Detection and Identification of Food-Borne Yeasts: An Overview of the Relevant Methods and Their Evolution
Abstract
1. Introduction
2. Yeasts in Foods
3. Detection of Yeasts from Food Matrices
3.1. Standards for the Enumeration of Yeasts from Food, Feed and Their Changes
3.2. Culturing Techniques and Culture Media Used for Detection of Yeasts
| Type of Culture Medium | Name of Culture Medium | Reference |
|---|---|---|
| Acidified culture media | Acidified Malt/Whey/Wort Agar | [76] |
| Acidified Potato Dextrose Agar | [77] | |
| Orange Serum Agar | [78] | |
| Antibiotic supplemented culture media | Dichloran Rose Bengal Chloramphenicol Agar | [79] |
| Rose Bengal Chloramphenicol Agar | [80] | |
| Oxytetracycline-Glucose-Yeast Extract Agar | [81] |
3.3. Detection of Yeasts by Non-Conventional Methods
3.3.1. Conductometry/Impedimetry
3.3.2. ATP Bioluminescence
3.3.3. Immunological Methods
3.3.4. Molecular Biological Methods for Yeast Detection
3.3.5. Application of Other Technologies in Detection of Foodborne Yeasts
3.4. Identification and Typing of Food-Borne Yeasts
3.4.1. Biochemical Identification
3.4.2. Rapid Tests for Identification
3.4.3. Applicability of MALDI-TOF MS for Yeast Identification
Approach to the Methodological Concept
Identification Reliability in the Case of Yeast
3.4.4. Nucleic Acid-Based Identification
Procedure of Nucleic Acid-Based Identification Techniques
Comparison of Culture-Dependent and Culture-Independent Molecular Identification Techniques
3.5. Future Trends in Investigation of Foodborne Yeasts
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NSYs | non-Saccharomyces yeasts |
| CFU | colony forming unit |
| DT, TTD | detection time, time to detection |
| ATP | adenosine triphosphate |
| RLU | relative light units |
| ELISA | enzyme-linked immunosorbent assay |
| rRNA | ribosomal ribonucleic acid |
| ITS | internal transcribed spacer |
| RT-PCR | reverse transcriptase PCR |
| PCR | polymerase chain reaction |
| SIP | surface-imprinted polymer |
| VBNC | viable but non-culturable |
| FT-IR | Fourier-transform infrared spectroscopy |
| GC | gas chromatography |
| MALDI-TOF MS | matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry |
| HCCA | α-cyano-4-hydroxycinnamic acid |
| m/z | mass-to-charge ratio |
| rDNA | ribosomal deoxyribonucleic acid |
| SSU | small subunit |
| LSU | large subunit |
| RFLP | restriction fragment length polymorphism |
| NGS | next-generation sequencing |
| OTUs | operational taxonomic units |
| ASVs | amplicon sequence variants |
| rep-PCR | repetitive sequence-based PCR |
| RAPD-PCR | randomly amplified polymorphic DNA PCR |
| BLAST | basic local alignment search tool |
| qPCR | quantitative real-time PCR |
| mtDNA | mitochondrial deoxyribonucleic acid |
| DGGE | denaturing gradient gel electrophoresis |
| RS | Raman spectroscopy |
| SERS | surface-enhanced Raman spectroscopy |
| SCRS | single-cell Raman spectroscopy |
| DL | deep learning |
| ANNs | artificial neural networks |
| DNNs | deep neural networks |
| CNNs | convolutional neural networks |
References
- Pouris, J.; Kolyva, F.; Bratakou, S.; Vogiatzi, C.A.; Chaniotis, D.; Beloukas, A. The Role of Fungi in Food Production and Processing. Appl. Sci. 2024, 14, 5046. [Google Scholar] [CrossRef]
- Rai, A.K.; Jeyaram, K. Role of Yeasts in Food Fermentation. In Yeast Diversity in Human Welfare, 1st ed.; Satyanarayana, T., Kunze, G., Eds.; Springer: Singapore, 2017; pp. 83–113. [Google Scholar] [CrossRef]
- Tamang, J.P.; Lama, S. Probiotic properties of yeasts in traditional fermented foods and beverages. J. Appl. Microbiol. 2022, 132, 3533–3542. [Google Scholar] [CrossRef]
- Moslehi-Jenabian, S.; Pedersen, L.L.; Jespersen, L. Beneficial effects of probiotic and food borne yeasts on human health. Nutrients 2010, 2, 449–473. [Google Scholar] [CrossRef]
- Parapouli, M.; Vasileiadis, A.; Afendra, A.S.; Hatziloukas, E. Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 2020, 6, 1–31. [Google Scholar] [CrossRef]
- Maicas, S. The Role of Yeasts in Fermentation Processes. Microorganisms 2020, 8, 1142. [Google Scholar] [CrossRef]
- Comitini, F.; Canonico, L.; Agarbati, A.; Ciani, M. Biocontrol and Probiotic Function of Non-Saccharomyces Yeasts: New Insights in Agri-Food Industry. Microorganisms 2023, 11, 1450. [Google Scholar] [CrossRef]
- Wahid, E.; Ocheja, O.B.; Marsili, E.; Guaragnella, C.; Guaragnella, N. Biological and technical challenges for implementation of yeast-based biosensors. Microb. Biotechnol. 2023, 16, 54–66. [Google Scholar] [CrossRef]
- Kernbach, S.; Kernbach, O.; Kuksin, I.; Kernbach, A.; Nepomnyashchiy, Y.; Dochow, T.; Bobrov, A.V. The biosensor based on electrochemical dynamics of fermentation in yeast Saccharomyces cerevisiae. Environ. Res. 2022, 213, 113535. [Google Scholar] [CrossRef]
- Liu, L.; Xie, M.; Wei, D. Biological Detoxification of Mycotoxins: Current Status and Future Advances. Int. J. Mol. Sci. 2022, 23, 1064. [Google Scholar] [CrossRef]
- Low, L.; Shaw, C.; Gerrard, A. The effect of Saccharomyces cerevisiae on the stability of the herbicide glyphosate during bread leavening. Lett. Appl. Microbiol. 2004, 40, 133–137. [Google Scholar] [CrossRef]
- Fan, C.; Zhang, D.; Mo, Q.; Yuan, J. High-performance Saccharomyces cerevisiae-based biosensor for heavy metal detection. bioRxiv 2021. [Google Scholar] [CrossRef]
- Gayda, G.; Stasyuk, N.; Smutok, O.; Gonchar, M.; Sibirny, A.A. Yeast-Based Biosensors for Clinical Diagnostics, Food Control, and Environmental Safety. In Biotechnology of Yeasts and Filamentous Fungi. Grand Challenges in Biology and Biotechnology, 2nd ed.; Sibirny, A.A., Ed.; Springer: Cham, Switzerland, 2025; Chapter 14; pp. 405–435. [Google Scholar] [CrossRef]
- Pop, O.L.; Nagy, C.C.; Gabianelli, R.; Coldea, T.E.; Pop, C.R.; Mudura, E.; Min, T.; Rangnoi, K.; Yamabhai, M.; Vlaic, R.; et al. Deciphering contaminants and toxins in fermented food for enhanced human health safeguarding. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13428. [Google Scholar] [CrossRef]
- Martin-Yken, H. Yeast-Based Biosensors: Current Applications and New Developments. Biosensors 2020, 10, 51. [Google Scholar] [CrossRef]
- Tullio, V. Probiotic Yeasts: A Developing Reality? J. Fungi 2024, 10, 489. [Google Scholar] [CrossRef]
- Kunyeit, L.; Rao, R.P.; Anu-Appaiah, K.A. Yeasts originating from fermented foods, their potential as probiotics and therapeutic implication for human health and disease. Crit. Rev. Food Sci. Nutr. 2024, 64, 6660–6671. [Google Scholar] [CrossRef]
- Ramos, C.L.; Magalhães-Guedes, K.T. Detection and Quantification of Yeast Species in Food Samples for Quality Control. In Detection and Enumeration of Bacteria, Yeast, Viruses, and Protozoan in Foods and Freshwater. Methods and Protocols in Food Science, 1st ed.; Magnani, M., Ed.; Humana: New York, NY, USA, 2021; Chapter 12; pp. 125–141. [Google Scholar] [CrossRef]
- Riesute, R.; Salomskiene, J.; Moreno, D.S.; Gustiene, S. Effect of yeasts on food quality and safety and possibilities of their inhibition. Trends Food Sci. Technol. 2021, 108, 1–10. [Google Scholar] [CrossRef]
- Fleet, G.H. Yeast Spoilage of Foods and Beverages. In The Yeasts, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 53–63. [Google Scholar] [CrossRef]
- Hernández, A.; Pérez-Nevado, F.; Ruiz-Moyano, S.; Serradilla, M.J.; Villalobos, M.C.; Martín, A.; Córdoba, M.G. Spoilage yeasts: What are the sources of contamination of foods and beverages? Int. J. Food Microbiol. 2018, 286, 98–110. [Google Scholar] [CrossRef]
- Bullerman, L.B. Spoilage. Fungi in Food—An Overview. In Encyclopedia of Food Sciences and Nutrition, 2nd ed.; Caballero, B., Ed.; Academic Press: Cambridge, MA, USA, 2003; pp. 5511–5522. [Google Scholar] [CrossRef]
- Salmanian, A.-H.; Siavoshi, F.; Beyrami, Z.; Latifi-Navid, S.; Tavakolian, A.; Sadjadi, A. Foodborne yeasts as reservoirs of Helicobacter pylori. J. Food Saf. 2012, 32, 152–160. [Google Scholar] [CrossRef]
- Carlino, N.; Blanco-Miguez, A.; Puncochar, M.; Cotter, P.D.; Segata, N.; Pasolli, E. Unexplored microbial diversity from 2,500 food metagenomes and links with the human microbiome. Cell 2024, 187, 5775–5795. [Google Scholar] [CrossRef]
- Boekhout, T.; Amend, A.S.; El Baidouri, F.; Gabaldon, T.; Geml, J.; Mittelbach, M.; Robert, V.; Tan, C.S.; Turchetti, B.; Vu, D.; et al. Trends in yeast diversity discovery. Fungal Divers. 2022, 114, 491–537. [Google Scholar] [CrossRef]
- Cadež, N.; Dlauchy, D.; Tome, M.; Péter, G. Novakomyces olei sp. nov., the first member of a novel Taphrinomycotina lineage. Microorganisms 2021, 9, 301. [Google Scholar] [CrossRef]
- Cadež, N.; Drumonde-Neves, J.; Sipiczki, M.; Dlauchy, D.; Lima, T.; Lachance, M.A.; Péter, G. Starmerella vitis f.a., sp. nov., a yeast species isolated from flowers and grapes. Antonie Van Leeuwenhoek 2020, 113, 1289–1298. [Google Scholar] [CrossRef]
- Nagy, E.; Dlauchy, D.; Medeiros, A.O.; Péter, G.; Rosa, C.A. Yarrowia porcina sp. nov. and Yarrowia bubula f.a. sp. nov., two yeast species from meat and river sediment. Antonie Van Leeuwenhoek 2014, 105, 697–707. [Google Scholar] [CrossRef]
- Péter, G.; Mounier, J.; Garnier, L.; Soós, D.; Dlauchy, D. Cutaneotrichosporon suis sp. nov., a lipolytic yeast species from food and food-related environment. Int. J. Syst. Evol. Microbiol. 2019, 69, 2367–2371. [Google Scholar] [CrossRef]
- Segal-Kischinevzky, C.; Romero-Aguilar, L.; Alcaraz, L.D.; López-Ortiz, G.; Martínez-Castillo, B.; Torres-Ramírez, N.; Sandoval, G.; González, J. Yeasts inhabiting extreme environments and their biotechnological applications. Microorganisms 2022, 10, 794. [Google Scholar] [CrossRef]
- Deák, T. Environmental factors influencing yeasts. In Biodiversity and Ecophysiology of Yeasts, 1st ed.; Rosa, C.A., Péter, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 1, pp. 155–174. [Google Scholar]
- Fleet, G.H. Microorganisms in food ecosystems. Int. J. Food Microbiol. 1999, 50, 101–117. [Google Scholar] [CrossRef]
- Galimberti, A.; Bruno, A.; Mezzasalma, V.; de Mattia, F.; Bruni, I.; Labra, M. Emerging DNA-based technologies to characterize food ecosystems. Food Res. Int. 2015, 69, 424–433. [Google Scholar] [CrossRef]
- Boekhout, T.; Robert, V. Yeasts in Food: Beneficial and Detrimental Aspects, 1st ed.; B.Behr’s Verlag GmbH: Hamburg, Germany, 2003; pp. 1–488. [Google Scholar]
- Querol, A.; Fleet, G.H. Yeasts in Food and Beverages, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 1–449. [Google Scholar]
