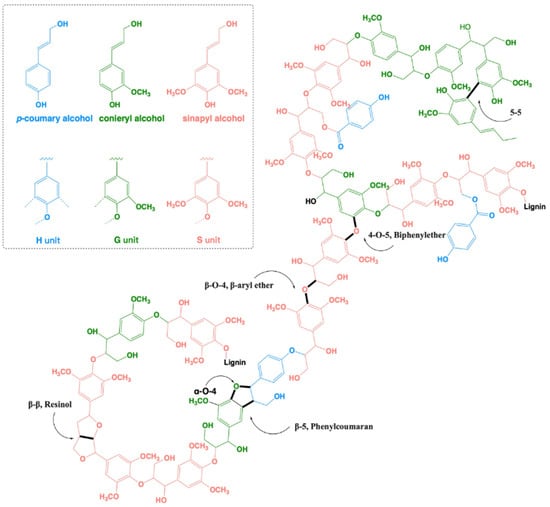
Figure 1.
A structural representation of the complex lignin polymeric network (adapted from [5,7]). The three basic units constituting lignin are p-coumaryl alcohol (blue), coniferyl alcohol (green), and sinapyl alcohol (red). These monomeric alcohols are linked to form lignin mainly by the following linkages: β-O-4 (β-aryl ether) linkages, β-β (resinol), β-5 (phenylcoumaran), 5-5 (biphenyl), and 4-O-5 (biphenylether) bonds.
With the advent in recent years of biorefineries, millions of tons of lignin are available annually as a side product of industrial lignocellulose hydrolysis and utilization [12]. In the pulp and paper industry alone, around 100 million tons of lignin are becoming available as valuable but limited-use feedstock [13,14]. Cellulose and hemicellulose fractions are readily used as feedstock in subsequent biorefinery and fermentation scale-ups for the biotechnological production of various biofuels and biochemicals. In contrast, most lignin cannot be utilized efficiently. The utilization of lignin still largely resides in heat and energy production through combustion and the implementation of raw lignin in the production of glues, resins, and asphalt [15,16,17]. The global lignin market size was estimated at USD 1.08 billion in 2023 and is expected to grow at a compound annual growth rate (CAGR) of 4.5% from 2024 to 2030 [18]. Consequently, a more efficient and valuable utilization of lignin has recently gained growing interest.
Lignin degradation is important for the recycling and valorization of plant biomass and plays a crucial role in carbon cycling and nutrient cycling in ecosystems. The first and most important step in the valorization of lignin is the efficient decomposition and depolymerization of the complex and recalcitrant polymer. The chemical decomposition of lignin has been described [19,20,21]; however, it yields a complex and toxic mixture with a difficult-to-define composition. Such chemical hydrolysates are not very useful in subsequent added-value utilization [22]. The Organosolv process, effectively reducing lignin molar mass and heterogeneity was recently published as an interesting example [23,24,25]. However, obtained fractions still contain relatively undefined lignin polymers. Combined and integrated chemical and biochemical approaches may be much more favorable [26]. Biological decomposition by microorganisms may provide a promising route toward added-value utilization of lignin. Microbial enzymes may specifically target lignin for efficient degradation into aromatic monomers and oligomers. These can subsequently be converted into valuable biochemicals through microbial metabolic pathways, providing a sustainable route for lignin utilization in biotechnology.
White-rot fungi have been known for several decades to naturally degrade lignin, whereas brown-rot fungi are only capable of modifying the lignin network to a limited extent [27,28]. These microbes produce oxidative enzymes like laccases and various types of peroxidases, which play a significant role in the aspecific breakdown of lignin [29,30,31]. In addition, many bacteria have been reported to degrade lignin. Moreover, bacterial lignin degradation involves enzymatic cleavage that is geared toward specific lignin linkages, suggesting a targeted approach to lignin breakdown [30]. In this critical review, we summarize and discuss the current knowledge and understanding of lignin-degrading microorganisms, their enzymes, reported to be active in lignin degradation, as well as aspects of proposed catalytic mechanisms. Despite the large volume of literature on microbial lignin degradation existing to date, relatively little insight has been gained on enzyme specificity and catalytic mechanisms operating to yield defined degradation products. We propose and discuss the potential of the robust soil bacterium Pseudomonas putida as a suitable host and cell factory for industrial lignin valorization, genetic and enzyme engineering strategies to enhance the synthesis of value-added bioproducts derived from lignin, and subsequent industrial applications of these bioproducts.
2. Microbial Degradation of Lignin
2.1. Lignin Degradation by Fungi
Various pioneering studies have identified fungi as effective lignin-degrading microorganisms, secreting a variety of non-specific but relatively efficient lignin-decomposing enzymes. Typical fungi implicated with lignin degradation are listed in Table 1.
White-rot fungi are commonly associated with hardwood and are renowned for their high potency to degrade lignin [32]. Lignin degradation by white-rot fungi leads to the bleaching of wood, exposing cellulose and hemicellulose fibers. This degradation enables more efficient enzymatic hydrolysis of polysaccharides, facilitating the subsequent conversion of these fibers into fermentable sugars for bioethanol production [33]. This selective degradation makes white-rot fungi interesting for many biotechnological applications since they remove lignin and leave the valuable cellulose intact.
Brown-rot fungi typically grow on softwoods and constitute only 7% of wood-rotting basidiomycetes [34]. In contrast to white-rot fungi, they preferentially hydrolyze the cellulose component of lignocellulose while only partially oxidizing lignin. This involves Fenton oxidation chemistry, during which hydroxyl radicals are produced that may partly be independent of specific enzyme activity [35].
In 1984, the white-rot fungus Phanerochaete chrysosporium, was found to produce an extracellular lignin-degrading enzyme, an oxygenase, which catalyzes several oxidations in the alkyl side chains of lignin-related compounds [36]. Subsequently, Phlebia radiata was reported to degrade lignin, and three peroxidases and one laccase were purified and characterized from this fungus. These enzymes were shown to modify kraft lignin and phenolic compounds containing hydroxyl and methoxy groups [37]. In addition, Pleurotus eryngii was shown to remove lignin from cereal straw [38], and two isoenzymes of manganese peroxidase were purified from this fungus [39]. Several other white-rot fungi have been reported to degrade different types of lignin. The white-rot fungus Trametes hirsuta has been shown to secrete several laccases and peroxidases to degrade kraft lignin. The decrease in kraft lignin molecular weight is clearly correlated with the activities of these enzymes [40]. Trametes versicolor [41] and Pycnoporus cinnabarius operated through laccase as the major phenoloxidase [42].

Table 1.
Lignin degradation by fungi: overview of strains, sources, and references. This table summarizes various fungi species known for their lignin-degrading capabilities. It includes the specific strains studied, the lignin sources they were tested on, the year of publication, and the corresponding references.
Table 1.
Lignin degradation by fungi: overview of strains, sources, and references. This table summarizes various fungi species known for their lignin-degrading capabilities. It includes the specific strains studied, the lignin sources they were tested on, the year of publication, and the corresponding references.
| Microorganism | Strain | Lignin Source | Year | Ref. |
|---|---|---|---|---|
| White-Rot Fungi | Phanerochaete chrysosporium BKM-1767 | Lignin model compounds | 1984 | [36] |
| White-Rot Fungi | Phlebia radiata | Kraft lignin | 1988 | [37] |
| White-Rot Fungi | Pleurotus eryngii | Cereal straw | 1994 | [38] |
| White-Rot Fungi | Trametes versicolor | Kraft lignin | 1995 | [41] |
| White-Rot Fungi | Pycnoporus cinnabarius | Pine wood | 1996 | [42] |
| White-Rot Fungi | Ceriporiopsis subvermispora | Pinus taeda Wood chips | 2004 | [43] |
| White-Rot Fungi | Ganoderma lucidum IBL-06 | Lignocellulosic substrates | 2010 | [44] |
| White-Rot Fungi | Phlebia sp. MG-60 | Oak wood | 2012 | [45] |
| White-Rot Fungi | Dichomytus squalens | Wheat straw Lignin | 2013 | [46] |
| White-Rot Fungi | Pleurotus ostreatus | Palm midrib | 2018 | [47] |
| White-Rot Fungi | Trametes hirsuta | Kraft lignin | 2021 | [40] |
| Brown-Rot Fungi | Gloeophyllum trabeum (Lenzites trabea) (Pers. ex Fr.) 83 | Lignin model compounds | 2008 | [48] |
| Brown-Rot Fungi | Postia placenta MAD-698-R | Aspen (Modification of Lignin) | 2009 | [49] |
| Brown-Rot Fungi | Fomitopsis pinicola | Wheat straw Lignin | 2013 | [46] |
| Soft-Rot Fungi | Aspergillus fumigatus | Kraft lignin | 1986 | [50] |
| Soft-Rot Fungi | Podospora anserina | Wheat straw Lignin | 2020 | [51] |
| Fungi | Aspergillus sp. | Alkali lignin | 2011 | [52] |
For the brown-rot fungus Gloeophyllum trabeum, a lignin degradation redox cycling process was proposed, involving two extracellularly produced quinones that reduce Fe3+ to Fe2+ [34]. This was supported by research on Postia placenta indicating up-regulation of genes associated with iron acquisition [53]. These Fe3+-reducing compounds play an important role since their low molecular weight enables them to access the cell wall structure in wood and initiate decay so that the larger lignin-degrading enzymes can access and act upon lignin [30]. Release of 14CO2 was observed when Gloeophyllum trabeum and Postia placenta were cultured with a non-phenolic, (O14CH3)-labeled lignin β-O-4 dimer model compound. Hence, these brown-rot fungi may produce enzymes that may specifically cleave the β-O-4 linkage in lignin (see Figure 1).
Apart from white-rot and brown-rot fungi, soft-rot fungi, such as Aspergillus flavus, Aspergillus fumigatus, and Aspergillus sp. LPB5, generally demonstrate only limited lignin degradation abilities and tend to be less efficient than other fungi proficient in lignin degradation [54]. However, Aspergillus fumigatus was reported to degrade kraft lignin through demethoxylation and dehydroxylation five times better compared to the white-rot fungus C. versicolor [50]. Moreover, the ascomycete Podospora anserina could cause 24% (w/w) of substantial lignin removal during the 7 days of growth [51], unambiguously confirming its ligninolytic activity.
2.2. Lignin Degradation by Bacteria
Several species of bacteria have been described to possess enzymes that can degrade lignin (Table 2). The following describes important examples of reported lignin-degrading bacterial species.
- Rhodococcus spp.
Certain strains of Rhodococcus bacteria, such as Rhodococcus jostii RHA1 [55], are known for their lignin-degrading capacity. R. jostii RHA1 degrades lignin in lignocellulose as well as kraft lignin to a low-molecular-weight phenolic byproduct, as monitored by spectrophotometric assays [55]. R. jostii RHA1 encodes two putative so-called dye-decolorizing peroxidases, or DyP peroxidases (see below for description of enzymes). One was characterized as lignin peroxidase DypB, active in lignin breakdown [56]. A genetically modified R. jostii RHA1 was able to produce 330 mg/L 2,4-PDCA (pyridine-dicarboxylic acid) in 40 h from 1% wheat straw lignocellulose, corresponding to a conversion yield of approximately 16% of the available lignin fraction [57]. Rhodococcus pyridinivorans CCZU-B16 [58], isolated from soil, could under optimized conditions degrade 30.2% of alkali lignin (4 g/L) in 72 h.
- Bacillus spp.
Bacteria of the genus Bacillus isolated from pulp and paper mill effluent exhibited the potential to degrade lignin [59]. For example, Bacillus altitudinis SL7 reduced lignin content by 44% when grown with alkali lignin [60]. Bacillus pumilus LSSC3 and Bacillus atrophaeus CL29 exhibited high oxidative laccase activity in kraft lignin degradation, measured by the oxidation of the lignin model compound guaiacol [61]. Bacillus flexus RMWW II showed lignin degradation by 20% at a lignin concentration of 400 mg L−1 [62]. Bacillus ligniniphilus L1 can utilize alkaline lignin as a sole carbon source, producing 15 types of aromatic compounds as identified via GC-MS analysis [63]. Transcriptomic data indicate at least four pathways putatively involved in lignin degradation and metabolization of breakdown products, including the Gentisate pathway, Benzoic acid pathway, and β-ketoadipate pathway. Bacillus sp. (CS-1 and CS-2) can degrade alkali lignin with high laccase activities detected in crude enzyme extracts [64]. Nevertheless, the specific lignin-degrading enzymes remain to be characterized.
- Pseudomonas spp.
Several Pseudomonas strains, including Pseudomonas putida and Pseudomonas fluorescens, have been found to possess lignin-degrading enzymes. These bacteria are often investigated for their applications in bioremediation and lignocellulosic biomass conversion. P. putida A514 was able to grow with alkali-insoluble lignin as the sole carbon source [65]. Recently, P. putida NX-1, isolated from leaf mold samples, could grow on kraft lignin and was engineered for PHA production [66,67]. Genome analysis of P. putida NX-1 revealed putative enzymes involved in lignin decomposition, including dyp-type peroxidases, versatile peroxidases, manganese peroxidases, and laccases. However, their functions and contributions to lignin decomposition have not yet been experimentally characterized. The ability to catabolize a wide range of natural aromatinds [68,69] indicates that P. putida KT2440 holds potential to be an excellent host for lignin degradation. P. putida KT2440 could utilize alkaline pretreated liquor (APL), primarily composed of lignin, to produce mcl-PHA in relatively good yield under nitrogen depletion [22]. Furthermore, outer membrane vesicles (OMVs) from P. putida KT2440 have been implicated in the biodegradation of lignin-derived aromatic compounds [68,69]. The copper-dependent oxidase CopA from P. putida KT2440 was shown to be involved in extracellular lignin oxidation [70]. Moreover, P. putida was recently shown to produce cis,cis-muconic acid from PCA, which is an intermediate product of lignin degradation [71,72]. Hence, P. putida appears highly promising as a biotechnology host strain to produce valuable compounds from lignin [73].
- Streptomyces spp.
Several actinobacterial species, such as members of the Streptomyces genus, have shown lignin-degrading potential. Streptomyces viridosporus T7A is an example of an actinobacterium with ligninolytic activity [74]. Streptomyces spp. F-6 and Streptomyces spp. F-7 can remove around 38% of lignin, after 12 days of culture [75]. Recently, Streptomyces thermocarboxydus DF3-3 was isolated for alkali lignin degradation [76], secreting ligninolytic enzymes, such as manganese peroxidase, laccase, and specific small laccases [77]. For this species, a total of seven lignin-based derivatives metabolic pathways were predicted: the β-ketoadipate pathway and peripheral reactions; the gentisate pathway; the anthranilate pathway; the homogentisic pathway; the catabolic pathway for resorcinol; the phenylacetate–CoA pathway; and the 2,3-dihydroxyphenylpropionic acid pathway [76]. Streptomyces sp. S6 isolated from a decaying oil palm empty fruit bunch can grow on kraft lignin as the sole carbon source. After 7 days of incubation with Streptomyces sp. S6, the loss of the molecular weight of kraft lignin was up to 55.3% [78].