- Deák, T. Handbook of Food Spoilage Yeasts, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2007; pp. 1–325. [Google Scholar] [CrossRef]
- Krisch, J.; Chandrasekaran, M.; Kadaikunnan, S.; Alharbi, N.S.; Vágvölgyi, C. Latest about spoilage by yeasts: Focus on the deterioration of beverages and other plant-derived products. J. Food Prot. 2016, 79, 825–829. [Google Scholar] [CrossRef]
- Fröhlich-Wyder, M.-T.; Arias-Roth, E.; Jakob, E. Cheese yeasts. Yeast 2019, 36, 129–141. [Google Scholar] [CrossRef]
- Zilelidou, E.A.; Nisiotou, A. Understanding wine through yeast interactions. Microorganisms 2021, 9, 1620. [Google Scholar] [CrossRef]
- Hittinger, C.T.; Steele, J.L.; Ryder, D.S. Diverse yeasts for diverse fermented beverages and foods. Curr. Opin. Biotechnol. 2018, 49, 199–206. [Google Scholar] [CrossRef]
- Hansen, A.; Schieberle, P. Generation of aroma compounds during sourdough fermentation: Applied and fundamental aspects. Trends Food Sci. Techol. 2005, 16, 85–94. [Google Scholar] [CrossRef]
- Tamang, J.P.; Cotter, P.D.; Endo, A.; Han, N.S.; Kort, R.; Liu, S.Q.; Mayo, B.; Westerik, N.; Hutkins, R. Fermented foods in a global age: East meets West. Compr. Rev. Food Sci. Food Saf. 2020, 19, 184–217. [Google Scholar] [CrossRef]
- Tamang, J.P.; Watanabe, K.; Holzapfel, W.H. Review: Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 2016, 7, 377. [Google Scholar] [CrossRef]
- Schmitt, M.J.; Breinig, F. The viral killer system in yeast: From molecular biology to application. FEMS Microbiol. Rev. 2002, 26, 257–276. [Google Scholar] [CrossRef]
- Golubev, W.I. Antagonistic interactions among yeasts. In Biodiversity and Ecophysiology of Yeasts, 1st ed.; Rosa, C., Péter, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 1, pp. 197–220. [Google Scholar]
- Billerbeck, S.; Walker, R.S.K.; Pretorius, I.S. Killer yeasts: Expanding frontiers in the age of synthetic biology. Trends Biotechnol. 2024, 42, 1081–1096. [Google Scholar] [CrossRef]
- Loureiro, V. Spoilage yeasts in foods and beverages: Characterisation and ecology for improved diagnosis and control. Food Res. Int. 2000, 33, 247–256. [Google Scholar] [CrossRef]
- Péter, G. Biodiversity of Zygosaccharomyces species in food systems. Acta Aliment. 2022, 51, 43–51. [Google Scholar] [CrossRef]
- de Melo Pereira, G.V.; Maske, B.L.; de Carvalho Neto, D.P.; Karp, S.G.; De Dea Lindner, J.; Martin, J.G.P.; de Oliveira Hosken, B.; Soccol, C.R. What Is Candida Doing in My Food? A Review and Safety Alert on Its Use as Starter Cultures in Fermented Foods. Microorganisms 2022, 10, 1855. [Google Scholar] [CrossRef]
- Horváth, B.O.; Sárdy, D.N.; Kellner, N.; Magyar, I. Effects of high sugar content on fermentation dynamics and some metabolites of wine-related yeast species Saccharomyces cerevisiae, S. uvarum and Starmerella bacillaris. Food Technol. Biotechnol. 2020, 58, 76–83. [Google Scholar] [CrossRef]
- Moreira, L.P.D.; Nadai, C.; da Silva Duarte, V.; Brearley-Smith, E.J.; Marangon, M.; Vincenzi, S.; Giacomini, A.; Corich, V. Starmerella bacillaris strains used in sequential alcoholic fermentation with Saccharomyces cerevisiae improves protein stability in white wines. Fermentation 2022, 8, 252. [Google Scholar] [CrossRef]
- Masneuf-Pomarede, I.; Bely, M.; Marullo, P.; Albertin, W. The genetics of non-conventional wine yeasts: Current knowledge and future challenges. Front. Microbiol. 2016, 11, 1563. [Google Scholar] [CrossRef]
- Bressani, A.P.P.; Martinez, S.J.; Evangelista, S.R.; Dias, D.R.; Schwan, R.F. Characteristics of fermented coffee inoculated with yeast starter cultures using different inoculation methods. LWT 2018, 92, 212–219. [Google Scholar] [CrossRef]
- Gutiérrez-Ríos, H.G.; Suárez-Quiroz, M.L.; Hernández-Estrada, Z.J.; Castellanos-Onorio, O.P.; Alonso-Villegas, R.; Rayas-Duarte, P.; Cano-Sarmiento, C.; Figueroa-Hernández, C.Y.; González-Rios, O. Yeasts as producers of flavor precursors during cocoa bean fermentation and their relevance as starter cultures: A review. Fermentation 2022, 8, 331. [Google Scholar] [CrossRef]
- de Boer, E.; Beumer, R.R. Methodology for detection and typing of foodborne microorganisms. Int. J. Food Microbiol. 1999, 50, 119–130. [Google Scholar] [CrossRef]
- Deák, T. Detection, enumeration and isolation of yeasts. In Yeasts in Food, 1st ed.; Boekhout, T., Roberts, V., Eds.; Woodhead Publishing Limited: Sawston, UK; Abington Hall: Abington Cambridge, UK, 2003; pp. 39–69. [Google Scholar]
- Beuchat, L.R. Selective media for detecting and enumerating foodborne yeasts. Int. J. Food Microbiol. 1993, 19, 1–14. [Google Scholar] [CrossRef]
- Tubia, I.; Prasad, K.; Pérez-Lorenzo, E.; Abadín, C.; Zumárraga, M.; Oyanguren, I.; Barbero, F.; Paredes, J.; Arana, S. Beverage spoilage yeast detection methods and control technologies: A review of Brettanomyces. Int. J. Food Microbiol. 2018, 283, 65–76. [Google Scholar] [CrossRef]
- Corbett, K.M.; de Smidt, O. Culture-dependent diversity profiling of spoilage yeasts species by PCR-RFLP comparative analysis. Food Sci. Technol. Int. 2019, 25, 671–679. [Google Scholar] [CrossRef]
- Aladhadh, M. A review of modern methods for the detection of foodborne pathogens. Microorganisms 2023, 11, 1111. [Google Scholar] [CrossRef]
- ISO 21527-1:2008; Microbiology of food and animal feeding stuffs — Horizontal method for the enumeration of yeasts and moulds. Part 1: Colony count technique in products with water activity greater than 0,95. International Organization for Standardisation: Geneva, Switzerland, 2008. Available online: https://www.iso.org/standard/38275.html (accessed on 15 March 2025).
- ISO 21527-2:2008; Microbiology of food and animal feeding stuffs — Horizontal method for the enumeration of yeasts and moulds. Part 2: Colony count technique in products with water activity less than or equal to 0,95. International Organization for Standardisation: Geneva, Switzerland, 2008. Available online: https://www.iso.org/standard/38276.html (accessed on 15 March 2025).
- ISO 6611:2004; Milk and Milk Products—Enumeration of Colony-Forming Units of Yeasts and/or Moulds—Colony-Count Technique at 25 Degrees C. International Organization for Standardisation: Geneva, Switzerland, 2004. Available online: https://www.iso.org/standard/40473.html (accessed on 15 March 2025).
- ISO/CD 21527; Microbiology of the Food Chain—Horizontal Method for the Enumeration of Yeasts and Moulds. International Organization for Standardisation: Geneva, Switzerland, Under development. Available online: https://www.iso.org/standard/79102.html (accessed on 15 March 2025).
- ISO 7954:1987; Microbiology—General Guidance for Enumeration of Yeasts and Moulds—Colony Count Technique at 25 Degrees C. International Organization for Standardization: Geneva, Switzerland, 1987.
- ISO 7698:1990; Cereals, Pulses and Derived Products—Enumeration of Bacteria, Yeasts and Moulds. International Organization for Standardization: Geneva, Switzerland, 1990.
- ISO 13681:1995; Meat and Meat Products—Enumeration of Yeasts and Moulds—Colony-Count Technique. International Organization for Standardization: Geneva, Switzerland, 1995.
- ISO 6611:1992; Milk and Milk Products—Enumeration of Colony-Forming Units of Yeasts and/or Moulds—Colony-Count Technique at 25 Degrees C. International Organization for Standardization: Geneva, Switzerland, 1992.
- Beuchat, L.R. Media for detecting and enumerating yeasts and moulds. Int. J. Food Microbiol. 1992, 17, 145–158. [Google Scholar] [CrossRef]
- Tournas, V.; Stack, M.E.; Mislivec, P.B.; Koch, H.A.; Bandler, R. Yeast, Moulds and Mycotoxins. In Bacteriological Analytical Manual (BAM); Tournas, V., Stack, M.E., Mislivec, P.B., Koch, H.A., Bandler, R., Eds.; United States Food and Drug Administration: Silver Spring, MD, USA, 2001. [Google Scholar]
- Jermini, M.F.G.; Geiges, O.; Schmidt-Lorenz, W. Detection, Isolation and Identification of Osmotolerant Yeasts from High-Sugar Products. J. Food Prot. 1987, 50, 468–472. [Google Scholar] [CrossRef]
- Rojo, M.C.; Torres Palazzolo, C.; Cuello, R.; González, M.; Guevara, F.; Ponsone, M.L.; Mercado, L.A.; Martínez, C.; Combina, M. Incidence of osmophilic yeasts and Zygosaccharomyces rouxii during the production of concentrate grape juices. Food Microbiol. 2017, 64, 7–14. [Google Scholar] [CrossRef]
- Péter, G.; Dlauchy, D.; Tóbiás, A.; Fülöp, L.; Podgoršek, M.; Čadež, N. Brettanomyces acidodurans sp. nov., a new acetic acid producing yeast species from olive oil. Antonie Van Leeuwenhoek 2017, 110, 657–664. [Google Scholar] [CrossRef]
- King, A.D.; Pitt, J.I.; Beuchat, L.R. Methods for the mycological examination of food. In NATO ASI Series A, 122; Plenum: New York, NY, USA, 1986; ISBN 0306424797. [Google Scholar]
- Samson, R.A.; Hocking, A.D.; Pitt, J.I.; King, A.D. Modern Methods in Food Mycology; Elsevier: Amsterdam, The Netherlands, 1992. [Google Scholar]
- Parfitt, E.H. The influence of media uponthe yeast and mold count of butter. J. Dairy Sci. 1933, 16, 141–147. [Google Scholar] [CrossRef]
- American Public Health Association. Standard Methods for the Examination of Dairy Products, 12th ed.; American Public Health Association: New York, NY, USA, 1967. [Google Scholar]
- Murdock, D.I.; Folinazzo, L.F.; Troy, V.S. Evaluation of plating media for citrus concentrates. Food Technol. 1952, 6, 181–185. [Google Scholar]
- King, A.D., Jr.; Hocking, A.D.; Pitt, J.I. Dichloran-rose bengal medium for enumeration and isolation of molds from foods. Appl. Environ. Microbiol. 1979, 37, 959–964. [Google Scholar] [CrossRef]
- Jarvis, B. Comparison of an improved rose-bengal-chlortetracycline agar with other media for the selective isolation and enumeration of moulds and yeasts in food. J. Appl. Bacteriol. 1973, 36, 723–727. [Google Scholar] [CrossRef]
- Mossel, D.A.; Kleynen-Semmeling, A.M.; Vincentie, H.M.; Beerens, H.; Catsaras, M. Oxytetracycline-glucose-yeast extract agar for selective enumeration of moulds and yeasts in foods and clinical material. J. Appl. Bacteriol. 1970, 33, 454–457. [Google Scholar] [CrossRef]
- Deák, T. Media for enumerating spoilage yeasts—A collaborative study. In Modern Methods in Food Mycology; Samson, R.A., Hocking, A.D., Pitt, J.I., King, A.D., Eds.; Elsevier: Amsterdam, The Netherlands, 1992; pp. 31–38. [Google Scholar]
- Methods of Analysis of the American Society of Brewing Chemists; 7th revised ed.; American Society of Brewing Chemists: St. Paul, MN, USA, 1976.
- Czarnecki, H.T.; Van Engel, E. The isolation and identification of diacetyl producing brewers yeast. Brew. Dig. 1959, 34, 52. [Google Scholar]
- Walters, L.S.; Thiselton, M.R. Utilization of lysine by yeasts. J. Inst. Brew. 1953, 59, 401–404. [Google Scholar] [CrossRef]
- Brenner, M.W.; Karpiscak, M.; Stern, H.; Hsu, W.P. A differential medium for detection of wild yeasts in the brewery. Am. Soc. Brew. Chem. Proc. 1970, 28, 79–88. [Google Scholar] [CrossRef]
- Kish, S.; Shaft, R.; Margalith, P. A note on a selective medium for wine yeasts. J. Appl. Bacteriol. 1983, 55, 177–179. [Google Scholar] [CrossRef]
- Heard, G.M.; Fleet, G.H. Evaluation of selective media for enumeration of yeasts during wine fermentation. J. Appl. Bacteriol. 1986, 60, 477–481. [Google Scholar] [CrossRef]
- Rodrigues, N.; Goncalves, G.; Pereira-da-Silva, S.; Malfeito-Ferreira, M.; Loureiro, V. Development and use of a new medium to detect yeast of the genera Dekkera/Brettanomyces sp. J. Appl. Microbiol. 2001, 90, 588–599. [Google Scholar] [CrossRef]
- Chatonnet, P.; Dubordieu, D.; Boidron, J.; Pons, M. The origin of ethylphenols in wines. J. Sci. Food. Agric. 1992, 60, 165–178. [Google Scholar] [CrossRef]
- Green, S.R.; Gray, P.P. A differential procedure applicable to bacteriological investigation in brewing. Wallerstein Lab. Commun. 1950, 13, 357–366. [Google Scholar] [CrossRef]
- Vasdinyei, R.; Simonics, T.; Mészáros, L.; Deák, T. Comparison of different media for isolation and enumeration of yeasts occurring in blue-veined cheese. Acta Aliment. 2003, 32, 205–212. [Google Scholar] [CrossRef]
- Mu, Z.; Yang, X.; Yuan, H. Detection and identification of wild yeast in Koumiss. Food Microbiol. 2012, 31, 301–308. [Google Scholar] [CrossRef]
- Greppi, A.; Rantsiou, K.; Padonou, W.; Hounhouigan, J.; Jespersen, L.; Jakobsen, M.; Cocolin, L. Determination of yeast diversity in ogi, mawè, gowé and tchoukoutou by using culture-dependent and -independent methods. Int. J. Food Microbiol. 2013, 165, 84–88. [Google Scholar] [CrossRef]
- Spangenberg, D.S.; Ingham, S.C. Comparison of methods for enumeration of yeasts and molds in shredded low-moisture, part-skim mozzarella cheese. J. Food Prot. 2000, 63, 529–533. [Google Scholar] [CrossRef]
- Gonçalves, M.T.P.; Benito, M.J.; Córdoba, M.G.; Egas, C.; Merchán, A.V.; Galván, A.I.; Ruiz-Moyano, S. Bacterial Communities in Serpa Cheese by Culture Dependent Techniques, 16S rRNA Gene Sequencing and High-throughput Sequencing Analysis. J. Food Sci. 2018, 83, 1333–1341. [Google Scholar] [CrossRef]
- Rale, B.V.; Vakil, J.R. A note on an improved molybdate agar for the selective isolation of yeasts from tropical fruits. J. Appl. Bacteriol. 1984, 56, 409–413. [Google Scholar] [CrossRef]
- Pitt, J.L.; Hocking, A.D. Fungi and Food Spoilage; Academic Press: Sydney, Australia, 1985. [Google Scholar]
- Hocking, A.D.; Pill, J.L. Dichloran glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Appl. Environ. Microbiol. 1980, 39, S8–S492. [Google Scholar] [CrossRef]
- Baird, R.M.; Corry, J.E.L.; Curtis, G.D.W. Pharmacopoeia of culture media for food microbiology. Int. J. Food Microbiol. 1987, 5, 187–300. [Google Scholar]
- Schuller, D.; Côrte-Real, M.; Leão, C.A. differential medium for the enumeration of the spoilage yeast Zygosaccharomyces bailii in wine. J. Food Prot. 2000, 63, 1570–1575. [Google Scholar] [CrossRef]
- Makdesi, A.K.; Beuchat, L.R. Performance of Selective Media for Enumerating Zygosaccharomyces bailii in Acidic Foods and Beverages. J. Food Prot. 1996, 59, 652–656. [Google Scholar] [CrossRef]
- Brysch-Herzberg, M.; Tobias, A.; Seidel, M.; Wittmann, R.; Wohlmann, E.; Fischer, R.; Dlauchy, D.; Peter, G. Schizosaccharomyces osmophilus sp. nov., an osmophilic fission yeast occurring in bee bread of different solitary bee species. FEMS Yeast Res. 2019, 19, foz038. [Google Scholar] [CrossRef]
- Čadež, N.; Fülöp, L.; Dlauchy, D.; Péter, G. Zygosaccharomyces favi sp. nov., an obligate osmophilic yeast species from bee bread and honey. Antonie Van Leeuwenhoek 2015, 107, 645–654. [Google Scholar] [CrossRef]
- Bolton, F.J.; Gibson, D.M. Automated electrical techniques in microbiological analysis. In Rapid Analysis Techniques in Food Microbiology; Patel, P., Ed.; Blackie Academic & Professional: London, UK; New York, NY, USA, 1994; pp. 131–169. [Google Scholar]
- Schaertel, B.J.; Tsang, I.V.; Firstenberg-Eden, R. Impedimetric detection of yeast and mould. Food Microbiol. 1987, 4, 155–163. [Google Scholar] [CrossRef]
- Firstenberg-Eden, R.; Eden, G. Impedance Microbiology; Research Studies Press: Letchworth, NY, USA, 1984. [Google Scholar]
- Connoly, P.J.; Lewis, S.J.; Corry, J.E.L. A medium for the detection of yeasts using a conductimetric method. Int. J. Food Microbiol. 1988, 7, 31–40. [Google Scholar] [CrossRef]
- Owens, J.D.; Thomas, D.J.; Thompson, R.S.; Timmerman, W. Indirect conductimetry: A novel approach to the conductimetric enumeration of microbial populations. Lett. Appl. Microbiol. 1989, 9, 245–249. [Google Scholar] [CrossRef]
- Bolton, F.J. An investigation of indirect conductimetry for detection of some food-borne bacteria. J. Appl. Bacteriol. 1990, 69, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Deak, T.; Beuchat, L.R. Evaluation of the indirect conductance method for the detection of yeasts in laboratory media and apple juice. Food Micobiol. 1993, 10, 255–262. [Google Scholar] [CrossRef]
- van Wyk, S.; Silva, F.V.M. Enumeration of Brettanomyces in wine using impedance. Appl. Microbiol. 2021, 1, 352–360. [Google Scholar] [CrossRef]
- McElroy, W.D. The energy source for bioluminescence in isolated systems. Proc. Natl. Acad. Sci. USA 1947, 33, 342–345. [Google Scholar] [CrossRef]
- Chollet, R.; Kukuczka, M.; Halter, N.; Romieux, M.; Marc, F.; Meder, H.; Beguin, V.; Ribault, S. Rapid Detection and Enumeration of Contaminants by ATP Bioluminescence Using the Milliflex® Rapid Microbiology Detection and Enumeration System. J. Rapid Met. Aut. Mic. 2008, 16, 256–272. [Google Scholar] [CrossRef]
- Narsaiah, K.; Jha, S.N.; Jaiswal, P.; Singh, A.K.; Gupta, M.; Bhardwaj, R. Estimation of total bacteria on mango surface by using ATP bioluminescence. Sci. Hortic. 2012, 146, 159–163. [Google Scholar] [CrossRef]
- Wang, H.Y.; Bhunia, A.K.; Lu, C. A microfluidic flow-through device for high throughput electrical lysis of bacterial cells based on continuous dc voltage. Biosens. Bioelectron. 2006, 22, 582–588. [Google Scholar] [CrossRef]
- Hysert, D.W.; Kovecses, F.; Morrison, N.M. A firefly bioluminescence ATP assay method for rapid detection and enumeration of brewery microorganisms. J. Am. Soc. Brew. Chem. 1976, 34, 145–150. [Google Scholar] [CrossRef]
- Ugarova, N.N.; Brovko, L.Y.; Trdatian, I.Y.; Rainina, E.I. Bioluminescent methods of analysis in microbiology. Appl. Biochem. Microbiol. 1987, 23, 11–20. [Google Scholar]
- Hattori, N.; Sakakibara, T.; Kajiyama, N.; Igarashi, T.; Maeda, M.; Murakami, S. Enhanced microbial biomass assay using mutant luciferase resistant to benzalkonium chloride. Anal. Biochem. 2003, 319, 287–295. [Google Scholar] [CrossRef] [PubMed]
- McElroy, W.D.; Green, A. Function of Adenosine Triphosphate in the Activation of Luciferin. Arch. Biochem. Biophys. 1956, 64, 257–271. [Google Scholar] [CrossRef]
- Chappelle, E.W.; Levin, G.V. Use of the firefly bioluminescent reaction for rapid detection and counting of bacteria. Biochem. Med. 1968, 2, 41–52. [Google Scholar] [CrossRef]
- Ugarova, N.N. Bioanalytical applications of firefly luciferase. Appl. Biochem. Microbiol. 1993, 29, 135–144. [Google Scholar]
- Bottari, B.; Santarelli, M.; Neviani, E. Determination of Microbial Load for Different Beverages and Foodstuff by Assessment of Intracellular ATP. Trends Food Sci. Technol. 2015, 44, 36–48. [Google Scholar] [CrossRef]
- Lundin, A. Use of firefly luciferase in ATP_related assays of biomass, enzymes, and metabolites. Method. Enzymol. 2000, 305, 346–370. [Google Scholar] [CrossRef]
- Lomakina, G.Y.; Modestova, Y.A.; Ugarova, N.N. Bioluminescence assay for cell viability. Biochemistry 2015, 80, 701–713. [Google Scholar] [CrossRef]
- Siro, M.-R.; Romar, H.; Lovgren, T. Continuous flow method for extraction and bioluminescence assay of ATP in baker’s yeast. Eur. J. Appl. Microbiol. Biotechnol. 1982, 15, 258–264. [Google Scholar] [CrossRef]
- Phillips, J.D.; Griffith, M.W. Bioluminescence and impedimetric methods for assessing shelf-life of pasteurized milk and cream. Food Microbiol. 1985, 2, 39–51. [Google Scholar] [CrossRef]
- Littel, K.J.; Rocco, K.A.L. ATP Screening Method for Presumptive Detection of Microbiologically Contaminated Carbonated Beverages. J. Food Sci. 1986, 51, 474–476. [Google Scholar] [CrossRef]
- Ogden, K. Practical experiences of hygiene control using ATP-bioluminescence. J. Inst. Brew. 1993, 99, 389–393. [Google Scholar] [CrossRef]
- Murphy, S.C.; Kozlowski, S.M.; Bandler, D.K.; Boor, K.J. Evaluation of adenosine triphosphate-bioluminescence hygiene monitoring for troubleshooting fluid milk shelf-life problems. J. Dairy Sci. 1998, 81, 817–820. [Google Scholar] [CrossRef]
- Lehto, M.; Kuisma, R.; Määttä, J.; Kymäläinen, H.R.; Mäki, M. Hygienic level and surface contamination in fresh-cut vegetable production plants. Food Control. 2011, 22, 469–475. [Google Scholar] [CrossRef]
- Efremenko, E.; Senko, O.; Stepanov, N.; Maslova, O.; Lomakina, G.Y.; Ugarova, N. Luminescent analysis of ATP: Modern objects and processes for sensing. Chemosensors 2022, 10, 493. [Google Scholar] [CrossRef]
- Hysert, D.W.; Morrison, N.M. Studies on ATP, ADP, and AMP concentrations in yeast and beer. J. Am. Soc. Brew. Chem. 1977, 35, 160–167. [Google Scholar] [CrossRef]
- Miller, R.; Galston, G. Rapid methods for the detection of yeast and Lactobacillus by ATP bioluminescence. J. Inst. Brew. 1989, 95, 317–319. [Google Scholar] [CrossRef]
- Miller, L.F.; Mabee, M.S.; Gress, H.S.; Jangaard, N.O. An ATP bioluminescence method for the quantification of viable yeast for fermenter pitching. J. Am. Soc. Brew. Chem. 1978, 36, 59–62. [Google Scholar] [CrossRef]
- Trudil, D.; Loomis, L.; Pabon, R.; Hasan, J.A.K.; Trudil, C.L. Rapid ATP method for the screening and identification of bacteria in food and water samples. Moscow Univ. Chem. Bull. 2000, 41, 27–29. [Google Scholar]
- Carrick, K.; Barney, M.; Navarro, A.; Ryder, D. The comparison of four bioluminometers and their swab kits for instant hygiene monitoring and detection of microorganisms in the brewery. J. Inst. Brew. 2001, 107, 31–37. [Google Scholar] [CrossRef]
- Mandl, K.; Mutz, C.; Silhavy-Richter, K. Use of ATP bioluminescence measuring devices for microbial screening filling lines, surfaces and barrels in cellars. CABI Databases 2018, 68, 141–147. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/20183245379 (accessed on 17 March 2025).
- Monica, S.; Bancalari, E.; Castellone, V.; Rijkx, J.; Wirth, S.; Jahns, A.; Bottari, B. ATP bioluminescence for rapid and selective detection of bacteria and yeasts in wine. Appl. Sci. 2021, 11, 4953. [Google Scholar] [CrossRef]
- Zhao, X.; Lin, C.W.; Wang, J.; Oh, D.H. Advances in rapid detection methods for foodborne Pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef]
- Law, J.W.-F.; Ab, M.N.-S.; Chan, K.-G.; Lee, L.-H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 2015, 5, 770. [Google Scholar] [CrossRef]
- Nogueira, F.; Istel, F.; Pereira, L.; Tscherner, M.; Kuchler, K. Immunological identification of fungal species. In Human Fungal Pathogen Identification: Methods and Protocols, 1st ed.; Lion, T., Ed.; Humana Press: New York, NY, USA, 2017; Volume 1508, pp. 1064–3745. [Google Scholar] [CrossRef]
- Shibata, N.; Kobayashi, H.; Suzuki, S. Immunochemistry of pathogenic yeast, Candida species, focusing on mannan. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2012, 88, 250–265. [Google Scholar] [CrossRef]
- Cherniak, R.; Sundstrom, J.B. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect. Immun. 1994, 62, 1507–1512. [Google Scholar] [CrossRef]
- Nishikawa, A.; Sekine, T.; Ikeda, R.; Shinoda, T.; Fukazawa, Y. Reassessment of antigenic determinant of Saccharomyces cerevisiae serotype Ia. Microbiol. Immunol. 1990, 34, 825–840. [Google Scholar] [CrossRef]
- Colombo, A.L.; Padovan, A.C.; Chaves, G.M. Current knowledge of Trichosporon spp. and Trichosporonosis. Clin. Microbiol. Rev. 2011, 24, 682–700. [Google Scholar] [CrossRef]
- Huang, H.-R.; Fan, L.-C.; Rajbanshi, B.; Xu, J.-F. Evaluation of a new Cryptococcal antigen lateral flow immunoassay in serum, cerebrospinal fluid and urine for the diagnosis of Cryptococcosis: A meta-analysis and systematic review. PLoS ONE 2015, 10, e0127117. [Google Scholar] [CrossRef]
- Middelhoven, W.J.; Notermans, S. Immuno-assay techniques for detecting yeasts in foods. Int. J. Food Microbiol. 1993, 19, 53–62. [Google Scholar] [CrossRef]
- Richard, M.; Cowland, T.W. The rapid detection of brewery contaminants belonging to the genus Saccharomyces by a serological technique. J. Inst. Brew. 1967, 73, 552–558. [Google Scholar] [CrossRef]