- Sphingomonas spp.
Sphingomonas species, and more specifically Sphingomonas paucimobilis SYK-6, have been shown to degrade lignin-related aromatic model compounds [79]. These bacteria are known for their ability to break down various lignin-related structures. SYK-6 was the first bacterium shown to harbor several functional lignin-degrading enzymatic routes, involving glutathione peroxidases and etherases (see below).
- Other Proteobacteria
Proteobacteria like Pandoraea sp., Enterobacter, or Ochrobactrum have been confirmed can utilize lignin or lignin model compounds. Pandoraea sp. B-6 secreted extracellular ligninolytic enzymes to degrade kraft lignin [80]. The low-molecular-weight compounds of kraft lignin were detected by GC-MS. Proteomics suggested Enterobacter lignolyticus SCF1 was able to use lignin in both assimilatory and dissimilatory pathways [81]. Ochrobactrum was first reported to depolymerize and utilize lignin in 2018 [58].

Table 2.
Lignin degradation by bacteria: overview of strains, sources, and references. This table summarizes various species of bacteria known for their lignin-degrading capabilities. It includes the specific strains studied, the lignin sources they were tested on, the year of publication, and the corresponding references.
Table 2.
Lignin degradation by bacteria: overview of strains, sources, and references. This table summarizes various species of bacteria known for their lignin-degrading capabilities. It includes the specific strains studied, the lignin sources they were tested on, the year of publication, and the corresponding references.
| Microorganism | Strain | Lignin Source | Year | Ref. |
|---|---|---|---|---|
| Actinobacteria | Rhodococcus jostii RHA1 | Kraft lignin | 2011 | [56] |
| Rhodococcus erythropolis | Alkali lignin | 2012 | [82] | |
| Rhodococcus opacus DSM 1069 | Lignin | 2013 | [83] | |
| Rhodococcus opacus PD630 | Alkali Corn Stover Lignin | 2017 | [84] | |
| Rhodococcus pyridinivorans CCZU-B16 | Alkali lignin | 2018 | [58] | |
| Amycolatopsis sp. 75iv2 | Acid-precipitable, polyphenolic, polymeric lignin (APPL) | 2011 | [85] | |
| Streptomyces viridosporus T7A | APPL | 1983 | [74] | |
| Streptomyces spp. F-6 | Alkali lignin | 2012 | [75] | |
| Streptomyces spp. F-7 | Alkali lignin | 2012 | [75] | |
| Streptomyces coelicolor A3(2) | Lignin model compounds | 2014 | [86] | |
| Streptomyces sp. S6 | Kraft lignin | 2020 | [78] | |
| Streptomyces thermocarboxydus DF3-3 | Alkali lignin | 2022 | [76] | |
| Micromonospora sp. | Kenaf | 2014 | [87] | |
| Thermobifida fusca YX | Untreated biomass | 2011 | [88] | |
| Anaerobic Microorganisms | Clostridium thermocellum | Populus Lignin | 2017 | [89] |
| Brevibacillus | Brevibacillus thermoruber | Lignin | 2021 | [90] |
| Caldicellulosiruptor bescii DSM 6725 | Untreated switchgrass | 2013 | [91] | |
| Bacteroidetes | Sphingobacterium sp. HY-H | Sodium lignosulfonate | 2013 | [92] |
| Sphingobacterium sp. T2 | Wheat straw Organosolv lignin, alkali kraft lignin | 2015 | [93] | |
| Sphingomonas paucimobilis SYK-6 | dimeric lignin compounds | 1999 | [79] | |
| Proteobacteria | Citrobacter sp. (HQ873619) | Black liquor | 2011 | [94] |
| Citrobacter sp. (FJ581023) | Black liquor | 2011 | [95] | |
| Citrobacter freundii (FJ581026) | Black liquor | 2011 | [95] | |
| Comamonas sp. B-9 | Kraft lignin | 2012 | [96] | |
| Comamonas testosterone KF-1 | Lignin-associated monomers | 2023 | [97] | |
| Klebsiella pneumoniae (GU193983) | Black liquor | 2011 | [94] | |
| Klebsiella pneumoniae NX-1 | Kraft lignin | 2018 | [66] | |
| Pseudomonas aeruginosa (DSMZ 03504) | Pulp mill effluents | 2010 | [98] | |
| Pseudochrobactrum glaciale | Pulp paper mill effluent | 2012 | [99] | |
| Pantoea sp. | Pulp paper mill effluent | 2012 | [99] | |
| Pseudomonas putida KT2440 | Alkaline pretreated liquor | 2014 | [22] | |
| Pseudomonas plecoglossicida ETLB-3 | Black liquor | 2015 | [100] | |
| Pseudomonas putida A514 | Alkali lignin | 2016 | [65] | |
| Pseudomonas strain | Alkaline insoluble lignin | 2016 | [65] | |
| Pseudomonas sp. Q18 | Alkali lignin | 2018 | [101] | |
| Pseudomonas putida NX-1 | Kraft lignin | 2018 | [66] | |
| Pseudomonas strain Hu109A | Lignin | 2023 | [102] | |
| Pandoraea sp. B-6 | Kraft lignin | 2013 | [80] | |
| Enterobacter soil sp. nov. | Alkali lignin | 2011 | [103] | |
| Enterobacter lignolyticus SCF1 | Alkali lignin | 2013 | [81] | |
| Ochrobactrum pseudogrignonense | Nitrated lignin | 2012 | [82] | |
| Ochrobactrum rhizosphaerae | Nitrated lignin | 2012 | [82] | |
| Ochrobactrum tritici NX-1 | Kraft lignin | 2018 | [66] | |
| Serratia marcescens (GU193982) | Black liquor | 2011 | [94] | |
| Serratia liquefaciens | Pulp paper mill effluent | 2012 | [99] | |
| Serratia liquefaciens LD-5 | Pulp paper mill effluent | 2016 | [104] | |
| Firmicutes | Aneurinibacillus aneurinilyticus (AY856831) | Kraft lignin | 2007 | [59] |
| Bacillus sp. (AY952465) | Kraft lignin | 2007 | [59] | |
| Bacillus sp. (accession no. AY 952465) | Kraft lignin | 2007 | [105] | |
| Bacillus cereus (DQ002384) | Kraft lignin | 2008 | [106] | |
| Bacillus atrophaeus LSSC3 | Kraft lignin | 2013 | [61] | |
| Bacillus pumilus CL29 | Kraft lignin | 2013 | [61] | |
| Bacillus sp. (CS-1 and CS-2) | Alkali lignin | 2014 | [64] | |
| Bacillus megaterium ETLB-1 | Black liquor | 2015 | [100] | |
| Bacillus ligniniphilus L1 | Alkali lignin | 2017 | [107] | |
| Bacillus endophyticus | Lignin | 2016 | [108] | |
| Bacillus subtilis | Lignin | 2016 | [108] | |
| Bacillus flexus RMWW II | Alkali lignin | 2019 | [62] | |
| Bacillus altitudinis SL7 | Purified synthetic alkali lignin | 2021 | [60] | |
| Paenibacillus sp. (AY952466) | Kraft lignin | 2008 | [106] | |
| Paenibacillus glucanolyticus SLM1 | Biochoice lignin | 2016 | [109] | |
| Paenibacillus glucanolyticus 5162 | Biochoice lignin | 2016 | [109] | |
| Paenibacillus sp. strain LD1 | Kraft lignin | 2014 | [110] | |
| Planococcus sp. TRC1 | Lignin | 2019 | [111] | |
| Extremophile bacteria | Arthrobacter sp. C2 | Sodium lignin sulfonate | 2022 | [112] |
3. Enzymes for Lignin Depolymerization
Several successful examples of P. putida converting lignin-derived compounds into valuable products have been reported [113]. However, despite these promising findings, achieving conversion starting from intact lignin remains challenging. Lignin degradation is a complex process that requires multiple enzymes and pathways. Many research efforts have been employed, trying to uncover the intricacies of these processes. These have yielded insights into fungal and bacterial enzymes with activity toward lignin degradation, with various bacterial enzymes putatively operating with higher specificity toward different lignin-specific linkages.
The initial and crucial step to effectively degrade lignin is to attack and depolymerize the complex lignin polymeric network into smaller phenoxy radical intermediates [114]. This step can be facilitated by external oxidoreductases, including laccase (Lac, EC 1.10.3.2), lignin peroxidase (LiP, EC 1.11.1.14), manganese peroxidase (MnP, EC 1.11.1.13), dye-decolorizing peroxidases (Dyp, EC 1.11.1.19), and versatile peroxidase (VP, EC 1.11.1.16) [115]. Typical enzymes capable of cleaving specific lignin linkages are summarized in Table 3. These enzymes have been extensively studied for their activity, however, almost exclusively on lignin model compounds [11]. They are known to target various linkages that occur within the lignin structure. These enzymes exhibit different substrate specificities and mechanisms of action; however, in most cases, their precise role in bioconversion of the lignin polymeric network remains elusive.

Table 3.
Enzymes capable of cleaving specific lignin linkages. This table summarizes the types of enzymes involved in cleaving specific lignin linkages, their classification, their names, their source strains, the substrates utilized, and the relevant references. The enzymes listed are crucial for understanding the biochemical pathways of lignin degradation and highlight the diverse microbial sources capable of lignin bioconversion.
3.1. The β-O-4 Bond
A number of enzymes are secreted by fungi and bacteria to degrade lignin or lignin-derived compounds [129]; however, there is limited evidence regarding their ability to cleave specific linkages within the complex lignin structure. Reported evidence for linkage specificity mostly stems from studies with relatively simple model compounds for each of the lignin linkages [11]. Among the various linkages present in lignin, 45–60% of the total linkages are β-O-4 aryl ether bonds [130]. Cleaving this bond presents an essential step in the efficient use of lignin. Hence, enzymes that can cleave β-O-4 aryl-ether bonds are highly interesting for application in lignin valorization.
3.1.1. Fungal Lignin Depolymerization Enzymes
- Lignin Peroxidases (LiPs)
Lignin peroxidase, a monomeric heme-containing enzyme, was the first enzyme found in P. chrysosporium that can degrade lignin [27,131]. Its proficiency lies in the effective degradation of non-phenolic lignin units by catalyzing oxidative breakdown in the presence of H2O2 [132]. Therefore, it can catalyze the cleavage of β-O-4 ether bonds and Cα-Cβ linkages. LiPs are considered strong biocatalyst in the bioremediation of lignin and are represented in Phanaerochaete chrysosporium, Trametes versicolor, Phanaerochaete sordida, and Phlebia radiata.
- Laccases
Laccases are widely found in plants, insects, fungi, and bacteria [133,134,135]. As a copper-containing enzyme of the polyphenol oxidases group, laccase catalyzes the oxidation of aromatic compounds, including phenols and phenolic derivatives during lignin degradation. Oxidation of these phenolic compounds leads to the formation of phenoxyl radicals, resulting in subsequent hydrolysis of C-C and β-aryl bonds in lignin’s aromatic rings [33], yielding various products such as syringaldehyde, 1-(3,5-dimethoxy-4-ethoxyphenyl)-2-hydroxyethanone, 1-(3,5-dimethoxy-4-ethoxyphenyl)-2-hydroxypropanal, and 2,6-dimethoxy-p-benzoquinone. Laccases are present in various fungi species such as Dichomitus squalens, Irpex lacteus, Lentinula edodes, Cerrena maxima, Trametes versicolor, Pleurotus ostreatus, and Phanaerochaete chrysosporium [132]. Interestingly, Peniophora lycii LE-BIN 2142 lacks ligninolytic peroxidases, which are typically considered key enzymes in white-rot fungi. Instead, this species primarily relies on multiple laccase isozymes and unique FAD-binding proteins, suggesting an alternative oxidative strategy for lignin modification [136].
- Versatile Peroxidases (VPs)
Versatile Peroxidase, a heme-containing ligninolytic peroxidase, was first found in white-rot fungi Pleurotus eryngii [137]. VPs have been characterized to have catalytic functions of LiP, capable of oxidizing high redox potential substrates, combined with MnP, which oxidizes Mn2+ to Mn3+, producing a diffusible oxidizing agent effective on low redox potential species. In the absence of mediators, they also oxidize azo-dyes and other non-phenolic compounds with high redox potentials [138]. Different from MnPs and LiPs, VPs have a wider range of substrates. Evidence shows that VPs could catalyze β-O-4 lignin dimer to monomeric products [139]. Additionally, the VPs from Physisporinus vitreus oxidized the β-O-4 dimer, guaiacylglycerol β-guaiacyl ether, by depolymerization to a monomer or polymerization to a tetramer concurrently [119]. VPs are found in Pleurotus, Bjerkandera sp., Panus, Calocybe, Trametes, Lepista, Dichomitous, and spongipelli fungi species [33].
- Manganese Peroxidases (MnPs)
Mangenese Peroxidase catalyzes the oxidation of a non-phenolic aromatic ring structure in lignin via oxidation of Mn2+ to Mn3+ as a redox mediator, leading to structural cleavage [140]. MnP from Phanerochaete chrysosporium was found able to cleave the β-O-4 of the phenolic lignin model dimer 1-(3,5-dimethoxy-4-hydroxyphenyl)-2-[4-(hydroxymethyl)-2-methoxyphenoxy]-1,3-dihydro-xypropane [120,141]. MnP was first discovered in P. chrysosporium but was also later detected in other Basidiomycota species, including Panus tigrinus, Lenzites betulinus, Agaricus bisporus, Bjerkandera sp., and Nematoloma frowardii [138].
- Dye-decolorizing Peroxidases (DyPs)
Lastly, dye-decolorizing peroxidases (DyPs) are evolutionarily not related to the classical LME peroxidases (LiPs, MnPs, and VPs) but are a new class of heme-containing peroxidases found in bacteria and fungi [33]. DyPs were first isolated in 1999 from the basidiomycetous fungus Bjerkandera adusta [142]. Some ligninolytic activity was found in Termitomyces albuminosus, Auricularia auricula-judae, and Irpex lacteus.
3.1.2. Bacterial Lignin Depolymerization Enzymes
In addition to harboring enzymes with characteristics comparable to fungal lignin-degrading enzymes, bacteria also have different lignin degradation mechanisms and enzymes. In the 1980s, Pseudomonas acidovorans had already been reported to degrade a β-aryl ether model compound [143]. Nevertheless, as of today, the actual number of functional bacterial enzymes well characterized in detail remains limited.