- García, T.; Mayoral, B.; González, I.; López-Calleja, I.; Sanz, A.; Hernández, P.E.; Martín, R. Enumeration of yeasts in dairy products: A comparison of immunological and genetic techniques. J. Food Prot. 2004, 67, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Debevere, J.; Uyttendaele, M. Validating detection techniques. In Detecting Pathogens in Food, 1st ed.; McMeekin, T.A., Ed.; Woodhead Publishing Limited: Cambridge, UK, 2003; pp. 69–92. [Google Scholar]
- Bossaert, S.; Winne, V.; Van Opstaele, F.; Buyse, J.; Verreth, C.; Herrera-Malaver, B.; Van Geel, M.; Verstrepen, K.J.; Crauwels, S.; De Rouck, G.; et al. Description of the temporal dynamics in microbial community composition and beer chemistry in sour beer production via barrel ageing of finished beers. Int. J. Food Microbiol. 2021, 339, 109030. [Google Scholar] [CrossRef]
- Tyakht, A.; Kopeliovich, A.; Klimenko, N.; Efimova, D.; Dovidchenko, N.; Odintsova, V.; Kleimenov, M.; Toshchakov, S.; Popova, A.; Khomyakova, M.; et al. Characteristics of bacterial and yeast microbiomes in spontaneous and mixed-fermentation beer and cider. Food Microbiol. 2021, 94, 103658. [Google Scholar] [CrossRef]
- Grassi, A.; Cristani, C.; Palla, M.; Di Giorgi, R.; Giovannetti, M.; Agnolucci, M. Storage time and temperature affect microbial dynamics of yeasts and acetic acid bacteria in a kombucha beverage. Int. J. Food Microbiol. 2022, 382, 109934. [Google Scholar] [CrossRef]
- Dorn-In, S.; Führer, L.; Gareis, M.; Schwaiger, K. Cold-tolerant microorganisms causing spoilage of vacuum-packed beef under time-temperature abuse determined by culture and qPCR. Food Microbiol. 2023, 109, 104147. [Google Scholar] [CrossRef]
- Sanz, A.; Martín, R.; Mayoral, M.B.; Hernández, P.E.; González, I.; Lacarra, T.G. Development of a PCR-culture technique for rapid detection of yeast species in vacuum packed ham. Meat Sci. 2005, 71, 230–237. [Google Scholar] [CrossRef]
- Makino, H.; Fujimoto, J.; Watanabe, K. Development and evaluation of a real-time quantitative PCR assay for detection and enumeration of yeasts of public health interest in dairy products. Int. J. Food Microbiol. 2010, 140, 76–83. [Google Scholar] [CrossRef]
- Vaitilingom, M.; Gendre, F.; Brignon, P. Direct Detection of Viable Bacteria, Molds, and Yeasts by Reverse Transcriptase PCR in Contaminated Milk Samples after Heat Treatment. Appl. Environ. Microbiol. 1998, 64, 1157–1160. [Google Scholar] [CrossRef]
- Bleve, G.; Rizzotti, L.; Dellaglio, F.; Torriani, S. Development of Reverse Transcription (RT)-PCR and Real-Time RT-PCR Assays for Rapid Detection and Quantification of Viable Yeasts and Molds Contaminating Yogurts and Pasteurized Food Products. Appl. Environ. Microbiol. 2003, 69, 4116–4122. [Google Scholar] [CrossRef]
- Casey, G.D.; Dobson, A.D.W. Molecular detection of Candida krusei contamination in fruit juice using the citrate synthase gene cs1 and a potential role for this gene in the adaptive response to acetic acid. J. Appl. Microbiol. 2003, 95, 13–22. [Google Scholar] [CrossRef]
- Nadai, C.; Campanaro, S.; Giacomini, A.; Corich, V. Selection and validation of reference genes for quantitative real-time PCR studies during Saccharomyces cerevisiae alcoholic fermentation in the presence of sulfite. Int. J. Food Microbiol. 2015, 215, 49–56. [Google Scholar] [CrossRef]
- Wu, Q.; Cui, K.; Lin, J.; Zhu, J.; Xu, Y. Urea production by yeasts other than Saccharomyces in food fermentation. FEMS Yeast Res. 2017, 17, fox072. [Google Scholar] [CrossRef]
- Chikano, M.; Takahashi, J.I. Complete mitochondrial DNA sequence of the yeast Zygosaccharomyces siamensis (Saccharomycetes: Saccharomycetales) from fermented honey of the Apis cerana japonica in Japan. Mitochondrial DNA B 2020, 5, 2645–2647. [Google Scholar] [CrossRef]
- Benito-Vazquez, I.; Belda, I.; Ruiz, J.; Vicente, J.; Navascués, E.; Marquina, D.; Santos, A. Direct detection of Brettanomyces bruxellensis in wine by PCR targeting the vinylphenol reductase gene. LWT 2021, 136, 110321. [Google Scholar] [CrossRef]
- Muyanlı, E.B.; Yılmaz, R. RT-qPCR based quantitative analysis of ARO and ADH genes in Saccharomyces cerevisiae and Metschnikowia pulcherrima strains growth white grape juice. Mol. Biol. Rep. 2024, 51, 547. [Google Scholar] [CrossRef]
- Kurtzman, C.P. Recognition of yeast species from gene sequence comparisons. Open Appl. Inf. J. 2011, 5 (Suppl. 1-M4), 20–29. [Google Scholar] [CrossRef]
- Wu, H.; Liu, S.; Li, M.; Zhao, L.; Zhu, Y.; Zhao, G.; Ma, Y.; Sun, L.; Liu, Y.; Liang, D. Isolation and purification of lactic acid bacteria and yeasts based on a multi-channel magnetic flow device and rapid qualitative and quantitative detection. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2025, 327, 125296. [Google Scholar] [CrossRef]
- Park, H.W.; Mason Earles, J.; Nitin, N. Deep learning enabled rapid classification of yeast species in food by imaging of yeast microcolonies. Food Res. Int. 2025, 201, 115604. [Google Scholar] [CrossRef]
- Stilman, W.; Yongabi, D.; Bakhshi Sichani, S.; Thesseling, F.; Deschaume, O.; Putzeys, T.; Pinto, T.C.; Verstrepen, K.; Bartic, K.; Wübbenhorst, M.; et al. Detection of yeast strains by combining surface-imprinted polymers with impedance-based readout. Sens. Actuators B Chem. 2021, 340, 129917. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, J.; Sun, P.; Ding, T.; Li, J.; Deng, Y. Formation and resuscitation of viable but non-culturable (VBNC) yeast in the food industry: A review. Int. J. Food Microbiol. 2025, 426, 110901. [Google Scholar] [CrossRef]
- Wang, Z.; Xiao, Y.; Li, X.; Li, Z.; Wang, Y.; He, W.; Wang, J.; Liu, L.; Ding, T.; Sun, P.; et al. POX1 regulates the formation of viable but non-culturable brewer’s yeast induced by iso-alpha acid from hops. Food Sci. Hum. Wellness 2024, 14. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, Z.; Sun, W.; Luan, Y.; Piao, M.; Deng, Y. Characterization and formation mechanisms of viable, but putatively non-culturable brewer’s yeast induced by isomerized hop extract. LWT 2022, 155, 112974. [Google Scholar] [CrossRef]
- Salma, M.; Rousseaux, S.; Sequeira-Le Grand, A.; Divol, B.; Alexandre, H. Characterization of the Viable but Nonculturable (VBNC) State in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e77600. [Google Scholar] [CrossRef]
- Ferrario, M.; Guerrero, S.; Alzamora, S.M. Study of pulsed light-induced damage on saccharomyces cerevisiae in apple juice by flow cytometry and transmission electron microscopy. Food Bioprocess. Technol. 2014, 7, 1001–1011. [Google Scholar] [CrossRef]
- Xie, M.; Xu, L.; Zhang, R.; Zhou, Y.; Xiao, Y.; Su, X.; Shen, C.; Sun, F.; Hashmi, M.Z.; Lin, H.; et al. Viable but Nonculturable State of Yeast Candida sp. Strain LN1 Induced by High Phenol Concentrations. Appl. Environ. Microbiol. 2021, 87, e01110-21. [Google Scholar] [CrossRef]
- He, Y.; Zhao, J.; Yin, H.; Deng, Y. Transcriptome Analysis of Viable but Non-Culturable Brettanomyces bruxellensis Induced by Hop Bitter Acids. Front. Microbiol. 2022, 13, 902110. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Boekhout, T.; Robert, V.; Fell, J.W.; Deak, T. Methods to identify yeasts. In Yeasts in Food. Beneficial and Detrimental Aspects; Boekhout, T., Robert, V., Eds.; Woodhead Publishing Limited: Cambridge, UK, 2003; pp. 69–122. [Google Scholar]
- Kümmerle, M.; Scherer, S.; Seiler, H. Rapid and reliable identification of food-borne yeasts by Fourier-transform infrared spectroscopy. Appl. Environ. Microbiol. 1998, 64, 2207–2214. [Google Scholar] [CrossRef]
- Botha, A.; Kock, J.L. Application of fatty acid profiles in the identification of yeasts. Int. J. Food Microbiol. 1993, 19, 39–51. [Google Scholar] [CrossRef]
- Aloklah, B.; Alhajali, A.; Yaziji, S. Identification of some yeasts by fatty acid profiles. Pol. J. Microbiol. 2014, 63, 467–472. [Google Scholar] [CrossRef]
- NerurKar, V.; KhaN, S.; KattuNgal, S.; Bhatia, S. Identifying Candida and other yeast-like fungi: Utility of an identification algorithm in resource limited setting. J. Clin. Diagn. Res. 2014, 8, DC01. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T.; Robert, V. Methods for isolation, phenotypic characterization and maintenance of yeast. In The Yeasts, a Taxanomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier Publishers: Amsterdam, The Netherlands, 2011; Volume 1, pp. 87–110. [Google Scholar]
- Sandven, P. Laboratory identification and sensitivity testing of yeast isolates. Acta Odontol. Scand. 1990, 48, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.M.D.C.; Salgado, A.M.; Coelho, M.A.Z. Urease activity. In Methods to Determine Enzymatic Activity; Vermelho, A.B., Couri, S., Eds.; Bentham Science Publishers: Honolulu, HI, USA, 2013; pp. 292–319. [Google Scholar]
- Kali, A.; Srirangaraj, S.; Charles, P.M.V. A cost-effective carbohydrate fermentation test for yeast using microtitre plate. Indian J. Med. Microbiol. 2015, 33, 293–295. [Google Scholar] [CrossRef] [PubMed]
- Fowler, Z.L.; Baron, C.M.; Panepinto, J.C.; Koffas, M.A. Melanization of flavonoids by fungal and bacterial laccases. Yeast 2011, 28, 181–188. [Google Scholar] [CrossRef]
- Bryer, P. A twist on measuring catalase. Sci. Teach. 2016, 83, 69–73. [Google Scholar]
- Zhang, P.; Zhang, R.; Sirisena, S.; Gan, R.; Fang, Z. Beta-glucosidase activity of wine yeasts and its impacts on wine volatiles and phenolics: A mini-review. Food Microbiol. 2021, 100, 103859. [Google Scholar] [CrossRef]
- Vecchione, A.; Florio, W.; Celandroni, F.; Barnini, S.; Lupetti, A.; Ghelardi, E. Comparative evaluation of six chromogenic media for presumptive yeast identification. J. Clin. Pathol. 2017, 70, 1074–1078. [Google Scholar] [CrossRef]
- Scharmann, U.; Kirchhoff, L.; Chapot, V.L.S.; Dziobaka, J.; Verhasselt, H.L.; Stauf, R.; Buer, J.; Steinmann, J.; Rath, P.M. Comparison of four commercially available chromogenic media to identify Candida albicans and other medically relevant Candida species. Mycoses 2020, 63, 823–831. [Google Scholar] [CrossRef]
- Ruiz-Gaitán, A.; Sigona-Giangreco, I.; Pérez-Royo, J.M.; Garcia-Bustos, V.; García-Hita, M.; Valentín-Gómez, E.; Almaraz, S.G.; de Groot, P.W.J.; Pemán, J. Usefulness of chromogenic media with fluconazole supplementation for presumptive identification of Candida auris. Diagnostics 2023, 13, 231. [Google Scholar] [CrossRef]
- McTaggart, L.; Richardson, S.E.; Seah, C.; Hoang, L.; Fothergill, A.; Zhang, S.X. Rapid identification of Cryptococcus neoformans var. grubii, C. neoformans var. neoformans, and C. gattii by use of rapid biochemical tests, differential media, and DNA sequencing. J. Clin. Microbiol. 2011, 49, 2522–2527. [Google Scholar] [CrossRef]
- Stevenson, L.G.; Drake, S.K.; Shea, Y.R.; Zelazny, A.M.; Murray, P.R. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 2010, 48, 3482–3486. [Google Scholar] [CrossRef]
- Wieser, A.; Schneider, L.; Jung, J.; Schubert, S. MALDI-TOF MS in microbiological diagnostics—Identification of microorganisms and beyond (mini review). Appl. Microbiol. Biotechnol. 2012, 93, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Biomérieux. Available online: www.biomerieux.com (accessed on 26 March 2025).
- Drug Delivery Systems. Available online: www.bd.com (accessed on 26 March 2025).
- Posteraro, B.; Ruggeri, A.; De Carolis, E.; Torelli, R.; Vella, A.; De Maio, F.; Ricciardi, W.; Posteraro, P.; Sanguinetti, M. Comparative evaluation of BD Phoenix and Vitek 2 systems for species identification of common and uncommon pathogenic yeasts. J. Clin. Microbiol. 2013, 51, 3841–3845. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, B.; López, R.; Muñoz, C.; Aguilar, K.; Pérez, F.; Laínez-Arteaga, I.; Chávez, F.; Galindo, C.; Rivera, L.; Ballesteros-Monrreal, M.G.; et al. First report of the emerging pathogen Kodamaea ohmeri in Honduras. J. Fungi. 2024, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.R.; Burns, J.K.; Friedrich, L.M.; Goodrich, R.M.; Parish, M.E. Yeast species associated with orange juice: Evaluation of different identification methods. Appl. Environ. Microbiol. 2002, 68, 1955–1961. [Google Scholar] [CrossRef]
- Abd El-Tawab, A.A.; Salem, R.; El-Diasty, E.M.; Abd El-Halem, R.H. Molecular identification of pathogenic yeast from yoghurt. Benha Vet. Med. J. 2019, 37, 131–136. [Google Scholar] [CrossRef]
- Utama, G.L.; Meliana, S.; Djali, M.; Yuliana, T.; Balia, R.L. Probiotic Candidates Yeast Isolated from Dangke—Indonesian Traditional Fermented Buffalo Milk. Acta Univ. Agric. Silvic. Mendel. Brun. 2019, 67, 179–187. [Google Scholar] [CrossRef]
- Musgrove, M.T.; Jones, D.R.; Hinton, A.; Ingram, K.D.; Northcutt, J.K. Identification of Yeasts Isolated from Commercial Shell Eggs Stored at Refrigerated Temperatures. J. Food Prot. 2008, 71, 1258–1261. [Google Scholar] [CrossRef]
- Soliman, N.; Aly, S. Occurrence and identification of yeast species isolated from Egyptian Karish cheese. J. Yeast Fungal Res. 2011, 2, 59–64. Available online: https://academicjournals.org/journal/JYFR/article-stat/A5B55FE3996 (accessed on 17 March 2025).