- β-Etherase
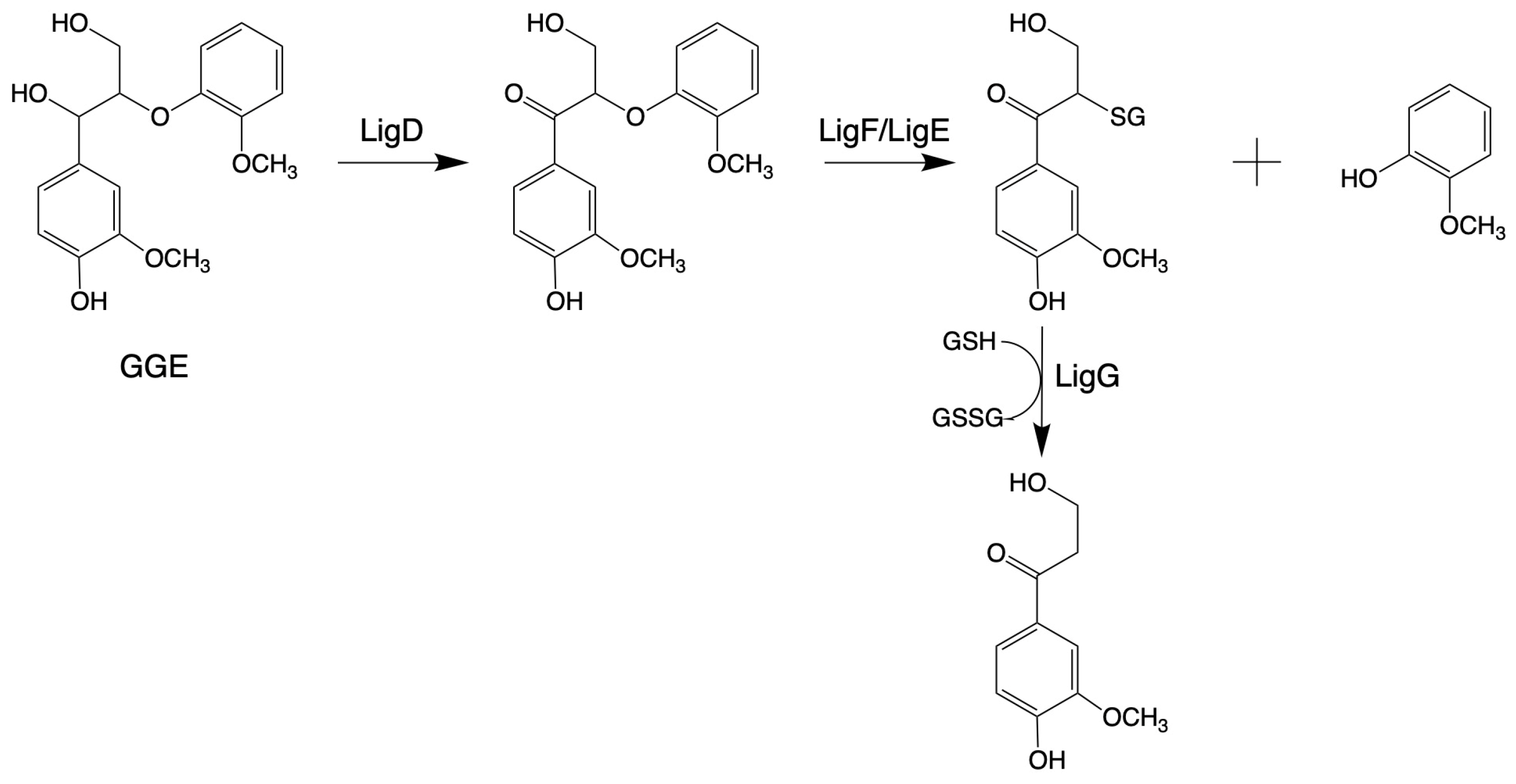
β-Etherase, belonging to the protein superfamily of glutathione-S-transferase (GST; EC 2.5.1.18), is the first bacterial gene reported to function specifically in lignin degradation in Sphingobium sp. SYK-6 [144,145,146]. β-Etherases exist especially in microorganisms that specialize in decomposing lignin [147]. The β-O-4 aryl-ether bond degradation pathway in Sphingobium sp. SYK-6 needs three steps, involving three enzymes: an NAD+-dependent Cα-dehydrogenase (LigD, LigL), a β-Etherase (LigE, LigF), and LigG, a glutathione-dependent lyase (LigG) [148,149]. First, LigD/LigL oxidizes the Cα in model substrates, like 1-(4-hydroxy-3-methoxyphenyl)-2-(2-methoxyphenyl) propane-1,3-diol (GGE), under consumption of NAD+. Only after this oxidation, LigE or LigF can cleave the Cβ ether bond, following the SN2-type mechanism with high stereoselectivity. While LigE cleaves ether bonds in substrates with (R)-configured β-carbon, resulting in the corresponding (S)-configured glutathione adducts, LigF converts the corresponding (S)-substrate enantiomers [150]. Finally, LigG catalyzes the thioether cleavage of the chiral glutathione adducts to produce oxidized glutathione (GSSG) [151,152], as shown in Figure 2.
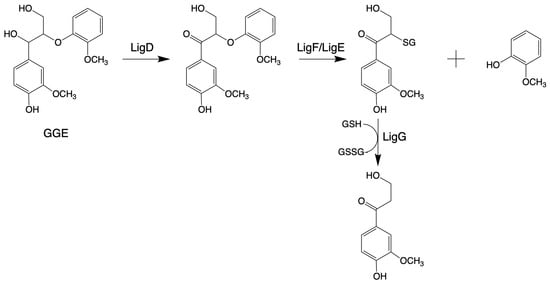
Figure 2.
Pathways for the cleavage of β-O-4 bond by β-Etherase in Sphingobium sp. SYK-6 (adapted from [34]).
- Dye-decolorizing Peroxidases (DyPs)
Dyps, heme-containing peroxidases are regarded as important enzymes involved in lignin degradation, since they can specifically cleave and degrade a list of such lignin model dye compounds [153]. Generally, peroxidase can catalyze the degradation reaction of hydrogen peroxide, leading to the generation of reactive oxygen species, which in turn participate in lignin degradation. Additionally, DyP enzymes also catalyze the oxidation of β-O-4 linkages, converting veratrylglycerol-β-guaiacyl ether into veratryl aldehyde and cleaving guaiacylglycerol-β-guaiacyl ether [154,155]. The DypB from Rhodococcus jostii RHA1 was the first bacterial lignin-degrading enzyme that has been characterized, which is capable of oxidizing polymeric lignin and lignin model compounds [56]. Novel research also found that Dyp1B from Pseudomonas fluorescens plays a significant role in lignin degradation [156].
- Laccase-like multicopper oxidases (LMCOs)
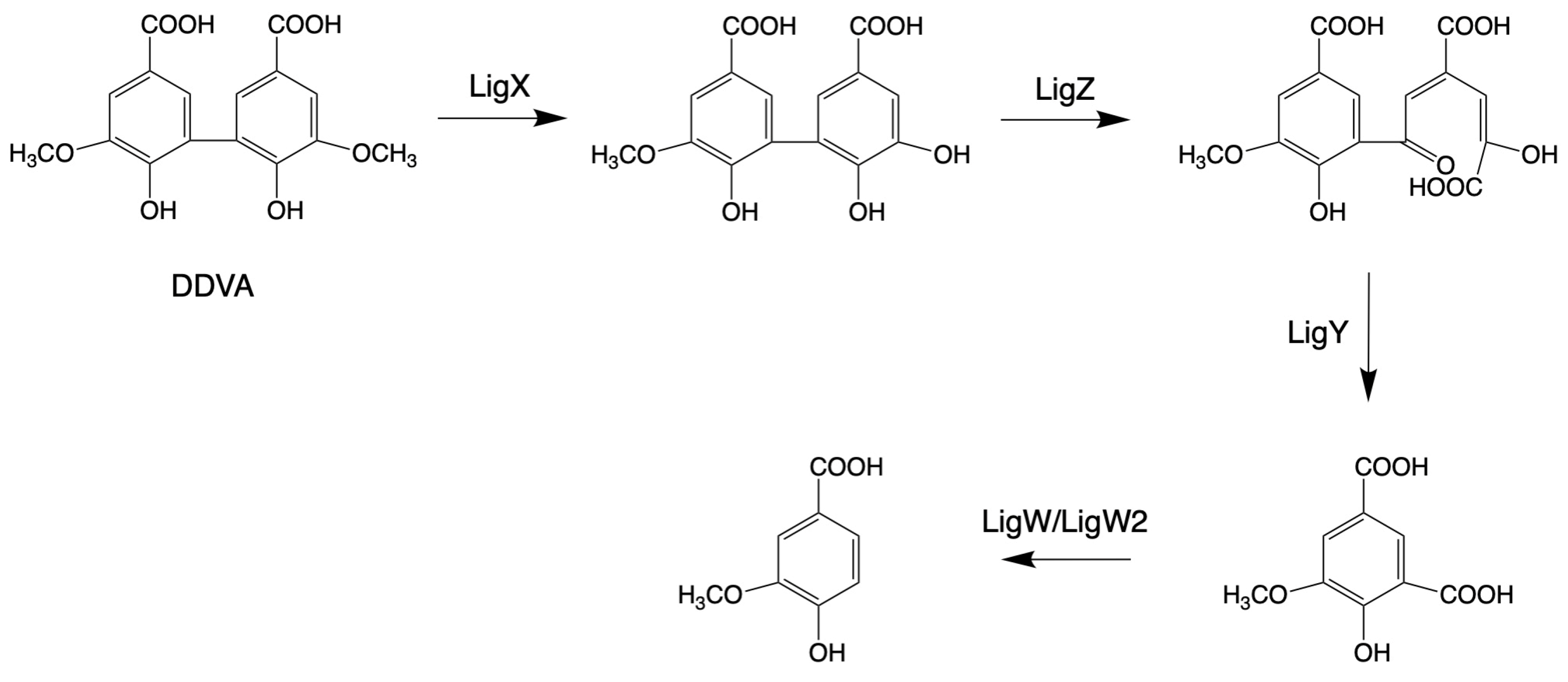
Laccase-like multicopper oxidases (LMCOs) are a diverse group of oxidoreductases found in bacteria, fungi, and plants [157]. CopA is a member of LMCOs or pseudo-laccases [158,159]. CopA enzymes from P. putida KT2440 and P. fluorescens Pf-5 catalyze the oxidization of the lignin model compound GGE (see above, Figure 2) and 2,2′-dihydroxy-3,3′-dimethoxy-5,5′-dicarboxybiphenyl (DDVA, see blew, Figure 3), producing oxidized dimerized products [70,160].
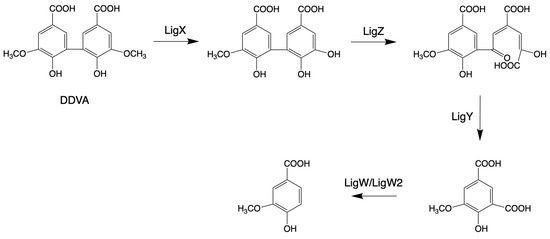
Figure 3.
Pathways for the structural cleavage of biphenyl moieties in Sphingobium sp. SYK-6 (adapted from [34]).
- Laccases
Laccase plays a crucial role in lignin biodepolymerization, but the reaction mechanism in bacteria remains incompletely elucidated.
Among bacterial laccases, small laccases (SLACs) are a type of laccase enzyme characterized by their smaller molecular size compared to traditional laccases [161]. The SLAC from Streptomyces can degrade a phenolic β-O-4 lignin model compound (LM-OH) [86]. Furthermore, SLAC variants have been functionally expressed in Aspergillus niger and are active in lignocellulose degradation [77].
The laccase from Bacillus ligniniphilus L1 was found to promote lignin degradation by oxidizing phenolic and non-phenolic structures in lignin [121]. In addition, this study highlights its potential role in cleaving key interunit linkages in lignin, including β-O-4, β-5, β-β, 4-O-5, and 5-5.
3.1.3. 5-5 Bond (Biphenyl Bond)
The proportion of 5-5 bonds in lignin is around 10% in softwood and 5% in hardwood [162]. Remarkably, it has been demonstrated that the cleavage of the biphenyl linkage plays a pivotal role in facilitating lignin degradation.
Amongst fungi, the versatile peroxidases (VPs) in Physisporinus vitreus, were also observed to cleave the 5-5 bond of dehydrodivanillic alcohol (5-5′ dimer) in vitro for the first time [119].
The bacterial biphenyl degradation pathway was also found in Sphingobium sp. SYK-6 by growing on 2,2′-dihydroxy-3,3′-dimethoxy-5,5′-dicarboxybiphenyl (DDVA) [163]. In the mechanism of degrading DDVA, four enzymes are involved: LigX (a non-heme iron-dependent demethylase), LigZ (an extradiol dioxygenase), LigY (a C-C hydrolase), and LigW/LigW2 (decarboxylases) [122,123,124]. In the catalytic progression of DDVA, the enzyme LigX catalyzes the elimination of a methoxy group, resulting in the formation of a hydroxyl group. Subsequently, the product generated by LigX serves as a substrate for oxidative meta-cleavage, facilitated by LigZ. Following this, LigY transforms the ring fission product into 4-carboxy-2-hydroxypentadienoic acid and 5-carboxyvanillic acid (5CVA). This sequence culminates with the participation of LigW and LigW2, which convert 5CVA into the pivotal metabolic intermediate, vanillic acid or vanillate, essential for the synthesis of various bioproducts, as shown in Figure 3.
3.1.4. β-β Bond (Resinol Bond)
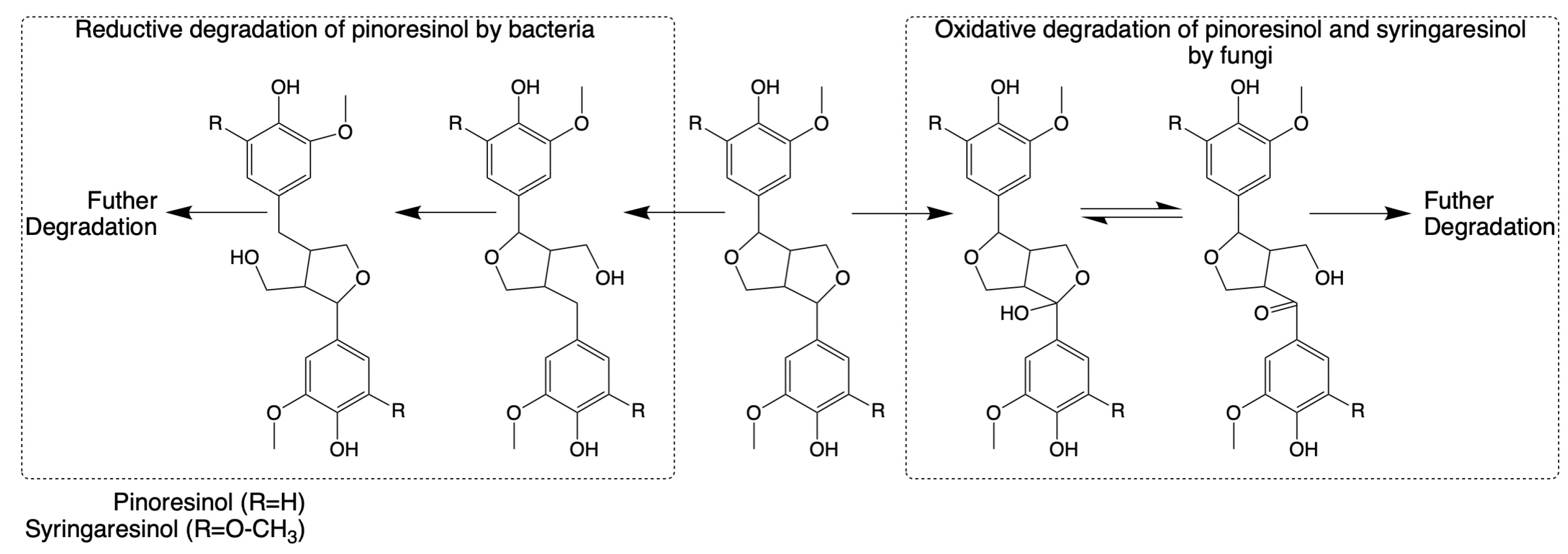
The breakdown of the pinoresinol lignin model compound has also been studied in Fusarium solani M-13-1 and S. paucimobilis SYK-6 [124,125]. The catabolic pathways for both heterocyclic lignin components appear to involve alpha-hydroxylation as an initial step. However, enzymes that participate in the reactions have not been characterized clearly. Until 2018, the isolation of the highly efficient (+)-pinoresinol-mineralizing Pseudomonas sp. strain SG-MS2 and its catabolic pathway were reported, highlighting a significant advancement in understanding the catabolism of pinoresinol lignin dimers, as shown in Figure 4 [164].
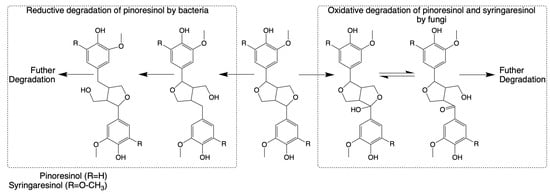
Figure 4.
Pathways for the cleavage and subsequent degradation of pinoresinol and syringaresinol by bacteria and fungi, respectively (adapted from [34,164]).
3.1.5. Other C-C Bonds
Fungal MnP possesses the capability to not only break β-O-4 linkages in phenolic structures but also disrupt Cα-Cβ and β-aryl ether bonds in non-phenolic substances [165,166]. Studies indicate that laccases could cleave Cα−Cβ bonds or aryl−Cα bonds and catalyze the oxidation of Cα−OH to Cα=O of lignin model compounds [167]. The laccase degradation mechanism may vary depending on the substrate, pH, temperature, and other environmental conditions [168]. In addition, different types of laccases may have different substrate specificity and degradation efficiency. A deep understanding of these mechanisms is needed for developing effective biotechnological applications for lignin degradation.
4. Pseudomonas putida as a Lignin-Degrading Cell Factory
Although the efficiency of lignin depolymerization by bacterial extracellular enzymes is less well studied than that of white-rot fungi, bacteria do provide a flexible platform for the heterologous expression of ligninolytic enzymes [169]. We and others have investigated endogenous lignin degradation with strains of Pseudomonas putida. Moreover, strains of P. putida have been implicated in bioconversion and biosynthesis of valuable products from lignin-derived compounds [71,72,170]. The direct and grand challenge now is to directly access and utilize lignin as a source of valuable products. P. putida may prove a promising vehicle for the expression of various specific ligninolytic enzymes to construct cell factories, enabling the direct conversion of lignin into high-value products (see Figure 5).
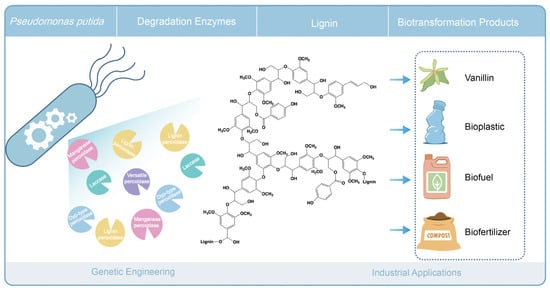
Figure 5.
Conceptual representation of lignin biodegradation and engineering in Pseudomonas putida.
4.1. Natural Capabilities and Metabolic Pathways
P. putida strains like S12 and KT2440 are recognized as highly promising industrial host strains [171]. These strains can adapt to diverse physiochemical and nutritional niches and possess robust metabolic redox power, enabling them to survive under high oxidative stress. Moreover, P. putida has been found in natural environments degrading various organic compounds, including lignin-derived molecules. For example, P. putida KT2440 has been identified to possess the ability to degrade p-hydroxybenzoate, benzene, and xylene, which are components of lignin-derived aromatic hydrocarbons [172]. P. putida has a wide range of metabolic functions, combined with its extensive catabolic pathways, enabling it to utilize lignin-derived aromatic compounds as carbon sources.
In recent years, the depolymerization of lignin has become a research hotspot [173,174]. Lignin-derived compounds like ferulic acid and vanillin have been given particular attention [175]. P. putida KT2440 can metabolize vanillin by conversion into vanillate, at a rate of 4.87 mmol (gCDW h)−1 [176]. Furthermore, P. putida KT2440 can degrade ferulic acid via a CoA-dependent non-β-oxidative pathway [177]. Additionally, this strain can metabolize benzoate and catechol through the native β-ketoadipate pathway, further demonstrating its ability to process lignin-derived compounds [172]. The β-ketoadipate pathway is a chromosomally encoded aromatic compound degradation pathway that is widespread among soil bacteria and fungi [178]. Such pathways are essential for lignin degradation and valorization, turning complex aromatic polymers into economically valuable products.
P. putida demonstrates exceptional performance in the degradation of lignin derivatives and other aromatic compounds, showing the potential applications of biotechnology in lignin valorization. Therefore, P. putida strains are promising suitable platforms for the bioconversion of exogenous toxic chemical streams into valuable products that derive from lignin degradation.
4.2. Genetic Engineering of Pseudomonas putida
4.2.1. Genomic Tools
To develop efficient cell factories for lignin degradation, a specifically robust bacterial chassis is needed. P. putida is a robust platform with advanced metabolic engineering applications [179]. The degradation capability of P. putida toward lignin can be further enhanced through genetic engineering. The introduction of genes encoding ligninolytic enzymes from other microorganisms can improve efficiency. Modifying the metabolic pathways of P. putida can convert the lignin degradation products into valuable compounds.
Over the years, dedicated genetic tools have been developed for the expression or deletion of genes in P. putida [180,181]; see Table 4. With the use of gene editing tools, it is highly possible to improve the lignin degradation of P. putida. For example, previous studies show that the expression of Pseudomonas fluorescens Dyp1B in P. putida KT2440 results in enhanced activity for the oxidation of 2,6-dichlorophenol (DCP) and polymeric lignin [156].

Table 4.
Typical genomic tools for cloning, insertions, and deletions in P. putida.
4.2.2. Secretion System
Lignin, as a polymeric network, is obviously too large and complex to be transported into the cell and be degraded intracellularly [191]. Hence, lignin-degrading enzymes for biotechnological applications should be extracellularly secreted with the microbial secretome. Several secretion systems have been studied in P. putida: Outer membrane vesicles (OMVs) are secreted by the bacterium to deliver enzymes [192]. Pioneering research has shown that OMVs in P. putida KT2440 can catabolize lignin-derived aromatic compounds [69]. This property can be exploited to deliver ligninolytic enzymes to lignin substrates, thereby enhancing the degradation process. A novel recombinant peroxidase secretion system has been constructed in P. putida KT-M2 [193]. A flagellar type III secretion system was used for the dye decolorization peroxidase of P. putida, resulting in efficient oxidative activity of cell-free supernatants against a variety of chemicals, including the lignin model compound. Additionally, the periplasmic expression of peroxidase Dyp1B has been explored for lignin valorization in P. putida [156]. The periplasmic expression strain shows higher lignin oxidation activity than the wide type.
These advancements in secretion systems, together with the genetic engineering tools, highlight P. putida’s potential as a powerful biotechnological platform for lignin degradation. By combining these approaches, it is possible to enhance the efficiency of lignin valorization processes, turning this complex and recalcitrant polymer into valuable bioproducts.
4.3. Biological Conversion and Reutilization
Biological funneling is a concept in bioconversion and metabolic engineering where a diverse array of complex molecules is funneled through a series of biological pathways to produce a single or a few specific valuable products [194,195]. Biological funnels can overcome the challenging heterogeneity of chemical mixtures as recently applied and demonstrated with low-molecular-weight lignin-derived aromatics [196,197]. Indeed, P. putida may convert a mixture of lignin degradation products into useful compounds. These compounds can be further utilized, such as biofuels, chemical raw materials, and other bio-based products.
To obtain low-molecular-weight lignin-derived molecules, we may rely on a combination of chemical and biological treatment. Recently, conversion from lignin to medium chain-length polyhydroxyalkanoates was achieved in P. putida by combining microbial treatment with chemical pretreatment [22]. In a comparable effort, lignin conversion to β-ketoadipate was achieved with engineered P. putida [198]. In this work, genes encoding enzymes mediating 4-hydroxybenzoate hydroxylation and vanillate O-demethylation were overexpressed to improve the yield, and the gene that could cause intermediate accumulation was deleted. Additionally, through genetic engineering, Altenbuchner et al. successfully introduced key enzymes involved in the conversion of lignin-derived ferulic acid to vanillin in P. putida KT2440 [170]. This research enhances vanillin production with up to 86% molar yields and few byproducts.
These studies further illustrate that engineering metabolic pathways in P. putida to funnel lignin breakdown products into desirable bioproducts can increase the economic value of lignin valorization processes.
5. Conclusions and Future Prospects
Lignin, the second most abundant terrestrial polymer found on Earth, constitutes an important part of plant fibers [199]. Lignin is composed of a network of aromatic compounds and is highly resistant to decomposition. Currently, this rich aromatic compounds resource is mainly separated as a waste stream, where 98% is used as a heat source in factories [200]. Only 2% is used in a chemical conversion to produce useful compounds like lignosulfonates [17].
Importantly, we here make a case for lignin to be utilized much more effectively through biotechnological and chemical processes. Using those techniques, separately or in combination, lignin can be degraded into hundreds of valuable derivatives [201]. In nature, bacteria and fungi can degrade lignin, with some differences in the degradation mechanism, substrate specificity, and product generation. Only recently, research on the enzymatic processes involved in bacterial lignin degradation has led to the identification and documentation of specific enzymes dedicated to this purpose. This review provides a list of microorganisms reported to utilize lignin and potential enzymes involved in specific lignin depolymerization.
Fungi, especially white-rot fungi, produce a variety of enzymes (such as lignin peroxidase, manganese peroxidase, laccase, etc.) that can directly oxidize and degrade lignin. Fungi typically work by producing multiple enzymes that work together to break down different lignin bonds and connections. The depolymerization of native lignin is facilitated by extracellular oxidative enzymes, including Lip, MnP, VP, and Lac, which have been extensively documented in fungi. The research on bacterial degradation of lignin is not as in-depth as that on fungi. It has only been a dozen years since the first bacterial enzyme that degrades lignin was discovered [56]. In bacteria, the lignin degradation enzyme systems are thought to be relatively simple and specific. Limited enzymes are involved, such as phenol oxidase. In bacteria like Cupriavidus basilensis B-8, Lac, and MnP activities were identified; however, no MnP or Lac genes were found [202]. Hence, bacteria are anticipated to possess distinctive lignin degradation mechanisms and novel types of peroxidases [169].
Typically, fungal lignin degradation spans 10–30 days, whereas in bacteria, it may be accomplished in as little as 2–7 days. From an industrial viewpoint, utilizing bacteria as a host strain to establish a lignin degradation and utilization cell factory would prove more cost-effective [203]. In bioreactors, bacteria have advantages over fungi due to their rapid growth, simpler cultivation requirements, higher metabolic rates, easier genetic manipulation, and simpler product recovery [204,205]. While research on the enzymatic processes of lignin-degrading bacteria is currently relatively limited, using bacteria for lignin degradation still holds great prospects. This review demonstrates the great potential of P. putida as a microbial cell factory in lignin degradation and valorization, providing a sustainable approach to converting lignin into valuable bioproducts. The plasmid-free strain P. putida KT2440 is particularly regarded as a microbial host for biotechnological applications due to its biosafety status [206] and is widely used in industrial production.
We believe that, through genetic engineering and process optimization, P. putida can be adapted to industrial needs and contribute to further developing the bioeconomy through sustainable industrial practices.
Funding
Q. Zhou was supported by a China Scholarship Council (CSC) fellowship.
Data Availability Statement
This review does not present any new data.