- Xu, X.L.; Feng, G.L.; Liu, H.W.; Li, X.F.; Zhao, G.L.; Xiao, X.L. Isolation, identification and control of osmophilic spoilage yeasts in sweetened condensed milk. Afr. J. Microbiol. Res. 2014, 8, 1032–1039. [Google Scholar] [CrossRef]
- Chandran, A.; Linsha, C.K.; Beena, A.K. Prevalence of Candida species in ‘thairu’a traditional fermented milk of Kerala, India. European J. Nutr. Food Saf. 2022, 14, 15–19. [Google Scholar] [CrossRef]
- Rodríguez, C.L.; Strub, C.; Fontana, A.; Verheecke-Vaessen, C.; Durand, N.; Beugré, C.; Guehi, T.; Medina, A.; Schorr-Galindo, S. Biocontrol activities of yeasts or lactic acid bacteria isolated from Robusta coffee against Aspergillus carbonarius growth and ochratoxin A production in vitro. Int. J. Food Microbiol. 2024, 415, 110638. [Google Scholar] [CrossRef] [PubMed]
- Gientka, I.; Kieliszek, M.; Jermacz, K.; Błażejak, S. Identification and characterization of oleaginous yeast isolated from kefir and its ability to accumulate intracellular fats in deproteinated potato wastewater with different carbon sources. BioMed Res. Int. 2017, 2017, 6061042. [Google Scholar] [CrossRef] [PubMed]
- Török, T.; King, A.D., Jr. Comparative study on the edentification of food-borne yeasts. Appl. Environ. Microbiol. 1991, 57, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Deák, T.; Beuchat, L.R. Evaluation of simplified and commercial systems for identification of foodborne yeasts. Int. J. Food Microbiol. 1998, 7, 135–145. [Google Scholar] [CrossRef]
- Middelhoven, W.J. Identification of yeasts present in sour fermented foods and fodders. Mol. Biotechnol. 2002, 21, 279–292. [Google Scholar] [CrossRef]
- Ceugniez, A.; Drider, D.; Jacques, P.; Coucheney, F. Yeast diversity in a traditional French cheese “Tomme d’orchies” reveals infrequent and frequent species with associated benefits. Food Microbiol. 2015, 52, 177–184. [Google Scholar] [CrossRef]
- Gotcheva, V.; Pandiella, S.S.; Angelov, A.; Roshkova, Z.G.; Webb, C. Microflora identification of the Bulgarian cereal-based fermented beverage boza. Process Biochem. 2000, 36, 127–130. [Google Scholar] [CrossRef]
- van der Aa Kühle, A.; Jespersen, A.L. Detection and identification of wild yeasts in lager breweries. Int. J. Food Microbiol. 1998, 43, 205–213. [Google Scholar] [CrossRef]
- Senses-Ergul, S.; Ágoston, R.; Belák, Á.; Deák, T. Characterization of some yeasts isolated from foods by traditional and molecular tests. Int. J. Food Microbiol. 2006, 108, 120–124. [Google Scholar] [CrossRef]
- Pavlovic, M.; Mewes, A.; Maggipinto, M.; Schmidt, W.; Messelhäußer, U.; Balsliemke, J.; Hörmansdorfer, S.; Busch, U.; Huber, I. MALDI-TOF MS based identification of food-borne yeast isolates. J. Microbiol. Methods. 2014, 106, 123–128. [Google Scholar] [CrossRef]
- Paludan-Müller, C.; Madsen, M.; Sophanodora, P.; Gram, L.; Møller, P.L. Fermentation and microflora of plaa-som, a Thai fermented fish product prepared with different salt concentrations. Int. J. Food Microbiol. 2002, 73, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Günay, M.; Genç, T.T. Molecular identification of yeasts from Turkish traditional cheeses: Extracellular enzyme activities and physiological properties important for dairy industry. Nusant. Biosci. 2023, 15, 1–11. [Google Scholar] [CrossRef]
- Ceylan, Z.; Urçar Gelen, S. Identification of some yeast species in traditional Turkish fermented sausage with Vitek 2 compact system. Turk. J. Vet. Res. 2023, 7, 15–18. [Google Scholar] [CrossRef]
- Chay, C.; Dizon, E.I.; Elegado, F.B.; Norng, C.; Hurtada, W.A.; Raymundo, L. Isolation and identification of mold and yeast in medombae, a rice wine starter culture from Kompong Cham Province, Cambodia. Food Res. 2017, 1, 213–220. [Google Scholar] [CrossRef]
- Al-Abedi, S.F. Comparative study for Candida spp isolated and identification from goats milk with mastitis between API Candida kit Test, Candida chromogenic agar and VITEK 2 system. Biochem. Cell. Arch. 2020, 20, 5329–5332. [Google Scholar]
- Velázquez, E.; Cruz-Sánchez, J.M.; Rivas-Palá, T.; Zurdo-Piñeiro, J.L.; Mateos, P.F.; Monte, E.; Martínez-Molina, E.; Chordi, A. YeastIdent-Food/ProleFood, a new system for the identification of food yeasts based on physiological and biochemical tests. Food Microbiol. 2001, 18, 637–646. [Google Scholar] [CrossRef]
- Lebano, I.; Fracchetti, F.; Vigni, M.L.; Mejía, J.F.; Felis, G.E.; Lampis, S. MALDI-TOF as a powerful tool for identifying and differentiating closely related microorganisms: The strange case of three reference strains of Paenibacillus polymyxa. Sci. Rep. 2024, 14, 2585. [Google Scholar] [CrossRef]
- Gunduz, C.P.B.; Agirman, B.; Erten, H. Identification of yeasts in fermented foods and beverages using MALDI-TOF MS. FEMS Yeast Res. 2022, 22, 1–8. [Google Scholar] [CrossRef]
- Haider, A.; Ringer, M.; Kotroczó, Z.; Mohácsi-Farkas, C.; Kocsis, T. The current level of MALDI-TOF MS applications in detecting microorganisms: A short review of benefits and limitations. Microbiol. Res. 2023, 14, 80–90. [Google Scholar] [CrossRef]
- Bizzini, A.; Greub, G. Matrix-assisted laser desorption ionisation time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 2010, 16, 1614–1619. [Google Scholar] [CrossRef]
- Satoh, T.; Tsuno, H.; Iwanaga, M.; Kammei, Y. The design and characteristic features of a new time-of-flight mass spectrometer with a spiral ion trajectory. J. Am. Soc. Mass Spectrom. 2005, 16, 1969–1975. [Google Scholar] [CrossRef] [PubMed]
- Carlsohn, E.; Carol, L.; Nilsson, E.C. Proteomic techniques for functional identification of bacterial adhesins. In Lectins; Nilsson, C.L., Ed.; Elsevier Science B.V.: Amsterdam, The Netherlands, 2007; pp. 299–325. [Google Scholar]
- Coryell, M.P.; Sava, R.L.; Hastie, J.L.; Carlson, P.E., Jr. Application of MALDI-TOF MS for enumerating bacterial constituents of defined consortia. Appl. Microbiol. Biotechnol. 2023, 107, 4069–4077. [Google Scholar] [CrossRef] [PubMed]
- Patel, R. A moldy application of MALDI: MALDI-TOF mass spectrometry for fungal identification. J. Fungi 2019, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Yang, Q.; Kudinha, T.; Sun, L.; Zhang, R.; Liu, C.; Xu, Y.C. An improved in-house MALDI-TOF MS protocol for direct, cost-effective identification of pathogens from blood cultures. Front. Microbiol. 2017, 8, 1824. [Google Scholar] [CrossRef]
- Penland, M.; Falentin, H.; Parayre, S.; Pawtowski, A.; Maillard, M.-B.; Thierry, A.; Mounier, J.; Coton, M.; Deutsch, S.-M. Linking Pélardon artisanal goat cheese microbial communities to aroma compounds during cheese-making and ripening. Int. J. Food Microbiol. 2021, 345, 109130. [Google Scholar] [CrossRef]
- Pereira, T.S.; Batista, N.N.; Pimenta, L.P.S.; Martinez, S.J.; Ribeiro, L.S.; Naves, J.A.O. Self-induced anaerobiosis coffee fermentation: Impact on microbial communities, chemical composition and sensory quality of coffee. Food Microbiol. 2022, 103, 103962. [Google Scholar] [CrossRef]
- Vermote, L.; De Roos, J.; Cnockaert, M.; Vandamme, P.; Weckx, S.; De Vuyst, L. New insights into the role of key microorganisms and wooden barrels during lambic beer fermentation and maturation. Int. J. Food Microbiol. 2023, 394, 110163. [Google Scholar] [CrossRef]
- van den Boom, D.; Beaulieu, M.; Oeth, P.; Roth, R.; Honisch, C.; Nelson, M.R.; Cantor, C. MALDI-TOF MS: A platform technology for genetic discovery. Int. J. Mass Spectrom. 2004, 238, 173–188. [Google Scholar] [CrossRef]
- Chao, Q.T.; Lee, T.F.; Teng, S.H.; Peng, L.Y.; Chen, P.H.; Teng, L.J.; Hsueh, P.R. Comparison of the accuracy of two conventional phenotypic methods and two MALDI-TOF MS systems with DNA sequencing analysis for correctly identifying clinically encountered yeasts. PLoS ONE 2014, 9, e109376. [Google Scholar] [CrossRef]
- Assi, F.; Jabri, B.; Melalka, L.; Sekhsokh, Y.; Zouhdi, M. Direct MALDI-TOF MS-based method for rapid identification of microorganisms and antibiotic susceptibility testing in urine specimens. Iran. J. Microbiol. 2025, 17, 92–98. [Google Scholar] [CrossRef]
- Câmara, A.A.; Margalho, L.P.; Lang, E.; Brexó, R.P.; Sant’Ana, A.S. Yeast diversity in Brazilian artisanal cheeses: Unveiling technologically relevant species to improve traditional cheese production. Food Res. Int. 2024, 196, 115107. [Google Scholar] [CrossRef] [PubMed]
- Traina, C.; Ferrocino, I.; Bonciolini, A.; Cardenia, V.; Lin, X.; Rantsiou, K.; Cocolin, L. Monitoring the yeasts ecology and volatiles profile throughout the spontaneous fermentation of Taggiasca cv. table olives through culture-dependent and independent methods. Int. J. Food Microbiol. 2024, 417, 110688. [Google Scholar] [CrossRef] [PubMed]
- Gounari, Z.; Bonatsou, S.; Ferrocino, I.; Cocolin, L.; Papadopoulou, O.S.; Panagou, E.Z. Exploring yeast diversity of dry-salted naturally black olives from Greek retail outlets with culture dependent and independent molecular methods. Int. J. Food Microbiol. 2023, 398, 110226. [Google Scholar] [CrossRef] [PubMed]
- Sampaolesi, S.; Pérez-Través, L.; Pérez, D.; Roldán-López, D.; Briand, L.E.; Pérez-Torrado, R.; Querol, A. Identification and assessment of non-conventional yeasts in mixed fermentations for brewing bioflavored beer. Int. J. Food Microbiol. 2023, 399, 110254. [Google Scholar] [CrossRef]
- Serafino, G.; Gianvito, P.D.; Giacosa, S.; Škrab, D.; Cocolin, L.; Englezos, V.; Rantsiou, K. Survey of the yeast ecology of dehydrated grapes and strain selection for wine fermentation. Food Res. Int. 2023, 170, 113005. [Google Scholar] [CrossRef]
- Sipiczki, M.; Hrabovszki, V. Galactomyces candidus diversity in the complex mycobiota of cow-milk bryndza cheese comprising antagonistic and sensitive strains. Int. J. Food Microbiol. 2023, 388, 110088. [Google Scholar] [CrossRef]
- Englezos, V.; Mota-Gutierrez, J.; Giacosa, S.; Río Segade, S.; Pollon, M.; Gambino, G.; Rolle, L.; Ferrocino, I.; Rantsiou, K. Effect of alternative fungicides and inoculation strategy on yeast biodiversity and dynamics from the vineyard to the winery. Food Res. Int. 2022, 162, 111935. [Google Scholar] [CrossRef]
- Onmaz, N.E.; Gungor, C.; Al, S.; Dishan, A.; Hizlisoy, H.; Yildirim, Y.; Tekinsen, F.K.; Disli, H.B.; Barel, M.; Karadal, F. Mycotoxigenic and phylogenetic perspective to the yeasts and filamentous moulds in mould-matured Turkish cheese. Int. J. Food Microbiol. 2021, 357, 109385. [Google Scholar] [CrossRef]
- Cocolin, L.; Bisson, L.F.; Mills, D.A. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol. Lett. 2000, 189, 81–87. [Google Scholar] [CrossRef]
- Jeong, D.M.; Yoo, S.J.; Jeon, M.-S.; Chun, B.H.; Han, D.M.; Jeon, C.O.; Eyun, S.-I.; Seo, Y.-J.; Kang, H.A. Genomic features, aroma profiles, and probiotic potential of the Debaryomyces hansenii species complex strains isolated from Korean soybean fermented food. Food Microbiol. 2022, 105, 104011. [Google Scholar] [CrossRef]
- Carbonero-Pacheco, J.; Rey, M.-D.; Moreno-García, J.; Moreno, J.; García-Martínez, T.; Mauricio, J.C. Microbial diversity in sherry wine biofilms and surrounding mites. Food Microbiol. 2023, 116, 104366. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, F.; Fernandes, T.; Tulha, J.; Bessa, D.; Pereira, J.; Schuller, D.; Sousa, M.J.; Sampaio, P.; Pais, C.; Franco-Duarte, R. Yeast mycobiome of fruit and vegetable biowastes revealed by culture-dependent and metabarcoding approaches: Screening for the production of succinic acid and single cell oils. Food Biosci. 2024, 62, 105340. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols a Guide to Methods and Applications; Innis, M.A., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- O’Donnell, K. Fusarium and its near relatives. In The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics; Reynolds, D.R., Taylor, J.W., Eds.; CAB International: Wallingford, UK, 1993; pp. 225–233. [Google Scholar]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburul, F.; QueroI, A. Identification of yeasts by RFLP analysis of the 5.85 rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 1999, 49, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Battistelli, N.; Perpetuini, G.; Piva, A.; Pepe, A.; Sidari, R.; Wache, Y.; Tofalo, R. Cultivable microbial ecology and aromatic profile of “mothers” for Vino cotto wine production. Food Res. Int. 2021, 143, 110311. [Google Scholar] [CrossRef]
- Lleixà, J.; Martínez-Safont, M.; Masneuf-Pomarede, I.; Magani, M.; Albertin, W.; Mas, A.; Portillo, M.C. Genetic and phenotypic diversity of Brettanomyces bruxellensis isolates from ageing wines. Food Biosci. 2021, 40, 100900. [Google Scholar] [CrossRef]
- Grijalva-Vallejos, N.; Aranda, A.; Matallana, E. Evaluation of yeasts from Ecuadorian chicha by their performance as starters for alcoholic fermentations in the food industry. Int. J. Food Microbiol. 2020, 317, 108462. [Google Scholar] [CrossRef]
- Aldrete-Tapia, J.A.; Escalante-Minakata, P.; Martínez-Peniche, R.A.; Tamplin, M.L.; Hernández-Iturriaga, M. Yeast and bacterial diversity, dynamics and fermentative kinetics during small-scale tequila spontaneous fermentation. Food Microbiol. 2020, 86, 103339. [Google Scholar] [CrossRef]
- Ruiz-Muñoz, M.; Cordero-Bueso, G.; Benítez-Trujillo, F.; Martínez, S.; Pérez, F.; Cantoral, F.P. Rethinking about flor yeast diversity and its dynamic in the “criaderas and soleras” biological aging system. Food Microbiol. 2020, 92, 103553. [Google Scholar] [CrossRef]
- Fazio, N.A.; Pino, A.; Foti, P.; Esteve-Zarzoso, B.; Randazzo, C.L.; Torija, M.-J.; Caggia, C. Screening and characterization of indigenous Saccharomyces cerevisiae and non-Saccharomyces yeasts isolated from Sicilian vineyards. Food Biosci. 2024, 62, 105282. [Google Scholar] [CrossRef]
- Parafati, L.; Palmeri, R.; Pitino, I.; Restuccia, C. Killer yeasts isolated from olive brines: Technological and probiotic aptitudes. Food Microbiol. 2022, 103, 103950. [Google Scholar] [CrossRef]
- Matraxia, M.; Alfonzo, A.; Prestianni, R.; Francesca, N.; Gaglio, R.; Todaro, A.; Alfeo, V.; Perretti, G.; Columba, P.; Settanni, L.; et al. Non-conventional yeasts from fermented honey by-products: Focus on Hanseniaspora uvarum strains for craft beer production. Food Microbiol. 2021, 99, 103806. [Google Scholar] [CrossRef] [PubMed]
- Ouattara, H.G.; Niamké, S.L. Mapping the functional and strain diversity of the main microbiota involved in cocoa fermentation from Cote d’Ivoire. Food Microbiol. 2021, 98, 103767. [Google Scholar] [CrossRef] [PubMed]
- Bo, B.; Kim, S.-A.; Han, N.S. Bacterial and fungal diversity in Laphet, traditional fermented tea leaves in Myanmar, analyzed by culturing, DNA amplicon-based sequencing, and PCR-DGGE methods. Int. J. Food Microbiol. 2020, 320, 108508. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Mills, D.A. Improved selection of Internal Transcribed Spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for Basidiomycetes: Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of Ascomycetes and Basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef]
- Mota-Gutierrez, J.; Ferrocino, I.; Rantsiou, K.; Cocolin, L. Metataxonomic comparison between internal transcribed spacer and 26S ribosomal large subunit (LSU) rDNA gene. Int. J. Food Microbiol. 2019, 290, 132–140. [Google Scholar] [CrossRef]
- Liu, S.; Jiang, Z.; Ma, D.; Liu, X.; Li, Y.; Ren, D.; Zhu, Y.; Zhao, H.; Qin, H.; Huang, M.; et al. Distance decay pattern of fermented-related microorganisms in the sauce-flavor Baijiu producing region. Food Biosci. 2023, 51, 102305. [Google Scholar] [CrossRef]
- Chen, G.; Li, W.; Yang, Z.; Liang, Z.; Chen, S.; Qiu, Y.; Lv, X.; Ai, L.; Ni, L. Insights into microbial communities and metabolic profiles in the traditional production of the two representative Hongqu rice wines fermented with Gutian Qu and Wuyi Qu based on single-molecule real-time sequencing. Food Res. Int. 2023, 173, 113488. [Google Scholar] [CrossRef]
- National Library of Medicine, National Center for Biotechnology Information. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 26 March 2025).
- Unite Community. Available online: https://unite.ut.ee/ (accessed on 26 March 2025).
- Silva, High Quality Ribosomal RNA Database. Available online: https://www.arb-silva.de/ (accessed on 26 March 2025).
- MycoBank Database. Available online: http://www.mycobank.org/ (accessed on 26 March 2025).
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- RPD Classifier. Available online: https://sourceforge.net/projects/rdp-classifier/ (accessed on 26 March 2025).
- Jia, X.; Wang, D.; Meng, A.-L.; Lin, Y.-J.; Huang, M.; Gao, P.; Xu, P.; Chen, H. Microbial composition of spoiled irradiated ready-to-eat chicken feet and their spoilage characteristics. Food Microbiol. 2024, 124, 104620. [Google Scholar] [CrossRef] [PubMed]
- Usal, M.; Özgölet, M.; Arici, M.; Törnük, F. Enzymatic and antimicrobial activities of lactic acid bacteria and yeasts isolated from boza, a traditional fermented grain based beverage. Food Biosci. 2024, 61, 104681. [Google Scholar] [CrossRef]
- Verni, M.; Torreggiani, A.; Patriarca, A.; Brasili, E.; Sciubba, F.; Rizzello, C.G. Sourdough fermentation for the valorization of sorghum flour: Microbiota characterization and metabolome profiling. Int. J. Food Microbiol. 2024, 421, 110805. [Google Scholar] [CrossRef]
- Chen, L.; Li, F.; Yang, F.; Chen, B.; Du, H.; Wang, L.; Xu, Y. Microbiome dynamics and environment driving factors throughout pit fermentation of Jiang-flavor Baijiu: A multi-omics study. Food Biosci. 2024, 60, 104363. [Google Scholar] [CrossRef]
- Kang, H.-Y.; Ao, X.-L.; Tang, Q.; Li, H.; Fan, Y.; Liu, A.-P.; Zou, L.-K.; Liu, S.-L.; Yang, Y.; Zhao, N.; et al. Effects of yeast screened from traditional fermented milk on commercial fermented milk as adjunct flavor culture. Food Biosci. 2024, 57, 103551. [Google Scholar] [CrossRef]
- Sonets, I.V.; Solovyev, M.A.; Ivanova, V.A.; Vasiluev, P.A.; Kachalkin, A.V.; Ochkalova, S.D.; Korobeynikov, A.I.; Razin, S.V.; Ulianov, S.V.; Tyakht, A.V. Hi-C metagenomics facilitate comparative genome analysis of bacteria and yeast from spontaneous beer and cider. Food Microbiol. 2024, 121, 104520. [Google Scholar] [CrossRef]
- Milanović, V.; Cardinali, F.; Boban, A.; Kljusurić, J.G.; Osimani, A.; Aquilanti, L.; Garofalo, C.; Budić-Leto, I. White grape variety Maraština as a promising source of non-Saccharomyces yeasts intended as starter cultures. Food Biosci. 2023, 55, 103033. [Google Scholar] [CrossRef]
- Vicente, J.; Ruiz, J.; Tomasi, S.; de Celis, M.; Ruiz-de-Villa, C.; Gombau, J.; Rozès, N.; Zamora, F.; Santos, A.; Marquina, D.; et al. Impact of rare yeasts in Saccharomyces cerevisiae wine fermentation performance: Population prevalence and growth phenotype of Cyberlindnera fabianii, Kazachstania unispora, and Naganishia globosa. Food Microbiol. 2023, 110, 104189. [Google Scholar] [CrossRef]
- Mi, T.; Jin, Y.; Che, Y.; Huang, J.; Zhou, R.; Wu, C. Profiling the composition and metabolic functions of microbial community in pellicle-forming radish paocai. Int. J. Food Microbiol. 2023, 388, 110087. [Google Scholar] [CrossRef]
- Aydın, F.; Özer, G.; Alkan, M.; Çakır, I. Start Codon Targeted (SCoT) markers for the assessment of genetic diversity in yeast isolated from Turkish sourdough. Food Microbiol. 2022, 107, 104081. [Google Scholar] [CrossRef]
- Nascimento, H.M.; do Prado-Silva, L.; Brandão, L.R.; Brexó, R.P.; Câmara, A.A., Jr.; Rosa, C.A.; Sant’Ana, A.S. Large scale survey of yeasts in frozen concentrated orange juice (FCOJ): Occurrence, diversity, and resistance to peracetic acid. Int. J. Food Microbiol. 2022, 367, 109589. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, M.; Xie, N.; Huang, M.; Feng, Y. Community structure of yeast in fermented soy sauce and screening of functional yeast with potential to enhance the soy sauce flavor. Int. J. Food Microbiol. 2022, 370, 109652. [Google Scholar] [CrossRef]
- Salazar, M.M.M.; Álvarez, O.L.M.; Castañeda, M.P.A.; Medina, P.X.L. Bioprospecting of indigenous yeasts involved in cocoa fermentation using sensory and chemical strategies for selecting a starter inoculum. Food Microbiol. 2022, 101, 103896. [Google Scholar] [CrossRef]
- Tzamourani, A.P.; Kasimati, A.; Karagianni, E.; Manthou, E.; Panagou, E.Z. Exploring microbial communities of Spanish-style green table olives of Conservolea and Halkidiki cultivars during modified atmosphere packaging in multi-layered pouches through culture-dependent techniques and metataxonomic analysis. Food Microbiol. 2022, 107, 104063. [Google Scholar] [CrossRef]
- Iacumin, L.; Colautti, A.; Comi, G. Zygosaccharomyces rouxii is the predominant species responsible for the spoilage of the mix base for ice cream and ethanol is the best inhibitor tested. Food Microbiol. 2022, 102, 103929. [Google Scholar] [CrossRef]
- Bao, Y.; Zhang, M.; Chen, W.; Chen, H.; Chen, W.; Zhong, Q. Screening and evaluation of suitable non-Saccharomyces yeast for aroma improvement of fermented mango juice. Food Biosci. 2021, 44, 101414. [Google Scholar] [CrossRef]
- Montaño, A.; Cortés-Delgado, A.; Sánchez, A.H.; Ruiz-Barba, J.L. Production of volatile compounds by wild-type yeasts in a natural olive-derived culture medium. Food Microbiol. 2021, 98, 103788. [Google Scholar] [CrossRef]
- Echeverrigaray, S.; Scariot, F.J.; Foresti, L.; Schwarz, L.V.; Rocha, R.K.M.; da Silva, G.P.; Moreira, J.P.; Delamare, A.P.L. Yeast biodiversity in honey produced by stingless bees raised in the highlands of southern Brazil. Int. J. Food Microbiol. 2021, 347, 109200. [Google Scholar] [CrossRef]
- Du, G.; Liu, L.; Guo, Q.; Cui, Y.; Chen, H.; Yuan, Y.; Wang, Z.; Gao, Z.; Sheng, Q.; Yue, T. Microbial community diversity associated with Tibetan kefir grains and its detoxification of Ochratoxin A during fermentation. Food Microbiol. 2021, 99, 103803. [Google Scholar] [CrossRef]
- Lin, M.M.-H.; Boss, P.K.; Walker, M.E.; Sumby, K.M.; Grbin, P.R.; Jiranek, V. Evaluation of indigenous non-Saccharomyces yeasts isolated from a South Australian vineyard for their potential as wine starter cultures. Int. J. Food Microbiol. 2020, 312, 108373. [Google Scholar] [CrossRef]
- Elhalis, H.; Cox, J.; Zhao, J. Ecological diversity, evolution and metabolism of microbial communities in the wet fermentation of Australian coffee beans. Int. J. Food Microbiol. 2020, 321, 108544. [Google Scholar] [CrossRef] [PubMed]
- Adepehin, J.O. Microbial diversity and pasting properties of finger millet (Eleusine coracana), pearl millet (Pennisetum glaucum) and sorghum (Sorghum bicolor) sourdoughs. Food Biosci. 2020, 37, 100684. [Google Scholar] [CrossRef]
- Cardinali, F.; Rampanti, G.; Paderni, G.; Milanović, V.; Ferrocino, I.; Reale, A.; Boscaino, F.; Raicevic, N.; Ilincic, M.; Osimani, A.; et al. A comprehensive study on the autochthonous microbiota, volatilome, physico-chemical, and morpho-textural features of Montenegrin Njeguški cheese. Food Res. Int. 2024, 197, 115169. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, D.; Mu, Y.; Zhu, Z.; Wu, Y.; Qi, Q.; Mu, Y.; Su, W. Exploring the heterogeneity of community and function and correspondence of “species-enzymes” among three types of Daqu with different fermentation peak-temperature via high-throughput sequencing and metagenomics. Food Res. Int. 2024, 176, 113805. [Google Scholar] [CrossRef]
- Li, Q.; Du, B.; Chen, X.; Zhao, Y.; Zhu, L.; Ma, H.; Sun, B.; Hao, J.; Li, L. Microbial community dynamics and spatial distribution of flavor compound metabolism during solid-state fermentation of Baijiu enhanced by Wickerhamomyces anomalus. Food Biosci. 2024, 59, 103909. [Google Scholar] [CrossRef]
- López-García, E.; Romero-Gil, V.; Arroyo-López, F.N.; Benítez-Cabello, A. Impact of lactic acid bacteria inoculation on fungal diversity during Spanish-style green table olive fermentations. Int. J. Food Microbiol. 2024, 417, 110689. [Google Scholar] [CrossRef]
- Lutin, J.; Dufrene, F.; Guyot, P.; Palme, R.; Achilleos, C.; Bouton, Y.; Buchin, S. Microbial composition and viability of natural whey starters used in PDO Comté cheese-making. Food Microbiol. 2024, 121, 104521. [Google Scholar] [CrossRef]
- Martins, V.; Teixeira, A.; Gerós, H. A comparison of microbiota isolation methods reveals habitat preferences for fermentative yeasts and plant pathogenic fungi in the grape berry. Food Microbiol. 2024, 118, 104408. [Google Scholar] [CrossRef]
- Cruz-O’Byrne, R.; Gamez-Guzman, A.; Piraneque-Gambasica, N.; Aguirre-Forero, S. Genomic sequencing in Colombian coffee fermentation reveals new records of yeast species. Food Biosci. 2023, 52, 102415. [Google Scholar] [CrossRef]
- Li, S.; Wang, S.; Wang, L.; Liu, X.; Wang, X.; Cai, R.; Yuan, Y.; Yue, T.; Wang, Z. Unraveling symbiotic microbial communities, metabolomics and volatilomics profiles of kombucha from diverse regions in China. Food Res. Int. 2023, 174, 113652. [Google Scholar] [CrossRef]
- Rampanti, G.; Raffo, A.; Melini, V.; Moneta, E.; Nardo, N.; Civitelli, E.S.; León, C.B.-D.; Portero, L.T.; Ferrocino, I.; Franciosa, I.; et al. Chemical, microbiological, textural, and sensory characteristics of pilot-scale Caciofiore cheese curdled with commercial Cynara cardunculus rennet and crude extracts from spontaneous and cultivated Onopordum tauricum. Food Res. Int. 2023, 173, 113459. [Google Scholar] [CrossRef] [PubMed]
- Osimani, A.; Belleggia, L.; Botta, C.; Ferrocino, I.; Milanović, V.; Cardinali, F.; Haouet, M.N.; Garofalo, C.; Mozzon, M.; Foligni, R.; et al. Journey to the morpho-textural traits, microbiota, and volatilome of Ciauscolo PGI salami. Food Biosci. 2023, 53, 102582. [Google Scholar] [CrossRef]
- Botello-Morte, L.; Moniente, M.; Gil-Ramírez, Y.; Virto, R.; García-Gonzalo, D.; Pagán, R. Identification by means of molecular tools of the microbiota responsible for the formation of histamine accumulated in commercial cheeses in Spain. Food Control. 2022, 133, 108595. [Google Scholar] [CrossRef]
- Benítez-Cabello, A.; Ramiro-García, J.; Romero-Gil, V.; Medina, E.; Arroyo-López, F.N. Fungal biodiversity in commercial table olive packages. Food Microbiol. 2022, 107, 104082. [Google Scholar] [CrossRef]
- Kang, J.; Sun, Y.; Huang, X.; Ye, L.; Chen, Y.; Chen, X.; Zheng, X.; Han, B.-Z. Unraveling the microbial compositions, metabolic functions, and antibacterial properties of Huangshui, a byproduct of Baijiu fermentation. Food Res. Int. 2022, 157, 111320. [Google Scholar] [CrossRef]