Acknowledgments
We thank Arthur Ram and Jo-anne Verschoor (Department of Molecular Biotechnology, Institute for Biology, Leiden University) and Nick Wierckx (Biotechnology Research Center, Juelich, Germany) for fruitful discussions.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Chauhan, P.S. Role of various bacterial enzymes in complete depolymerization of lignin: A review. Biocatal. Agric. Biotechol. 2020, 23, 101498. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [PubMed]
- Karthäuser, J.; Biziks, V.; Mai, C.; Militz, H. Lignin and lignin-derived compounds for wood applications—A review. Molecules 2021, 26, 2533. [Google Scholar] [CrossRef]
- Palazzolo, M.A.; Kurina-Sanz, M. Microbial utilization of lignin: Available biotechnologies for its degradation and valorization. World J. Microb. Biot. 2016, 32, 173. [Google Scholar] [CrossRef]
- Faix, O. Classification of Lignins from Different Botanical Origins by FT-IR Spectroscopy. Holzforschung 1991, 45, 21–28. [Google Scholar] [CrossRef]
- Mansfield, S.D.; Kim, H.; Lu, F.; Ralph, J. Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 2012, 7, 1579–1589. [Google Scholar] [CrossRef]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin Biosynthesis and Structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef]
- Parthasarathi, R.; Romero, R.A.; Redondo, A.; Gnanakaran, S. Theoretical Study of the Remarkably Diverse Linkages in Lignin. J. Phys. Chem. Lett. 2011, 2, 2660–2666. [Google Scholar] [CrossRef]
- Picart, P.; Müller, C.; Mottweiler, J.; Wiermans, L.; Bolm, C.; Domínguez de María, P.; Schallmey, A. From gene towards selective biomass valorization: Bacterial β-etherases with catalytic activity on lignin-like polymers. ChemSusChem 2014, 7, 3164–3171. [Google Scholar] [CrossRef]
- Kai, D.; Tan, M.J.; Chee, P.L.; Chua, Y.K.; Yap, Y.L.; Loh, X.J. Towards lignin-based functional materials in a sustainable world. Green Chem. 2016, 18, 1175–1200. [Google Scholar] [CrossRef]
- Lahive, C.W.; Kamer, P.C.J.; Lancefield, C.S.; Deuss, P.J. An Introduction to Model Compounds of Lignin Linking Motifs; Synthesis and Selection Considerations for Reactivity Studies. ChemSusChem 2020, 13, 4238–4265. [Google Scholar] [CrossRef] [PubMed]
- A Guide to Lignin Valorization in Biorefineries: Traditional, Recent, and Forthcoming Approaches to Convert Raw Lignocellulose into Valuable Materials and Chemicals. Available online: https://pubs.rsc.org/en/content/articlehtml/2023/su/d3su00140g (accessed on 14 April 2025).
- Beckham, G.T.; Johnson, C.W.; Karp, E.M.; Salvachúa, D.; Vardon, D.R. Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 2016, 42, 40–53. [Google Scholar] [CrossRef]
- Zhang, S.; Xiao, J.; Wang, G.; Chen, G. Enzymatic hydrolysis of lignin by ligninolytic enzymes and analysis of the hydrolyzed lignin products. Bioresour. Technol. 2020, 304, 122975. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D. Lignin as a base material for materials applications: Chemistry, application and economics. Ind. Crop. Prod. 2008, 27, 202–207. [Google Scholar] [CrossRef]
- Lora, J.H.; Glasser, W.G. Recent Industrial Applications of Lignin: A Sustainable Alternative to Nonrenewable Materials. J. Polym. Environ. 2002, 10, 39–48. [Google Scholar] [CrossRef]
- Demuner, I.F.; Colodette, J.L.; Demuner, A.J.; Jardim, C.M. Biorefinery Review: Wide-Reaching Products Through Kraft Lignin. BioResources 2019, 14, 7543–7581. [Google Scholar] [CrossRef]
- Research, G.V. Lignin Market Size, Share & Trends Analysis Report by Produc. 2024. Available online: https://www.grandviewresearch.com/industry-analysis/lignin-market (accessed on 14 April 2025).
- Kleine, T.; Buendia, J.; Bolm, C. Mechanochemical degradation of lignin and wood by solvent-free grinding in a reactive medium. Green Chem. 2013, 15, 160–166. [Google Scholar] [CrossRef]
- vom Stein, T.; Weigand, T.; Merkens, C.; Klankermayer, J.; Leitner, W. Trimethylenemethane-Ruthenium(II)-Triphos Complexes as Highly Active Catalysts for Catalytic C-O Bond Cleavage Reactions of Lignin Model Compounds. ChemCatChem 2013, 5, 439–441. [Google Scholar] [CrossRef]
- van Erven, G.; Boerkamp, V.J.P.; van Groenestijn, J.W.; Gosselink, R.J.A. Choline and lactic acid covalently incorporate into the lignin structure during deep eutectic solvent pulping. Green Chem. 2024, 26, 7101–7112. [Google Scholar] [CrossRef]
- Linger, J.G.; Vardon, D.R.; Guarnieri, M.T.; Karp, E.M.; Hunsinger, G.B.; Franden, M.A.; Johnson, C.W.; Chupka, G.; Strathmann, T.J.; Pienkos, P.T.; et al. Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. USA 2014, 111, 12013–12018. [Google Scholar] [CrossRef]
- Smit, A.; Huijgen, W. Effective fractionation of lignocellulose in herbaceous biomass and hardwood using a mild acetone organosolv process. Green Chem. 2017, 19, 5505–5514. [Google Scholar] [CrossRef]
- Smit, A.T.; Hoek, M.; Bonouvrie, P.A.; van Zomeren, A.; Riddell, L.A.; Bruijnincx, P.C.A. Semicontinuous Aqueous Acetone Organosolv Fractionation of Lignocellulosic Biomass: Improved Biorefinery Processing and Output. ACS Sustain. Chem. Eng. 2024, 12, 4731–4742. [Google Scholar] [CrossRef]
- Smit, A.T.; Dezaire, T.; Riddell, L.A.; Bruijnincx, P.C.A. Reductive Partial Depolymerization of Acetone Organosolv Lignin to Tailor Lignin Molar Mass, Dispersity, and Reactivity for Polymer Applications. ACS Sustain. Chem. Eng. 2023, 11, 6070–6080. [Google Scholar] [CrossRef]
- Bugg, T.D.H.; Williamson, J.J.; Rashid, G.M.M. Bacterial enzymes for lignin depolymerisation: New biocatalysts for generation of renewable chemicals from biomass. Curr. Opin. Chem. Biol. 2020, 55, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Tien, M.; Kirk, T.K. Lignin-Degrading Enzyme from the Hymenomycete Phanerochaete chrysosporium Burds. Science 1983, 221, 661–663. [Google Scholar] [CrossRef]
- Kirk, T.K.; Farrell, R.L. Enzymatic “combustion”: The microbial degradation of lignin. Annu. Rev. Microbiol. 1987, 41, 465–505. [Google Scholar] [CrossRef] [PubMed]
- Martínez, A.T.; Speranza, M.; Ruiz-Dueñas, F.J.; Ferreira, P.; Camarero, S.; Guillén, F.; Martínez, M.J.; Gutiérrez, A.; del Río, J.C. Biodegradation of lignocellulosics: Microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 2005, 8, 195–204. [Google Scholar] [PubMed]
- Atiwesh, G.; Parrish, C.C.; Banoub, J.; Le, T.-A.T. Lignin degradation by microorganisms: A review. Biotechnol. Prog. 2022, 38, e3226. [Google Scholar] [CrossRef]
- Weng, C.; Peng, X.; Han, Y. Depolymerization and conversion of lignin to value-added bioproducts by microbial and enzymatic catalysis. Biotechnol. Biofuels 2021, 14, 84. [Google Scholar] [CrossRef]
- Nurul-Aliyaa, Y.A.; Awang, N.A.; Mohd, M.H. Characterization of white rot fungi from wood decayed for lignin degradation. Lett. Appl. Microbiol. 2023, 76, ovad118. [Google Scholar] [CrossRef]
- Paul, M.; Pandey, N.K.; Banerjee, A.; Shroti, G.K.; Tomer, P.; Gazara, R.K.; Thatoi, H.; Bhaskar, T.; Hazra, S.; Ghosh, D. An insight into omics analysis and metabolic pathway engineering of lignin-degrading enzymes for enhanced lignin valorization. Bioresour. Technol. 2023, 379, 129045. [Google Scholar] [CrossRef] [PubMed]
- Bugg, T.D.H.; Ahmad, M.; Hardiman, E.M.; Rahmanpour, R. Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 2011, 28, 1883–1896. [Google Scholar] [CrossRef] [PubMed]
- Arantes, V.; Milagres, A.M.F.; Filley, T.R.; Goodell, B. Lignocellulosic polysaccharides and lignin degradation by wood decay fungi: The relevance of nonenzymatic Fenton-based reactions. J. Ind. Microbiol. Biotechnol. 2011, 38, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Tien, M.; Kirk, T.K. Lignin-degrading enzyme from Phanerochaete chrysosporium: Purification, characterization, and catalytic properties of a unique H(2)O(2)-requiring oxygenase. Proc. Natl. Acad. Sci. USA 1984, 81, 2280–2284. [Google Scholar] [CrossRef]
- Niku-Paavola, M.L.; Karhunen, E.; Salola, P.; Raunio, V. Ligninolytic enzymes of the white-rot fungus Phlebia radiata. Biochem. J. 1988, 254, 877–883. [Google Scholar] [CrossRef]
- Martínez, A.T.; Camarero, S.; Guillén, F.; Gutiérrez, A.; Muñoz, C.; Varela, E.; Martínez, M.J.; Barrasa, J.; Ruel, K.; Pelayo, J. Progress in biopulping of non-woody materials: Chemical, enzymatic and ultrastructural aspects of wheat straw delignification with ligninolytic fungi from the genus Pleurotus. FEMS Microbiol. Rev. 1994, 13, 265–273. [Google Scholar] [CrossRef]
- Martínez, M.J.; Ruiz-Dueñas, F.J.; Guillén, F.; Martínez, Á.T. Purification and Catalytic Properties of Two Manganese Peroxidase Isoenzymes from Pleurotus eryngii. Eur. J. Biochem. 1996, 237, 424–432. [Google Scholar] [CrossRef]
- Moiseenko, K.V.; Glazunova, O.A.; Savinova, O.S.; Vasina, D.V.; Zherebker, A.Y.; Kulikova, N.A.; Nikolaev, E.N.; Fedorova, T.V. Relation between lignin molecular profile and fungal exo-proteome during kraft lignin modification by Trametes hirsuta LE-BIN 072. Bioresour. Technol. 2021, 335, 125229. [Google Scholar] [CrossRef]
- Bourbonnais, R.; Paice, M.G.; Reid, I.D.; Lanthier, P.; Yaguchi, M. Lignin oxidation by laccase isozymes from Trametes versicolor and role of the mediator 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) in kraft lignin depolymerization. Appl. Environ. Microbiol. 1995, 61, 1876–1880. [Google Scholar] [CrossRef]
- Eggert, C.; Temp, U.; Eriksson, K.E. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: Purification and characterization of the laccase. Appl. Environ. Microbiol. 1996, 62, 1151–1158. [Google Scholar] [CrossRef]
- Guerra, A.; Mendonça, R.; Ferraz, A.; Lu, F.; Ralph, J. Structural characterization of lignin during Pinus taeda wood treatment with Ceriporiopsis subvermispora. Appl. Environ. Microbiol. 2004, 70, 4073–4078. [Google Scholar] [CrossRef] [PubMed]
- Asgher, M.; Sharif, Y.; Bhatti, H.N. Enhanced Production of Ligninolytic Enzymes by Ganoderma lucidum IBL-06 Using Lignocellulosic Agricultural Wastes. Int. J. Chem. Reactor Eng. 2010, 8, 1. [Google Scholar] [CrossRef]
- Kamei, I.; Hirota, Y.; Meguro, S. Integrated delignification and simultaneous saccharification and fermentation of hard wood by a white-rot fungus, Phlebia sp. MG-60. Bioresour. Technol. 2012, 126, 137–141. [Google Scholar] [CrossRef]
- Knežević, A.; Milovanović, I.; Stajić, M.; Lončar, N.; Brčeski, I.; Vukojević, J.; Cilerdžić, J. Lignin degradation by selected fungal species. Bioresour. Technol. 2013, 138, 117–123. [Google Scholar] [CrossRef]
- Metri, Y.; Warly, L.; Suyitman, S. Biodegradation of lignin by white rot fungi (Pleurotus ostreatus) to decrease the fibre components in the palm midrib. Pak. J. Nutr. 2018, 17, 71–75. [Google Scholar] [CrossRef]
- Niemenmaa, O.; Uusi-Rauva, A.; Hatakka, A. Demethoxylation of [O14CH3]-labelled lignin model compounds by the brown-rot fungi Gloeophyllum trabeum and Poria (Postia) placenta. Biodegradation 2008, 19, 555–565. [Google Scholar] [CrossRef]
- Martinez, D.; Challacombe, J.; Morgenstern, I.; Hibbett, D.; Schmoll, M.; Kubicek, C.P.; Ferreira, P.; Ruiz-Duenas, F.J.; Martinez, A.T.; Kersten, P.; et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc. Natl. Acad. Sci. USA 2009, 106, 1954–1959. [Google Scholar] [CrossRef]
- Kadam, K.L.; Drew, S.W. Study of lignin biotransformation by Aspergillus fumigatus and white-rot fungi using 14C-labeled and unlabeled kraft lignins. Biotechnol. Bioeng. 1986, 28, 394–404. [Google Scholar] [CrossRef]
- van Erven, G.; Kleijn, A.F.; Patyshakuliyeva, A.; Di Falco, M.; Tsang, A.; de Vries, R.P.; van Berkel, W.J.H.; Kabel, M.A. Evidence for ligninolytic activity of the ascomycete fungus Podospora anserina. Biotechnol. Biofuels 2020, 13, 75. [Google Scholar] [CrossRef]
- Yang, Y.S.; Zhou, J.T.; Lu, H.; Yuan, Y.L.; Zhao, L.H. Isolation and characterization of a fungus Aspergillus sp. strain F-3 capable of degrading alkali lignin. Biodegradation 2011, 22, 1017–1027. [Google Scholar] [CrossRef]
- Wymelenberg, A.V.; Gaskell, J.; Mozuch, M.; Sabat, G.; Ralph, J.; Skyba, O.; Mansfield, S.D.; Blanchette, R.A.; Martinez, D.; Grigoriev, I.; et al. Comparative Transcriptome and Secretome Analysis of Wood Decay Fungi Postia placenta and Phanerochaete chrysosporium. Appl. Environ. Microbiol. 2010, 76, 3599–3610. [Google Scholar] [CrossRef] [PubMed]
- Betts, W.B.; Dart, R.K. The Degradation of Lignin-related Compounds by Aspergillus flavus. Microbiology 1988, 134, 2413–2420. [Google Scholar] [CrossRef]
- Ahmad, M.; Taylor, C.R.; Pink, D.; Burton, K.; Eastwood, D.; Bending, G.D.; Bugg, T.D. Development of novel assays for lignin degradation: Comparative analysis of bacterial and fungal lignin degraders. Mol. Biosyst. 2010, 6, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Roberts, J.N.; Hardiman, E.M.; Singh, R.; Eltis, L.D.; Bugg, T.D.H. Identification of DypB from Rhodococcus jostii RHA1 as a Lignin Peroxidase. Biochemistry 2011, 50, 5096–5107. [Google Scholar] [CrossRef] [PubMed]
- Spence, E.M.; Calvo-Bado, L.; Mines, P.; Bugg, T.D.H. Metabolic engineering of Rhodococcus jostii RHA1 for production of pyridine-dicarboxylic acids from lignin. Microb. Cell Fact. 2021, 20, 15. [Google Scholar] [CrossRef]
- Chong, G.G.; Huang, X.J.; Di, J.H.; Xu, D.Z.; He, Y.C.; Pei, Y.N.; Tang, Y.J.; Ma, C.L. Biodegradation of alkali lignin by a newly isolated Rhodococcus pyridinivorans CCZU-B16. Bioprocess Biosyst. Eng. 2018, 41, 501–510. [Google Scholar] [CrossRef]
- Chandra, R.; Raj, A.; Purohit, H.J.; Kapley, A. Characterisation and optimisation of three potential aerobic bacterial strains for kraft lignin degradation from pulp paper waste. Chemosphere 2007, 67, 839–846. [Google Scholar] [CrossRef]
- Khan, S.I.; Zarin, A.; Ahmed, S.; Hasan, F.; Belduz, A.O.; Çanakçi, S.; Khan, S.; Badshah, M.; Farman, M.; Shah, A.A. Degradation of lignin by Bacillus altitudinis SL7 isolated from pulp and paper mill effluent. Water Sci. Technol. 2021, 85, 420–432. [Google Scholar] [CrossRef]
- Huang, X.-F.; Santhanam, N.; Badri, D.V.; Hunter, W.J.; Manter, D.K.; Decker, S.R.; Vivanco, J.M.; Reardon, K.F. Isolation and characterization of lignin-degrading bacteria from rainforest soils. Biotechnol. Bioeng. 2013, 110, 1616–1626. [Google Scholar] [CrossRef]
- Kumar, A.; Priyadarshinee, R.; Singha, S.; Sengupta, B.; Roy, A.; Dasgupta, D.; Mandal, T. Biodegradation of alkali lignin by Bacillus flexus RMWW II: Analyzing performance for abatement of rice mill wastewater. Water Sci. Technol. 2019, 80, 1623–1632. [Google Scholar] [CrossRef]
- Zhu, D.; Zhang, P.; Xie, C.; Zhang, W.; Sun, J.; Qian, W.-J.; Yang, B. Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnol. Biofuels 2017, 10, 44. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Choi, D.; Takamizawa, K.; Kikuchi, S. Isolation of Bacillus sp. strains capable of decomposing alkali lignin and their application in combination with lactic acid bacteria for enhancing cellulase performance. Bioresour. Technol. 2014, 152, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Cheng, Y.; Pu, Y.; Sun, S.; Li, X.; Jin, M.; Pierson, E.A.; Gross, D.C.; Dale, B.E.; Dai, S.Y.; et al. Systems biology-guided biodesign of consolidated lignin conversion. Green Chem. 2016, 18, 5536–5547. [Google Scholar] [CrossRef]
- Xu, Z.; Qin, L.; Cai, M.; Hua, W.; Jin, M. Biodegradation of kraft lignin by newly isolated Klebsiella pneumoniae, Pseudomonas putida, and Ochrobactrum tritici strains. Environ. Sci. Pollut. Res. 2018, 25, 14171–14181. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Xu, M.; Cai, C.; Chen, S.; Jin, M. Microbial polyhydroxyalkanoate production from lignin by Pseudomonas putida NX-1. Bioresour. Technol. 2021, 319, 124210. [Google Scholar] [CrossRef]
- Eberlein, C.; Baumgarten, T.; Starke, S.; Heipieper, H.J. Immediate response mechanisms of Gram-negative solvent-tolerant bacteria to cope with environmental stress: Cis-trans isomerization of unsaturated fatty acids and outer membrane vesicle secretion. Appl. Microbiol. Biotechnol. 2018, 102, 2583–2593. [Google Scholar] [CrossRef]
- Salvachúa, D.; Werner, A.Z.; Pardo, I.; Michalska, M.; Black, B.A.; Donohoe, B.S.; Haugen, S.J.; Katahira, R.; Notonier, S.; Ramirez, K.J. Outer membrane vesicles catabolize lignin-derived aromatic compounds in Pseudomonas putida KT2440. Proc. Natl. Acad. Sci. USA 2020, 117, 9302–9310. [Google Scholar] [CrossRef] [PubMed]
- Granja-Travez, R.S.; Bugg, T.D.H. Characterization of multicopper oxidase CopA from Pseudomonas putida KT2440 and Pseudomonas fluorescens Pf-5: Involvement in bacterial lignin oxidation. Arch. Biochem. Biophys. 2018, 660, 97–107. [Google Scholar] [CrossRef]
- He, S.; Wang, W.; Wang, W.; Hu, H.; Xu, P.; Tang, H. Microbial production of cis,cis-muconic acid from aromatic compounds in engineered Pseudomonas. Synth. Syst. Biotechnol. 2023, 8, 536–545. [Google Scholar] [CrossRef]
- Johnson, C.W.; Salvachúa, D.; Khanna, P.; Smith, H.; Peterson, D.J.; Beckham, G.T. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 2016, 3, 111–119. [Google Scholar] [CrossRef]
- Belda, E.; Van Heck, R.G.; José Lopez-Sanchez, M.; Cruveiller, S.; Barbe, V.; Fraser, C.; Klenk, H.P.; Petersen, J.; Morgat, A.; Nikel, P.I. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ. Microbiol. 2016, 18, 3403–3424. [Google Scholar] [CrossRef] [PubMed]
- Crawford, D.L.; Pometto, A.L.; Crawford, R.L. Lignin Degradation by Streptomyces viridosporus: Isolation and Characterization of a New Polymeric Lignin Degradation Intermediate. Appl. Environ. Microbiol. 1983, 45, 898–904. [Google Scholar] [CrossRef]
- Yang, Y.S.; Zhou, J.T.; Lu, H.; Yuan, Y.L.; Zhao, L.H. Isolation and characterization of Streptomyces spp. strains F-6 and F-7 capable of decomposing alkali lignin. Environ. Technol. 2012, 33, 2603–2609. [Google Scholar] [CrossRef]
- Tan, F.; Cheng, J.; Zhang, Y.; Jiang, X.; Liu, Y. Genomics analysis and degradation characteristics of lignin by Streptomyces thermocarboxydus strain DF3-3. Biotechnol. Biofuels Bioprod. 2022, 15, 78. [Google Scholar] [CrossRef]
- Sidar, A.; Voshol, G.P.; El-Masoudi, A.; Vijgenboom, E.; Punt, P.J. Streptomyces small laccase expressed in Aspergillus Niger as a new addition for the lignocellulose bioconversion toolbox. Fungal Biol. Biotechnol. 2024, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Riyadi, F.A.; Tahir, A.A.; Yusof, N.; Sabri, N.S.A.; Noor, M.J.M.M.; Akhir, F.N.M.D.; Othman, N.a.; Zakaria, Z.; Hara, H. Enzymatic and genetic characterization of lignin depolymerization by Streptomyces sp. S6 isolated from a tropical environment. Sci. Rep. 2020, 10, 7813. [Google Scholar] [CrossRef] [PubMed]
- Masai, E.; Katayama, Y.; Nishikawa, S.; Fukuda, M. Characterization of Sphingomonas paucimobilis SYK-6 genes involved in degradation of lignin-related compounds. J. Ind. Microbiol. Biotechnol. 1999, 23, 364–373. [Google Scholar] [CrossRef]
- Shi, Y.; Chai, L.; Tang, C.; Yang, Z.; Zheng, Y.; Chen, Y.; Jing, Q. Biochemical investigation of kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips. Bioprocess Biosyst. Eng. 2013, 36, 1957–1965. [Google Scholar] [CrossRef]
- DeAngelis, K.; Sharma, D.; Varney, R.; Simmons, B.; Isern, N.; Markillie, L.M.; Nicora, C.; Norbeck, A.; Taylor, R.; Aldrich, J.; et al. Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1. Front. Microbiol. 2013, 4, 280. [Google Scholar] [CrossRef]
- Taylor, C.R.; Hardiman, E.M.; Ahmad, M.; Sainsbury, P.D.; Norris, P.R.; Bugg, T.D.H. Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J. Appl. Microbiol. 2012, 113, 521–530. [Google Scholar] [CrossRef]
- Kosa, M.; Ragauskas, A.J. Lignin to lipid bioconversion by oleaginous Rhodococci. Green Chem. 2013, 15, 2070–2074. [Google Scholar] [CrossRef]
- He, Y.; Li, X.; Ben, H.; Xue, X.; Yang, B. Lipid Production from Dilute Alkali Corn Stover Lignin by Rhodococcus Strains. ACS Sustain. Chem. Eng. 2017, 5, 2302–2311. [Google Scholar] [CrossRef]
- Brown, M.E.; Walker, M.C.; Nakashige, T.G.; Iavarone, A.T.; Chang, M.C.Y. Discovery and Characterization of Heme Enzymes from Unsequenced Bacteria: Application to Microbial Lignin Degradation. J. Am. Chem. Soc. 2011, 133, 18006–18009. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, S.; Lukk, T.; Solbiati, J.O.; Bauer, S.; Nair, S.K.; Cronan, J.E.; Gerlt, J.A. Roles of Small Laccases from Streptomyces in Lignin Degradation. Biochemistry 2014, 53, 4047–4058. [Google Scholar] [CrossRef]
- Brzonova, I.; Kozliak, E.; Kubátová, A.; Chebeir, M.; Qin, W.; Christopher, L.; Ji, Y. Kenaf biomass biodecomposition by basidiomycetes and actinobacteria in submerged fermentation for production of carbohydrates and phenolic compounds. Bioresour. Technol. 2014, 173, 352–360. [Google Scholar] [CrossRef]
- Deng, Y.; Fong, S.S. Metabolic engineering of Thermobifida fusca for direct aerobic bioconversion of untreated lignocellulosic biomass to 1-propanol. Metab. Eng. 2011, 13, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Akinosho, H.O.; Yoo, C.G.; Dumitrache, A.; Natzke, J.; Muchero, W.; Brown, S.D.; Ragauskas, A.J. Elucidating the Structural Changes to Populus Lignin during Consolidated Bioprocessing with Clostridium thermocellum. ACS Sustain. Chem. Eng. 2017, 5, 7486–7491. [Google Scholar] [CrossRef]
- Niu, J.; Li, X.; Qi, X.; Ren, Y. Pathway analysis of the biodegradation of lignin by Brevibacillus thermoruber. Bioresour. Technol. 2021, 341, 125875. [Google Scholar] [CrossRef]
- Kataeva, I.; Foston, M.B.; Yang, S.-J.; Pattathil, S.; Biswal, A.K.; Poole, F.L., II; Basen, M.; Rhaesa, A.M.; Thomas, T.P.; Azadi, P.; et al. Carbohydrate and lignin are simultaneously solubilized from unpretreated switchgrass by microbial action at high temperature. Energy Environ. Sci. 2013, 6, 2186–2195. [Google Scholar] [CrossRef]
- Wang, D.; Lin, Y.; Du, W.; Liang, J.; Ning, Y. Optimization and characterization of lignosulfonate biodegradation process by a bacterial strain, Sphingobacterium sp. HY-H. Int. Biodeterior. Biodegrad. 2013, 85, 365–371. [Google Scholar] [CrossRef]
- Rashid, G.M.M.; Taylor, C.R.; Liu, Y.; Zhang, X.; Rea, D.; Fülöp, V.; Bugg, T.D.H. Identification of Manganese Superoxide Dismutase from Sphingobacterium sp. T2 as a Novel Bacterial Enzyme for Lignin Oxidation. ACS Chem. Biol. 2015, 10, 2286–2294. [Google Scholar] [CrossRef]
- Chandra, R.; Abhishek, A.; Sankhwar, M. Bacterial decolorization and detoxification of black liquor from rayon grade pulp manufacturing paper industry and detection of their metabolic products. Bioresour. Technol. 2011, 102, 6429–6436. [Google Scholar] [CrossRef]
- Chandra, R.; Abhishek, A. Bacterial decolorization of black liquor in axenic and mixed condition and characterization of metabolites. Biodegradation 2011, 22, 603–611. [Google Scholar] [CrossRef]
- Chen, Y.H.; Chai, L.Y.; Zhu, Y.H.; Yang, Z.H.; Zheng, Y.; Zhang, H. Biodegradation of kraft lignin by a bacterial strain Comamonas sp. B-9 isolated from eroded bamboo slips. J. Appl. Microbiol. 2012, 112, 900–906. [Google Scholar] [CrossRef]
- Wilkes, R.A.; Waldbauer, J.; Caroll, A.; Nieto-Domínguez, M.; Parker, D.J.; Zhang, L.; Guss, A.M.; Aristilde, L. Complex regulation in a Comamonas platform for diverse aromatic carbon metabolism. Nat. Chem. Biol. 2023, 19, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Tiku, D.K.; Kumar, A.; Chaturvedi, R.; Makhijani, S.D.; Manoharan, A.; Kumar, R. Holistic bioremediation of pulp mill effluents using autochthonous bacteria. Int. Biodeterior. Biodegrad 2010, 64, 173–183. [Google Scholar] [CrossRef]
- Chandra, R.; Singh, R.; Yadav, S. Effect of bacterial inoculum ratio in mixed culture for decolourization and detoxification of pulp paper mill effluent. J. Chem. Technol. Biotechnol. 2012, 87, 436–444. [Google Scholar] [CrossRef]
- Paliwal, R.; Uniyal, S.; Rai, J.P.N. Evaluating the potential of immobilized bacterial consortium for black liquor biodegradation. Environ. Sci. Pollut. Res. 2015, 22, 6842–6853. [Google Scholar] [CrossRef]
- Yang, C.; Yue, F.; Cui, Y.; Xu, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Lü, X. Biodegradation of lignin by Pseudomonas sp. Q18 and the characterization of a novel bacterial DyP-type peroxidase. J. Ind. Microbiol. Biotechnol. 2018, 45, 913–927. [Google Scholar] [CrossRef]