- Milanović, V.; Cardinali, F.; Ferrocino, I.; Boban, A.; Franciosa, I.; Kljusurić, J.G.; Mucalo, A.; Osimani, A.; Aquilanti, L.; Garofalo, C.; et al. Croatian white grape variety Maraština: First taste of its indigenous mycobiota. Food Res. Int. 2022, 162, 111917. [Google Scholar] [CrossRef]
- Wang, X.; Schlatter, D.C.; Glawe, D.A.; Edwards, C.G.; Weller, D.M.; Paulitz, T.C.; Abatzoglou, J.T.; Okubara, P.A. Native yeast and non-yeast fungal communities of Cabernet Sauvignon berries from two Washington State vineyards, and persistence in spontaneous fermentation. Int. J. Food Microbiol. 2021, 350, 109225. [Google Scholar] [CrossRef]
- Cruz-O’Byrne, R.; Piraneque-Gambasica, N.; Aguirre-Forero, S. Microbial diversity associated with spontaneous coffee bean fermentation process and specialty coffee production in northern Colombia. Int. J. Food Microbiol. 2021, 354, 109282. [Google Scholar] [CrossRef]
- Manthou, E.; Coeuret, G.; Chaillou, S.; Nychas, G.-J.E. Evolution of fungal community associated with ready-to-eat pineapple during storage under different temperature conditions. Food Microbiol. 2021, 97, 103736. [Google Scholar] [CrossRef]
- Cardinali, F.; Ferrocino, I.; Milanović, V.; Belleggia, L.; Corvaglia, M.R.; Garofalo, C.; Foligni, R.; Mannozzi, C.; Mozzon, M.; Cocolin, L.; et al. Microbial communities and volatile profile of Queijo de Azeitão PDO cheese, a traditional Mediterranean thistle-curdled cheese from Portugal. Food Res. Int. 2021, 147, 110537. [Google Scholar] [CrossRef]
- Michailidou, S.; Pavlou, E.; Pasentsis, K.; Rhoades, J.; Likotrafiti, E.; Argiriou, A. Microbial profiles of Greek PDO cheeses assessed with amplicon metabarcoding. Food Microbiol. 2021, 99, 103836. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Xiang, Q.; Sun, W.; Wang, X.; Lin, J.; Che, Z.; Ma, P. Correlation between microbial communities and key flavors during postfermentation of Pixian broad bean paste. Food Res. Int. 2020, 137, 109513. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jiang, D.; Liu, W.; Yang, Y.; Zhang, Y.; Jin, C.; Sun, S. Comparison of fermentation behaviors and properties of raspberry wines by spontaneous and controlled alcoholic fermentations. Food Res. Int. 2020, 128, 108801. [Google Scholar] [CrossRef]
- Balzan, S.; Carraro, L.; Merlanti, R.; Lucatello, L.; Capolongo, F.; Fontana, F.; Novelli, E.; Larini, I.; Vitulo, N.; Cardazzo, B. Microbial metabarcoding highlights different bacterial and fungal populations in honey samples from local beekeepers and market in northeastern Italy. Int. J. Food Microbiol. 2020, 334, 108806. [Google Scholar] [CrossRef]
- Prada, P.; Brunel, B.; Reffuveille, F.; Gangloff, S.C. Technique evolutions for microorganism detection in complex samples: A Review. Appl. Sci. 2022, 12, 5892. [Google Scholar] [CrossRef]
- Rebrosova, K.; Samek, O.; Kizovsky, M.; Bernatova, S.; Hola, V.; Ruzicka, F. Raman spectroscopy-A novel method for identification and characterization of microbes on a single-cell level in clinical settings. Front. Cell Infect. Microbiol. 2022, 12, 866463. [Google Scholar] [CrossRef]
- Li, N.; Hussain, N.; Ding, Z.; Qu, C.; Li, Y.; Chu, L.; Liu, H. Guidelines for Raman spectroscopy and imaging techniques in food safety analysis. Food Saf. Health 2024, 2, 221–237. [Google Scholar] [CrossRef]
- Udupa, R.; Peralam-Yegneswaran, P.; Lukose, J.; Chidangil, S. Utilization of Raman spectroscopy for identification and characterization of fungal pathogens. Fungal Biol. Rev. 2024, 47, 100339. [Google Scholar] [CrossRef]
- Maquelin, K.; Kirschner, C.; Choo-Smith, L.P.; Ngo-Thi, N.A.; van Vreeswijk, T.; Stämmler, M.; Endtz, H.P.; Bruining, H.A.; Naumann, D.; Puppels, G.J. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 2003, 41, 324–329. [Google Scholar] [CrossRef]
- Lemma, T.; Wang, J.; Arstila, K.; Hytönen, V.P.; Toppari, J.J. Identifying yeasts using surface enhanced Raman spectroscopy. Spectrochim. Acta A. Mol. Biomol. Spectrosc. 2019, 218, 299–307. [Google Scholar] [CrossRef]
- Wang, J.; Meng, S.; Lin, K.; Yi, X.; Sun, Y.; Xu, X.; He, N.; Zhang, Z.; Hu, H.; Qie, X.; et al. Leveraging single-cell Raman spectroscopy and single-cell sorting for the detection and identification of yeast infections. Anal. Chim. Acta 2023, 1239, 340658. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, S.; Thornton, M.; Thornton, R. Raman spectroscopy and chemometrics for identification and strain discrimination of the wine spoilage yeasts Saccharomyces cerevisiae, Zygosaccharomyces bailii, and Brettanomyces bruxellensis. Appl. Environ. Microbiol. 2013, 79, 01886. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, J.; Martiniuk, J.; Ma, X.; Li, Q.; Measday, V.; Lu, X. Species identification and strain discrimination of fermentation yeasts Saccharomyces cerevisiae and Saccharomyces uvarum using Raman spectroscopy and convolutional neural networks. Appl. Environ. Microbiol. 2023, 89, 0167323. [Google Scholar] [CrossRef] [PubMed]
- Zieliński, B.; Sroka-Oleksiak, A.; Rymarczyk, D.; Piekarczyk, A.; Brzychczy-Włoch, M. Deep learning approach to describe and classify fungi microscopic images. PLoS ONE 2020, 15, e0234806. [Google Scholar] [CrossRef]
- da Silva Neto, S.R.; Tabosa Oliveira, T.; Teixeira, I.V.; Aguiar de Oliveira, S.B.; Souza Sampaio, V.; Lynn, T.; Endo, P.T. Machine learning and deep learning techniques to support clinical diagnosis of arboviral diseases: A systematic review. PLoS Negl. Trop. Dis. 2022, 16, e0010061. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, H.; Ye, T.; Juhas, M. Deep learning for imaging and detection of microorganisms. Trends Microbiol. 2021, 29, 569–572. [Google Scholar] [CrossRef]

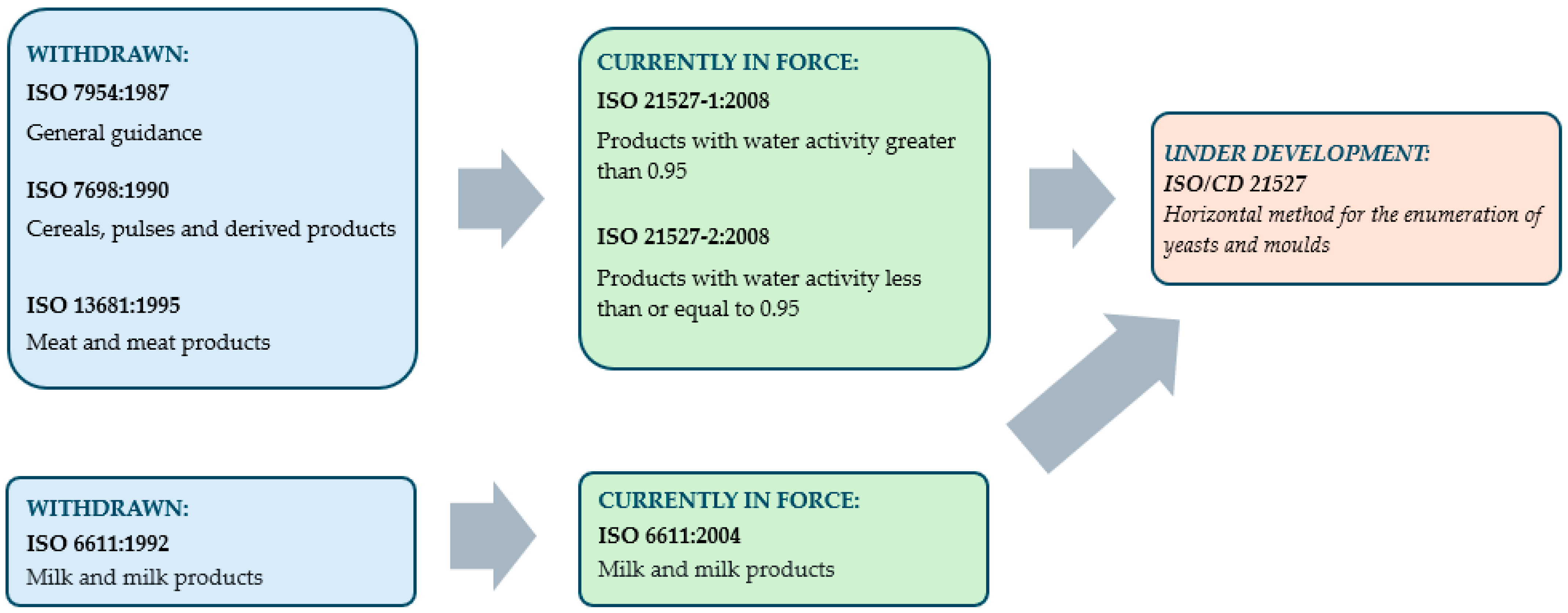

| Advantages | Disadvantages or Limitations |
|---|---|
| Viability Assessment: only live, viable yeasts will grow on culture media, allowing for an accurate assessment of contamination risks | Low sensitivity: may result in the failure to detect viable but non-culturable (VBNC) yeasts, or those present in low numbers |
| Cost-effective: relatively inexpensive in comparison to molecular methods, due to the relatively low cost of equipment and reagents, and the requirement of only rudimentary laboratory facilities | Time-consuming: requires incubation periods that can extend from 24 to 72 h or more, thereby delaying the delivery of results |
| Isolation and characterisation: enables isolation of pure cultures for further phenotypic and genotypic characterisation (e.g., morphology, biochemical tests, antifungal susceptibility) | Lack of specificity: morphologically similar colonies may not be distinguishable without further testing or molecular confirmation |
| Quantification: colony counts (e.g., CFU/mL or CFU/g) provide quantitative data on levels of yeast contamination | Selective media limitations: some yeast strains being susceptible to inhibition by the selective agents or exhibiting impaired growth on standard media |
| Simplicity and accessibility: straightforward protocols that are widely available and do not require specialised expertise | Labor-intensive: requires the implementation of manual plating, the undertaking of incubation, and the execution of colony counting, therefore these procedures can be arduous and prone to human error |
| Standardized protocols: established international guidelines for common food matrices, e.g., in the form of ISO methods, which serve to ensure the standardisation of protocols | Underestimation in mixed populations: in heterogeneous population structures, the proliferation of faster-growing or more robust yeast strains may result in the underestimation of diversity or abundance |
| Food Type/Occurrence | Detected Yeast(s) | Culture Medium | Selective-Differential Factors a | References |
|---|---|---|---|---|
| Beer | most culture yeasts, nonculture yeasts and Gram-negative bacteria commonly found in breweries | Universal Beer Agar | cycloheximide | [83,84] |
| most culture yeasts, nonculture yeasts and Gram-negative bacteria commonly found in breweries | Brewers’ Tomato Juice Medium | cycloheximide | ||
| wild yeasts—other yeast genera than Saccharomyces | Lysine Agar | lysine as sole nitrogen source | [85] | |
| wild yeasts (Saccharomyces pastorianus) | Schwarz Differential Medium (SDM) | Fuchsin–sulphite mixture | [86] | |
| inhibits, or markedly restricts, growth of brewery culture yeasts while permitting growth of many wild yeasts | Lin’s Wild Yeast Differential Agar (LWYD) | chrystal violet | [83] | |
| Alcoholic fermentation beverages (wine, cider) | enumerating Saccharomyces during the early stages of fermentation | Ethanol Sulphite Yeast Extract (ESY) agar | ethanol, sodium metabisulfite | [87] |
| monitoring growth of Kloeckera apiculata, Saccharomycodes ludwigii, and Candida stellata | Lysine Agar | lysine as sole nitrogen source | [88] | |
| Brettanomyces | Brettanomyces Selective Medium (BSM) | cycloheximide, chloramphenicol, gentamicin, chlorotetracycline | [89] | |
| Dekkera/Brettanomyces Differential Medium (DBDM) | cycloheximide, ethanol, p-coumaric acid, bromocresol green | |||
| Brettanomyces/Dekkera specific medium (DHSA) | cycloheximide, penicillin, gentamicin, ethanol, trehalose, saccharose, sorbic acid, bromocresol green, nutrients | [90] | ||
| Wallerstein Differential Agar (WLD Agar, WL Differential Agar) | cycloheximide, bromocresol green | [91] | ||
| Blue-veined cheese | isolation of yeasts | Dichloran Rose Bengal Chloramphenicol Agar (DRBC) | dichloran, chloramphenicol | [92] |
| Rose Bengal Chloramphenicol Agar (RBC) | chloramphenicol | |||
| Koumiss (spontaneously fermented mare’s milk drink) | native populations of Saccharomyces | Ethanol Sulphite Agar supplemented with antibiotic | ethanol, sodium metabisulfite, chloramphenicol | [93] |
| Ogi, mawè, gowé and tchoukoutou (traditional African fermented food products) | Candida krusei, Clavispora lusitaniae, Saccharomyces cerevisiea, Candida tropicalis, Kluyveromyces marxianus, Candida rugosa | Malt extract Yeast extract Glucose Pepton Agar (MYGP) supplemented with antibiotics | chloramphenicol and chlortetracycline | [94] |
| Part-skim mozzarella cheese | yeast detection | Potato dextrose agar (PDA) supplemented with antibiotic, | chlortetracycline, | [95] |
| Dichloran Rose Bengal Chloramphenicol Agar (DRBC) | dichloran, chloramphenicol | |||
| Serpa (Portuguese cheese) | yeast detection | Rose Bengal Chloramphenicol Agar (RBC) | chloramphenicol | [96] |