- Nawaz, M.Z.; Shang, H.; Sun, J.; Geng, A.; Ali, S.S.; Zhu, D. Genomic insights into the metabolic potential of a novel lignin-degrading and polyhydroxyalkanoates producing bacterium Pseudomonas sp. Hu109A. Chemosphere 2023, 310, 136754. [Google Scholar] [CrossRef]
- Manter, D.K.; Hunter, W.J.; Vivanco, J.M. Enterobacter soli sp. nov.: A Lignin-Degrading γ-Proteobacteria Isolated from Soil. Curr. Microbiol. 2011, 62, 1044–1049. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.; Kumar, S.; Kumari, V.; Singh, S.K.; Raj, A. Evaluation of bioremediation potentiality of ligninolytic Serratia liquefaciens for detoxification of pulp and paper mill effluent. J. Hazard. Mater. 2016, 305, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Krishna Reddy, M.M.; Chandra, R. Identification of low molecular weight aromatic compounds by gas chromatography–mass spectrometry (GC–MS) from kraft lignin degradation by three Bacillus sp. Int. Biodeterior. Biodegrad. 2007, 59, 292–296. [Google Scholar] [CrossRef]
- Chandra, R.; Singh, S.; Krishna Reddy, M.M.; Patel, D.K.; Purohit, H.J.; Kapley, A. Isolation and characterization of bacterial strains Paenibacillus sp. and Bacillus sp. for kraft lignin decolorization from pulp paper mill waste. J. Gen. Appl. Microbiol. 2008, 54, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Gücyeter, S.; Erpelding, R.; Schmidt, M.S. Review: Chemical approaches toward catalytic lignin degradation. Chem. Pap. 2022, 76, 1899–1922. [Google Scholar] [CrossRef]
- Ojha, A.K.; Tiwari, M. Lignin Decolorization and Degradation of Pulp and Paper Mill Effluent by Ligninolytic Bacteria. Iran. J. Energy Environ. 2016, 7, 282–293. [Google Scholar]
- Mathews, S.L.; Grunden, A.M.; Pawlak, J. Degradation of lignocellulose and lignin by Paenibacillus glucanolyticus. Int. Biodeterior. Biodegrad. 2016, 110, 79–86. [Google Scholar] [CrossRef]
- Raj, A.; Kumar, S.; Haq, I.; Singh, S.K. Bioremediation and toxicity reduction in pulp and paper mill effluent by newly isolated ligninolytic Paenibacillus sp. Ecol. Eng. 2014, 71, 355–362. [Google Scholar] [CrossRef]
- Majumdar, S.; Priyadarshinee, R.; Kumar, A.; Mandal, T.; Dasgupta Mandal, D. Exploring Planococcus sp. TRC1, a bacterial isolate, for carotenoid pigment production and detoxification of paper mill effluent in immobilized fluidized bed reactor. J. Clean. Prod. 2019, 211, 1389–1402. [Google Scholar] [CrossRef]
- Jiang, C.; Yan, H.; Shen, X.; Zhang, Y.; Wang, Y.; Sun, S.; Jiang, H.; Zang, H.; Zhao, X.; Hou, N.; et al. Genome Functional Analysis of the Psychrotrophic Lignin-Degrading Bacterium Arthrobacter sp. C2 and the Role of DyP in Catalyzing Lignin Degradation. Front. Microbiol. 2022, 13, 921549. [Google Scholar] [CrossRef]
- Liu, H.; Tao, X.; Ntakirutimana, S.; Liu, Z.-H.; Li, B.-Z.; Yuan, Y.-J. Engineering Pseudomonas putida for lignin bioconversion into cis-cis muconic acid. Chem. Eng. J. 2024, 495, 153375. [Google Scholar] [CrossRef]
- Zhu, D.; Qaria, M.A.; Zhu, B.; Sun, J.; Yang, B. Extremophiles and extremozymes in lignin bioprocessing. Renew. Sustain. Energy Rev. 2022, 157, 112069. [Google Scholar]
- Pollegioni, L.; Tonin, F.; Rosini, E. Lignin-degrading enzymes. FEBS J. 2015, 282, 1190–1213. [Google Scholar] [CrossRef]
- Gall, D.L.; Ralph, J.; Donohue, T.J.; Noguera, D.R. A Group of Sequence-Related Sphingomonad Enzymes Catalyzes Cleavage of β-Aryl Ether Linkages in Lignin β-Guaiacyl and β-Syringyl Ether Dimers. Environ. Sci. Technol. 2014, 48, 12454–12463. [Google Scholar] [CrossRef] [PubMed]
- Marinović, M.; Nousiainen, P.; Dilokpimol, A.; Kontro, J.; Moore, R.; Sipilä, J.; de Vries, R.P.; Mäkelä, M.R.; Hildén, K. Selective Cleavage of Lignin β-O-4 Aryl Ether Bond by β-Etherase of the White-Rot Fungus Dichomitus squalens. ACS Sustain. Chem. Eng. 2018, 6, 2878–2882. [Google Scholar] [CrossRef]
- Vignali, E.; Tonin, F.; Pollegioni, L.; Rosini, E. Characterization and use of a bacterial lignin peroxidase with an improved manganese-oxidative activity. Appl. Microbiol. Biotechnol. 2018, 102, 10579–10588. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Fu, X.; Wang, L.; Alhujaily, A.; Zhang, J.; Ma, F.; Zhang, X.; Yu, H. A novel and efficient fungal delignification strategy based on versatile peroxidase for lignocellulose bioconversion. Biotechnol. Biofuels 2017, 10, 218. [Google Scholar] [CrossRef]
- Wariishi, H.; Valli, K.; Gold, M.H. Oxidative cleavage of a phenolic diarylpropane lignin model dimer by manganese peroxidase from Phanerochaete chrysosporium. Biochemistry 1989, 28, 6017–6023. [Google Scholar] [CrossRef]
- Zhu, D.; Liang, N.; Zhang, R.; Ahmad, F.; Zhang, W.; Yang, B.; Wu, J.; Geng, A.; Gabriel, M.; Sun, J. Insight into Depolymerization Mechanism of Bacterial Laccase for Lignin. ACS Sustain. Chem. Eng. 2020, 8, 12920–12933. [Google Scholar] [CrossRef]
- Sonoki, T.; Obi, T.; Kubota, S.; Higashi, M.; Masai, E.; Katayama, Y. Coexistence of Two Different O Demethylation Systems in Lignin Metabolism by Sphingomonas paucimobilis SYK-6: Cloning and Sequencing of the Lignin Biphenyl-Specific O-Demethylase (LigX) Gene. Appl. Environ. Microbiol. 2000, 66, 2125–2132. [Google Scholar] [CrossRef]
- Peng, X.; Egashira, T.; Hanashiro, K.; Masai, E.; Nishikawa, S.; Katayama, Y.; Kimbara, K.; Fukuda, M. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 1998, 64, 2520–2527. [Google Scholar] [CrossRef] [PubMed]
- Masai, E.; Katayama, Y.; Fukuda, M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotechnol. Biochem. 2007, 71, 1–15. [Google Scholar] [CrossRef]
- Kamaya, Y.; Nakatsubo, F.; Higuchi, T.; Iwahara, S. Degradation of d,l-syringaresinol, a β-β′ linked lignin model compound, by Fusarium solani M-13-1. Arch. Microbiol. 1981, 129, 305–309. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Goradia, B.; Jha, B. Optimization and up scaling of ionic liquid tolerant and thermo-alkali stable laccase from a marine Staphylococcus arlettae S1-20 using tea waste. J. Taiwan Inst. Chem. Eng. 2018, 86, 1–8. [Google Scholar] [CrossRef]
- Ramachandra, M.; Crawford, D.L.; Hertel, G. Characterization of an extracellular lignin peroxidase of the lignocellulolytic actinomycete Streptomyces viridosporus. Appl. Environ. Microbiol. 1988, 54, 3057–3063. [Google Scholar] [CrossRef] [PubMed]
- Asgher, M.; Iqbal, H.M.N.; Irshad, M. Characterization of purified and Xerogel immobilized Novel Lignin Peroxidase produced from Trametes versicolor IBL-04 using solid state medium of Corncobs. BMC Biotechnol. 2012, 12, 46. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Sulej, J.; Swiderska-Burek, U.; Jarosz-Wilkolazka, A.; Paszczynski, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef]
- Adler, E. Lignin chemistry—Past, present and future. Wood Sci. Technol. 1977, 11, 169–218. [Google Scholar] [CrossRef]
- Glenn, J.K.; Morgan, M.A.; Mayfield, M.B.; Kuwahara, M.; Gold, M.H. An extracellular H2O2-requiring enzyme preparation involved in lignin biodegradation by the white rot basidiomycete Phanerochaete chrysosporium. Biochem. Biophys. Res. Commun. 1983, 114, 1077–1083. [Google Scholar] [CrossRef]
- Dashora, K.; Gattupalli, M.; Tripathi, G.D.; Javed, Z.; Singh, S.; Tuohy, M.; Sarangi, P.K.; Diwan, D.; Singh, H.B.; Gupta, V.K. Fungal Assisted Valorisation of Polymeric Lignin: Mechanism, Enzymes and Perspectives. Catalysts 2023, 13, 149. [Google Scholar] [CrossRef]
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Thurston, C.F. The structure and function of fungal laccases. Microbiology 1994, 140, 19–26. [Google Scholar] [CrossRef]
- Dwivedi, U.N.; Singh, P.; Pandey, V.P.; Kumar, A. Structure–function relationship among bacterial, fungal and plant laccases. J. Mol. Catal. B Enzym. 2011, 68, 117–128. [Google Scholar] [CrossRef]
- Shabaev, A.V.; Moiseenko, K.V.; Glazunova, O.A.; Savinova, O.S.; Fedorova, T.V. Comparative Analysis of Peniophora lycii and Trametes hirsuta Exoproteomes Demonstrates “Shades of Gray” in the Concept of White-Rotting Fungi. Int. J. Mol. Sci. 2022, 23, 10322. [Google Scholar] [CrossRef]
- Manavalan, T.; Manavalan, A.; Heese, K. Characterization of Lignocellulolytic Enzymes from White-Rot Fungi. Curr. Microbiol. 2015, 70, 485–498. [Google Scholar] [CrossRef]
- Civzele, A.; Stipniece-Jekimova, A.A.; Mezule, L. Fungal Ligninolytic Enzymes and Their Application in Biomass Lignin Pretreatment. J. Fungi 2023, 9, 780. [Google Scholar] [CrossRef]
- Zeng, J.; Mills, M.J.L.; Simmons, B.A.; Kent, M.S.; Sale, K.L. Understanding factors controlling depolymerization and polymerization in catalytic degradation of β-ether linked model lignin compounds by versatile peroxidase. Green Chem. 2017, 19, 2145–2154. [Google Scholar] [CrossRef]
- Hofrichter, M. Review: Lignin conversion by manganese peroxidase (MnP). Enzyme Microb. Technol. 2002, 30, 454–466. [Google Scholar] [CrossRef]
- Tuor, U.; Wariishi, H.; Schoemaker, H.E.; Gold, M.H. Oxidation of phenolic arylglycerol beta-aryl ether lignin model compounds by manganese peroxidase from Phanerochaete chrysosporium: Oxidative cleavage of an alpha-carbonyl model compound. Biochemistry 1992, 31, 4986–4995. [Google Scholar] [CrossRef]
- Yoshida, T.; Sugano, Y. Unexpected diversity of dye-decolorizing peroxidases. Biochem. Biophys. Rep. 2023, 33, 101401. [Google Scholar] [CrossRef]
- Vicuña, R.; González, B.; Mozuch, M.D.; Kirk, T.K. Metabolism of Lignin Model Compounds of the Arylglycerol-beta-Aryl Ether Type by Pseudomonas acidovorans D(3). Appl. Environ. Microbiol. 1987, 53, 2605–2609. [Google Scholar] [CrossRef] [PubMed]
- Masai, E.; Katayama, Y.; Kawai, S.; Nishikawa, S.; Yamasaki, M.; Morohoshi, N. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves beta-aryl ether. J. Bacteriol. 1991, 173, 7950–7955. [Google Scholar] [CrossRef] [PubMed]
- Masai, E.; Katayama, Y.; Nishikawa, S.; Yamasaki, M.; Morohoshi, N.; Haraguchi, T. Detection and localization of a new enzyme catalyzing the β-aryl ether cleavage in the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett. 1989, 249, 348–352. [Google Scholar] [CrossRef]
- Sheehan, D.; Meade, G.; Foley, V.M.; Dowd, C.A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001, 360, 1–16. [Google Scholar] [CrossRef]
- Voß, H.; Heck, C.A.; Schallmey, M.; Schallmey, A. Database Mining for Novel Bacterial β-Etherases, Glutathione-Dependent Lignin-Degrading Enzymes. Appl. Environ. Microbiol. 2020, 86, e02026-19. [Google Scholar] [CrossRef]
- Rosini, E.; Allegretti, C.; Melis, R.; Cerioli, L.; Conti, G.; Pollegioni, L.; D’Arrigo, P. Cascade enzymatic cleavage of the β-O-4 linkage in a lignin model compound. Catal. Sci. Technol. 2016, 6, 2195–2205. [Google Scholar] [CrossRef]
- Reiter, J.; Strittmatter, H.; Wiemann, L.O.; Schieder, D.; Sieber, V. Enzymatic cleavage of lignin β-O-4 aryl ether bonds via net internal hydrogen transfer. Green Chem. 2013, 15, 1373–1381. [Google Scholar] [CrossRef]
- Helmich, K.E.; Pereira, J.H.; Gall, D.L.; Heins, R.A.; McAndrew, R.P.; Bingman, C.; Deng, K.; Holland, K.C.; Noguera, D.R.; Simmons, B.A.; et al. Structural Basis of Stereospecificity in the Bacterial Enzymatic Cleavage of β-Aryl Ether Bonds in Lignin. J. Biol. Chem. 2016, 291, 5234–5246. [Google Scholar] [CrossRef]
- Masai, E.; Ichimura, A.; Sato, Y.; Miyauchi, K.; Katayama, Y.; Fukuda, M. Roles of the enantioselective glutathione S-transferases in cleavage of beta-aryl ether. J. Bacteriol. 2003, 185, 1768–1775. [Google Scholar] [CrossRef]
- Sato, Y.; Moriuchi, H.; Hishiyama, S.; Otsuka, Y.; Oshima, K.; Kasai, D.; Nakamura, M.; Ohara, S.; Katayama, Y.; Fukuda, M.; et al. Identification of three alcohol dehydrogenase genes involved in the stereospecific catabolism of arylglycerol-beta-aryl ether by Sphingobium sp. strain SYK-6. Appl. Environ. Microbiol. 2009, 75, 5195–5201. [Google Scholar] [CrossRef]
- Colpa, D.I.; Fraaije, M.W.; van Bloois, E. DyP-type peroxidases: A promising and versatile class of enzymes. J. Ind. Microbiol. Biotechnol. 2014, 41, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Min, K.; Gong, G.; Woo, H.M.; Kim, Y.; Um, Y. A dye-decolorizing peroxidase from Bacillus subtilis exhibiting substrate-dependent optimum temperature for dyes and β-ether lignin dimer. Sci. Rep. 2015, 5, 8245. [Google Scholar] [CrossRef] [PubMed]
- Rahmanpour, R.; Rea, D.; Jamshidi, S.; Fülöp, V.; Bugg, T.D. Structure of Thermobifida fusca DyP-type peroxidase and activity towards Kraft lignin and lignin model compounds. Arch. Biochem. Biophys. 2016, 594, 54–60. [Google Scholar] [CrossRef]
- Ehibhatiomhan, A.O.; Pour, R.R.; Farnaud, S.; Bugg, T.D.H.; Mendel-Williams, S. Periplasmic expression of Pseudomonas fluorescens peroxidase Dyp1B and site-directed mutant Dyp1B enzymes enhances polymeric lignin degradation activity in Pseudomonas putida KT2440. Enzyme Microb. Technol. 2023, 162, 110147. [Google Scholar] [CrossRef]
- Reiss, R.; Ihssen, J.; Richter, M.; Eichhorn, E.; Schilling, B.; Thöny-Meyer, L. Laccase versus laccase-like multi-copper oxidase: A comparative study of similar enzymes with diverse substrate spectra. PLoS ONE 2013, 8, e65633. [Google Scholar] [CrossRef]
- Rydén, L.G.; Hunt, L.T. Evolution of protein complexity: The blue copper-containing oxidases and related proteins. J. Mol. Evol. 1993, 36, 41–66. [Google Scholar] [CrossRef]
- Solano, F.; Lucas-Elío, P.; López-Serrano, D.; Fernández, E.; Sanchez-Amat, A. Dimethoxyphenol oxidase activity of different microbial blue multicopper proteins. FEMS Microbiol. Lett. 2001, 204, 175–181. [Google Scholar] [CrossRef]
- Xu, Z.; Peng, B.; Kitata, R.B.; Nicora, C.D.; Weitz, K.K.; Pu, Y.; Shi, T.; Cort, J.R.; Ragauskas, A.J.; Yang, B. Understanding of bacterial lignin extracellular degradation mechanisms by Pseudomonas putida KT2440 via secretomic analysis. Biotechnol. Biofuels Bioprod. 2022, 15, 117. [Google Scholar] [CrossRef] [PubMed]
- Machczynski, M.C.; Vijgenboom, E.; Samyn, B.; Canters, G.W. Characterization of SLAC: A small laccase from Streptomyces coelicolor with unprecedented activity. Protein Sci. 2004, 13, 2388–2397. [Google Scholar] [CrossRef]
- Pandey, M.P.; Kim, C.S. Lignin Depolymerization and Conversion: A Review of Thermochemical Methods. Chem. Eng. Technol. 2011, 34, 29–41. [Google Scholar] [CrossRef]
- Sonoki, T.; Masai, E.; Sato, K.; Kajita, S.; Katayama, Y. Methoxyl groups of lignin are essential carbon donors in C1 metabolism of Sphingobium sp. SYK-6. J. Basic Microbiol. 2009, 49 (Suppl. S1), S98–S102. [Google Scholar] [CrossRef]
- Shettigar, M.; Balotra, S.; Cahill, D.; Warden, A.C.; Lacey, M.J.; Kohler, H.E.; Rentsch, D.; Oakeshott, J.G.; Pandey, G. Isolation of the (+)-Pinoresinol-Mineralizing Pseudomonas sp. Strain SG-MS2 and Elucidation of Its Catabolic Pathway. Appl. Environ. Microbiol. 2018, 84, e02531-17. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Fukushima, Y.; Jensen, K.A., Jr.; Moen, M.A.; Hammel, K.E. Oxidative degradation of non-phenolic lignin during lipid peroxidation by fungal manganese peroxidase. FEBS Lett. 1994, 354, 297–300. [Google Scholar] [CrossRef]
- Kapich, A.; Hofrichter, M.; Vares, T.; Hatakka, A. Coupling of manganese peroxidase-mediated lipid peroxidation with destruction of nonphenolic lignin model compounds and 14C-labeled lignins. Biochem. Biophys. Res. Commun. 1999, 259, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Kawai, S.; Umezawa, T.; Higuchi, T. Degradation mechanisms of phenolic beta-1 lignin substructure model compounds by laccase of Coriolus versicolor. Arch. Biochem. Biophys. 1988, 262, 99–110. [Google Scholar] [CrossRef]
- Agustin, M.B.; de Carvalho, D.M.; Lahtinen, M.H.; Hilden, K.; Lundell, T.; Mikkonen, K.S. Laccase as a Tool in Building Advanced Lignin-Based Materials. ChemSusChem 2021, 14, 4615–4635. [Google Scholar] [CrossRef] [PubMed]
- Bugg, T.D.; Ahmad, M.; Hardiman, E.M.; Singh, R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 2011, 22, 394–400. [Google Scholar] [CrossRef]
- Graf, N.; Altenbuchner, J. Genetic engineering of Pseudomonas putida KT2440 for rapid and high-yield production of vanillin from ferulic acid. Appl. Microbiol. Biotechnol. 2014, 98, 137–149. [Google Scholar] [CrossRef]
- Ankenbauer, A.; Schäfer, R.A.; Viegas, S.C.; Pobre, V.; Voß, B.; Arraiano, C.M.; Takors, R. Pseudomonas putida KT2440 is naturally endowed to withstand industrial-scale stress conditions. Microb. Biotechnol. 2020, 13, 1145–1161. [Google Scholar] [CrossRef]
- Jiménez, J.I.; Miñambres, B.; García, J.L.; Díaz, E. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 2002, 4, 824–841. [Google Scholar] [CrossRef]
- Zakzeski, J.; Bruijnincx, P.C.A.; Jongerius, A.L.; Weckhuysen, B.M. The Catalytic Valorization of Lignin for the Production of Renewable Chemicals. Chem. Rev. 2010, 110, 3552–3599. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, X.; Wang, A.; Huber, G.W.; Zhang, T. Catalytic Transformation of Lignin for the Production of Chemicals and Fuels. Chem. Rev. 2015, 115, 11559–11624. [Google Scholar] [CrossRef] [PubMed]
- Fache, M.; Boutevin, B.; Caillol, S. Vanillin Production from Lignin and Its Use as a Renewable Chemical. ACS Sustain. Chem. Eng. 2016, 4, 35–46. [Google Scholar] [CrossRef]
- Ravi, K.; García-Hidalgo, J.; Gorwa-Grauslund, M.F.; Lidén, G. Conversion of lignin model compounds by Pseudomonas putida KT2440 and isolates from compost. Appl. Microbiol. Biotechnol. 2017, 101, 5059–5070. [Google Scholar] [CrossRef] [PubMed]
- Plaggenborg, R.; Overhage, J.; Steinbüchel, A.; Priefert, H. Functional analyses of genes involved in the metabolism of ferulic acid in Pseudomonas putida KT2440. Appl. Microbiol. Biotechnol. 2003, 61, 528–535. [Google Scholar] [CrossRef]
- Harwood, C.S.; Parales, R.E. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996, 50, 553–590. [Google Scholar] [CrossRef]
- Nikel, P.I.; de Lorenzo, V. Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism. Metab. Eng. 2018, 50, 142–155. [Google Scholar] [CrossRef]
- Cook, T.B.; Rand, J.M.; Nurani, W.; Courtney, D.K.; Liu, S.A.; Pfleger, B.F. Genetic tools for reliable gene expression and recombineering in Pseudomonas putida. J. Ind. Microbiol. Biotechnol. 2018, 45, 517–527. [Google Scholar] [CrossRef]
- Martin-Pascual, M.; Batianis, C.; Bruinsma, L.; Asin-Garcia, E.; Garcia-Morales, L.; Weusthuis, R.A.; van Kranenburg, R.; Martins dos Santos, V.A.P. A navigation guide of synthetic biology tools for Pseudomonas putida. Biotechnol. Adv. 2021, 49, 107732. [Google Scholar] [CrossRef]
- Zobel, S.; Benedetti, I.; Eisenbach, L.; de Lorenzo, V.; Wierckx, N.; Blank, L.M. Tn7-Based Device for Calibrated Heterologous Gene Expression in Pseudomonas putida. ACS Synth. Biol. 2015, 4, 1341–1351. [Google Scholar] [CrossRef]
- Calero, P.; Jensen, S.I.; Nielsen, A.T. Broad-Host-Range ProUSER Vectors Enable Fast Characterization of Inducible Promoters and Optimization of p-Coumaric Acid Production in Pseudomonas putida KT2440. ACS Synth. Biol. 2016, 5, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Bagdasarian, M.M.; Amann, E.; Lurz, R.; Rückert, B.; Bagdasarian, M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene 1983, 26, 273–282. [Google Scholar] [CrossRef]
- Martínez-García, E.; Calles, B.; Arévalo-Rodríguez, M.; de Lorenzo, V. pBAM1: An all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol. 2011, 11, 38. [Google Scholar] [CrossRef]
- Volke, D.C.; Turlin, J.; Mol, V.; Nikel, P.I. Physical decoupling of XylS/Pm regulatory elements and conditional proteolysis enable precise control of gene expression in Pseudomonas putida. Microb. Biotechnol. 2020, 13, 222–232. [Google Scholar] [CrossRef]
- Aparicio, T.; de Lorenzo, V.; Martínez-García, E. CRISPR/Cas9-Based Counterselection Boosts Recombineering Efficiency in Pseudomonas putida. Biotechnol. J. 2018, 13, 1700161. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, T.; de Lorenzo, V.; Martínez-García, E. CRISPR/Cas9-enhanced ssDNA recombineering for Pseudomonas putida. Microb. Biotechnol. 2019, 12, 1076–1089. [Google Scholar] [CrossRef] [PubMed]
- Lammens, E.-M.; Volke, D.C.; Schroven, K.; Voet, M.; Kerremans, A.; Lavigne, R.; Hendrix, H. A SEVA-based, CRISPR-Cas3-assisted genome engineering approach for Pseudomonas with efficient vector curing. Microbiol. Spectr. 2023, 11, e02707–e02723. [Google Scholar] [CrossRef]
- Lammens, E.-M.; Volke, D.C.; Kerremans, A.; Aerts, Y.; Boon, M.; Nikel, P.I.; Lavigne, R. Engineering a phi15-based expression system for stringent gene expression in Pseudomonas putida. Commun. Biol. 2025, 8, 171. [Google Scholar] [CrossRef]
- Bugg, T.D.H. The chemical logic of enzymatic lignin degradation. Chem. Commun. 2024, 60, 804–814. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Lee, S.; Kang, M.; Jung, C.-D.; Bae, J.-H.; Lee, J.Y.; Park, Y.-K.; Joo, J.C.; Kim, H.; Sohn, J.-H.; Sung, B.H. Development of novel recombinant peroxidase secretion system from Pseudomonas putida for lignin valorisation. Bioresour. Technol. 2023, 388, 129779. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Wittmann, C. Advanced biotechnology: Metabolically engineered cells for the bio-based production of chemicals and fuels, materials, and health-care products. Angew. Chem. Int. Ed. Engl. 2015, 54, 3328–3350. [Google Scholar] [CrossRef]
- Weiland, F.; Kohlstedt, M.; Wittmann, C. Guiding stars to the field of dreams: Metabolically engineered pathways and microbial platforms for a sustainable lignin-based industry. Metab. Eng. 2022, 71, 13–41. [Google Scholar] [CrossRef]
- Abdelaziz, O.Y.; Brink, D.P.; Prothmann, J.; Ravi, K.; Sun, M.; García-Hidalgo, J.; Sandahl, M.; Hulteberg, C.P.; Turner, C.; Lidén, G.; et al. Biological valorization of low molecular weight lignin. Biotechnol. Adv. 2016, 34, 1318–1346. [Google Scholar] [CrossRef]
- Kamimura, N.; Takahashi, K.; Mori, K.; Araki, T.; Fujita, M.; Higuchi, Y.; Masai, E. Bacterial catabolism of lignin-derived aromatics: New findings in a recent decade: Update on bacterial lignin catabolism. Environ. Microbiol. Rep. 2017, 9, 679–705. [Google Scholar] [CrossRef]
- Werner, A.Z.; Cordell, W.T.; Lahive, C.W.; Klein, B.C.; Singer, C.A.; Tan, E.C.D.; Ingraham, M.A.; Ramirez, K.J.; Kim, D.H.; Pedersen, J.N.; et al. Lignin conversion to β-ketoadipic acid by Pseudomonas putida via metabolic engineering and bioprocess development. Sci. Adv. 2023, 9, eadj0053. [Google Scholar] [CrossRef]
- Huang, K.; Fasahati, P.; Maravelias, C.T. System-Level Analysis of Lignin Valorization in Lignocellulosic Biorefineries. Iscience 2020, 23, 100751. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Wang, Y.; Liu, J.; Xu, M.; Ma, P.; Ji, J.; You, Z. Review on Applications of Lignin in Pavement Engineering: A Recent Survey. Front. Mater. 2022, 8, 803524. [Google Scholar] [CrossRef]
- Zhou, N.; Thilakarathna, W.P.D.W.; He, Q.S.; Rupasinghe, H.P.V. A Review: Depolymerization of Lignin to Generate High-Value Bio-Products: Opportunities, Challenges, and Prospects. Front. Energy Res. 2022, 9, 758744. [Google Scholar] [CrossRef]
- Shi, Y.; Chai, L.; Tang, C.; Yang, Z.; Zhang, H.; Chen, R.; Chen, Y.; Zheng, Y. Characterization and genomic analysis of kraft lignin biodegradation by the beta-proteobacterium Cupriavidus basilensis B-8. Biotechnol. Biofuels 2013, 6, 1. [Google Scholar] [CrossRef]
- Asina, F.; Brzonova, I.; Voeller, K.; Kozliak, E.; Kubátová, A.; Yao, B.; Ji, Y. Biodegradation of lignin by fungi, bacteria and laccases. Bioresour. Technol. 2016, 220, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Cuebas-Irizarry, M.F.; Grunden, A.M. Streptomyces spp. as biocatalyst sources in pulp and paper and textile industries: Biodegradation, bioconversion and valorization of waste. Microb. Biotechnol. 2024, 17, e14258. [Google Scholar] [CrossRef] [PubMed]
- Eng, T.; Banerjee, D.; Lau, A.K.; Bowden, E.; Herbert, R.A.; Trinh, J.; Prahl, J.-P.; Deutschbauer, A.; Tanjore, D.; Mukhopadhyay, A. Engineering Pseudomonas putida for efficient aromatic conversion to bioproduct using high throughput screening in a bioreactor. Metab. Eng. 2021, 66, 229–238. [Google Scholar] [CrossRef]
- Weimer, A.; Kohlstedt, M.; Volke, D.C.; Nikel, P.I.; Wittmann, C. Industrial biotechnology of Pseudomonas putida: Advances and prospects. Appl. Microbiol. Biotechnol. 2020, 104, 7745–7766. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).