| Spoiled olive oil | Brettanomyces acidodurans | Rose Bengal Chloramphenicol Agar (RBC) | chloramphenicol | [73] |
| Tropical fruits | Saccharomyces, Debaryomyces, Rhodotorula and Cryptococcus species | Modified Molybdate agar | the medium to contain 0.125% calcium propionate and 3% agar | [97] |
| Preservative sensitive yeasts | Zygosaccharomyces bailii and, to a lesser extent, Z. rouxii | malt acetic acid agar | standard malt extract agar to which 0.5% acetic acid is added just before pouring | [98] |
| Osmophilic yeasts | Zygosaccharomyces rouxii, Debaryomyces hansenii | Dichloran 18% Glycerol Agar (DG18) | dichloran, chloramphenicol, glycerol | [99,100] |
| Zygosaccharomyces bailii in wine | Zygosaccharomyces bailii Differential Medium (ZBD), Zygosaccharomyces bailii Selective Agar (ZBA) | mineral medium supplemented with vitamins, oligoelements, formic acid-glucose mixture, pH 4.5 | [101] | |
| Zygosaccharomyces rouxii in high-sugar products | Yeast Extract Glucosa Agar (YEG50 Agar) | glucose (50%) aw 0.909, pH 4.5 | [71] | |
| osmophilic yeast count in grape juice | Malt Extract Yeast Extract Glucose (50%) Agar (MY50G) | glucose (50%) | [72] | |
| Zygosaccharomyces bailii from spoiled commercially processed food | Acidified Yeast Extract Malt Agar (AYMA) | pH 3.8 | [102] | |
| Acidified Tryptone Glucose Yeast Extract Agar (TGYA) | pH 3.9 | |||
| modified Zygosaccharomyces bailii Selective Agar (ZBA) | pH 4.0 | |||
| Schizosaccharomyces osmophilus in bee bread | Peptone Yeast extract Glucose and Fructose medium | glucose 5% (w/w) and fructose 55% (w/w) | [103] | |
| Zygosaccharomyces favi in bee bread | Rose Bengal Chloramphenicol Agar (RBC) | chloramphenicol | [104] | |
| 50% Glucose Agar | glucose (50%) |
| Name of the Kit | Application Field | Principle of Mode of Action | Target Microorganisms | Time Requirement | Producer |
|---|---|---|---|---|---|
| AuxaColor 2 Kit | identification | sugar assimilation | 33 yeast species (most frequently isolated in human medical mycology) | 48–72 h | BioRad |
| BioLumix Rapid Simplified Assay with YM vials | detection | CO2 absorption | yeasts and moulds | 6–48 h | Neogen |
| BioTrace 4250 and BacTrac 4300 | detection | impedance | yeasts and moulds, osmotolerant yeasts | within hours | Sy-Lab |
| D-COUNT® | detection | flow cytometry | yeast detection in fruit preparations, fruit juices and beverages yeast and moulds detection in non-filterable beverages and fruit juice yeast detection in yoghurts and fermented milk products | 90 min | BioMérieux |
| Foodproof® Spoilage Yeast Detection 1 LyoKit | detection and quantification | real-time PCR technology | major beverage spoilage yeasts (Dekkera/Brettanomyces, Zygosaccharomyces and Saccharomyces species) | 5 h | Hygiena |
| HybriScan® test | detection and identification | sandwich hybridisation | yeasts in foods, beverages, and water | <3 h | Merck |
| QuickGEN PCR Kit Yeast | quantitative detection | real-time PCR technology | contaminating yeasts from beverages | not specified | Gen-IAL |
| RapID™ YEAST PLUS System | identification | enzyme technology | 40 different yeasts | 4 h | Thermo Fisher Scientific |
| SCANRDI® | detection | combination of universal cell labelling and solid-phase cytometry | yeasts and moulds– both in vegetative and sporulated forms | less than 4 h | BioMérieux |
| VIT® Spoilage Yeasts | detection and differentiation | gene probe technology | obligate and potentially fermentative yeasts | <3 h | Vermicon |
| VITEK® MS and VITEK® MS PRIME | identification | Matrix-Assisted Laser Desorption Ionisation-Time of Flight (MALDI-TOF) and Mass Spectrometry | yeasts and fungi from food and beverage | within a few minutes | BioMérieux |
| Yeast HCP ELISA Kits | detection | ELISA | Pichia pastoris and Saccharomyces cerevisiae | 1 h 45 min–3 h 50 min | Sygnus Technologies |
| Identification Method | Source of Yeast | Reference |
|---|---|---|
| API 20C AUX | citrus juice | [199] |
| commercial shell eggs | [202] | |
| Egyptian Karish cheese | [203] | |
| condensed milk | [204] | |
| fermented milk | [205] | |
| robusta coffee | [206] | |
| kefir | [207] | |
| API 20C | partially and fully processed fruits and vegetables | [208] |
| fresh sweet corn | [209] | |
| ID32C | sour fermented foods and fodders | [210] |
| traditional French cheese (“Tomme d’orchies”) | [211] | |
| traditional Bulgarian cereal-based beverage (boza) | [212] | |
| lager breweries | [213] | |
| commercial shell eggs | [202] | |
| fermented and non-fermented home-made carrot juice, irradiated radish sprout, homogenized black currant, French-type soft cheese | [214] | |
| seafood, beef, sushi, raw chicken | [215] | |
| fermented fish | [216] | |
| cheese | [217] | |
| RapID Yeast Plus system | citrus juice | [199] |
| Indonesian traditional fermented buffalo milk | [201] | |
| yoghurt | [200] | |
| VITEK 2 system with ID-YST database | traditional Turkish fermented sausage | [218] |
| medombae (rice wine) | [219] | |
| goat milk | [220] | |
| YeastIdent-Food kit/ProleFood system | foodborne yeasts | [221] |
| Phoenix Yeast ID | carrot, parsley | [215] |
| rDNA | Primer | Feature | Sequence (5′-3′) | Reference |
|---|---|---|---|---|
| SSU | BITS | forward | ACCTGCGGARGGATCA | [262] |
| ITS1 | forward | TCCGTAGGTGAACCTGCGG | [249] | |
| ITS1-F | forward | CTTGGTCATTTAGAGGAAGTAA | [263] | |
| ITS1-F_KY02 | forward | TAGAGGAAGTAAAAGTCGTAA | [264] | |
| ITS5 | forward | GGAAGTAAAAGTCGTAACAAGG | [249] | |
| 5.8S | B58S3 | reverse | GAGATCCRTTGYTRAAAGTT | [262] |
| ITS2 | reverse | GCTGCGTTCTTCATCGATGC | [249] | |
| ITS2_KY02 | reverse degenerate | TTYRCTRCGTTCTTCATC | [264] | |
| ITS3 | forward | GCATCGATGAAGAACGCAGC | [249] | |
| ITS3_KY02 | forward degenerate | GATGAAGAACGYAGYRAA | [264] | |
| LSU | ITS4 | reverse | TCCTCCGCTTATTGATATGC | [249] |
| ITS4_KY01 | reverse degenerate | TCCTCCGCTTWTTGWTWTGC | [264] | |
| NL1 | forward | GCATATCAATAAGCGGAGGAAAAG | [250] | |
| NL4 | reverse | GGTCCGTGTTTCAAGACGG | [250] | |
| LS2 | reverse | ATTCCCAAACAACTCGACTC | [245] | |
| LS2-MF | forward | GAGTCGAGTTGTTTGGGAAT | [265] |
| Year | Food | DNA Extraction | PCR: Amplified Region (Used Primer Pair: Forward-Reverse) | Sequencing Technique | Identification Database | Pre-Grouping and/or Typing Techniques | Reference |
|---|---|---|---|---|---|---|---|
| 2024 | fruit and vegetable biowastes | ns-ref a | ITS1-5.8S-ITS2 region (ITS1-ITS4) | Sanger | NCBI database, MycoBank database | np b | [248] |
| musts and grapes | conventional nucleic acid extraction techniques | ITS1-5.8S-ITS2 region (ITS1-ITS4) | Sanger | Basic Local Alignment Search Tool (wnm c) | Pre-grouping: ITS-RFLP | [257] | |
| cheese | conventional nucleic acid extraction techniques | ITS1-5.8S-ITS2 region (ITS1-ITS4) | Sanger | NCBI database | Pre-grouping: RAPD-PCR (primers: P1, P2) | [237] | |
| irradiated ready-to-eat chicken feet | TIANcombi DNA Lyse & Det PCR Kit (Tiangen Biotechnology Co., Ltd.) | ITS1-5.8S-ITS2 region (ITS1-ITS4) | scs d | NCBI database | np | [274] | |
| boza | EasyPure® Genomic DNA Kit (TransGen Biotech Co., Ltd.) | 26S rDNA gene (F63-LR3) | ns e | NCBI database | np | [275] | |
| sourdough | Wizard® Genomic DNA Purification Kit (Promega Corp.) | D1/D2 domain (NL1-NL4) | scs | NCBI database | np | [276] | |
| baijiu | ALFA-Soil DNA Extraction Mini Kit (Guangzhou Cellgene Biotechnology Co., Ltd.) | ITS2 (ITS2-ITS3) | Illumina Nova6000 platform | NCBI database | np | [277] | |
| fermented milk | Fungal Genomic DNA Extraction Kit (Tiangen Biotechnology Co., Ltd.) | D1/D2 domain (NL1-NL4) | scs | NCBI database | np | [278] | |
| beer, cider | conventional nucleic acid extraction techniques | ITS (ns-ref) | Genetic Analyser (ABI 3130xl) | NCBI database, MycoBank database | np | [279] | |
| table olives | conventional nucleic acid extraction techniques | D1/D2 domain (NL1-NL4) | scs | NCBI database | Pre-grouping: rep-PCR (primer: (GTG)5) | [238] | |
| 2023 | black olives | ns-ref | ITS1-5.8S-ITS2 region (ITS1-ITS4) | scs | NCBI database | Pre-grouping: rep-PCR (primer: (GTG)5); ITS-RFLP | [239] |
| beer | ns-ref | ITS1-5.8S-ITS2 region (ITS1-ITS4); D1/D2 domain (NL1-NL4) | scs | NCBI database | Interspecies hybrids identification: PCR-RFLP (analysed genes: GSY1, UBP7, MAG2) (Saccharomyces sp.); ITS-RFLP (Meyerozyma sp.) Typing: mtDNA-RFLP (Saccharomyces sp.) | [240] | |
| grape | ns-ref | ITS1-5.8S-ITS2 region (ITS1-ITS4) | scs | NCBI database | np | [280] | |
| grape | ns-ref | D1-D2 loop (ns-ref) | scs | NCBI database | Pre-grouping: ITS-RFLP Typing: rep-PCR (primer: (GTG)5); Interdelta analysis (δ-PCR) (S. cerevisiae) | [241] | |
| grape | ns | D1/D2 domain (NL1-NL4) | Sanger | NCBI database | np | [281] | |
| cow-milk bryndza cheese | ns-ref | ITS1-5.8S-ITS2 region (ITS1-ITS4) D1/D2 domain (NL1-NL4) | ns | NCBI database | Interspecies diversity: MLST analysis (analysed genes: ALA1, CDC19, GLN4, PGI1, PGM2, YSP3) (G. candidus) | [242] | |
| Sherry wine | conventional nucleic acid extraction techniques | ITS1 and ITS2 regions, ITS5 region, D1/D2 domain (ns-ref, provided in the original article) | scs | NCBI database | np | [247] | |
| pellicle-forming radish paocai | Fungal DNA Extraction Kit (Tsingke Biotechnology Co., Ltd.) | ITS1-5.8S-ITS2 region (ITS1-ITS4) D1/D2 domain (NL1-NL4) | scs | NCBI database | np | [282] | |
| 2022 | kombucha | MasterPure™ Yeast DNA Purification Kit (Biosearch Technologies) | D1/D2 domain (NL1-NL4) | scs | NCBI database | Pre-grouping: ITS1-5.8S-ITS2 region size; ITS-RFLP (Saccharomyces sp.) | [154] |
| sourdough | conventional nucleic acid extraction techniques | ITS1-5.8S-ITS2 region (ITS1-ITS4) | scs | NCBI database | Pre-grouping: Start Codon Targeted (SCoT) Polymorphism marker system | [283] | |
| olive brine | Fungi/Yeast Genomic DNA Isolation Kit (Norgen Biotek Corp.) | D1/D2 domain (NL1-NL4) | ns | NCBI database | Pre-grouping: ITS-RFLP | [258] | |
| orange juice | conventional nucleic acid extraction techniques | D1/D2 domain (NL1-NL4) | Genetic Analyzer (ABI 3130) | NCBI database | Pre-grouping: PCR fingerprinting (primers: (GTG)5, M13) | [284] | |
| grape; must | ns-ref | ITS-RFLP; D1/D2 domain (ns-ref) | ns (D1/D2) | ns | Typing: Interdelta analysis (δ-PCR) (S. cerevisiae) | [243] | |
| soy sauce | ns-ref | ITS-RFLP; D1/D2 domain (NL1-NL4) | ns (D1/D2) | NCBI database | np | [285] | |
| fermenting cocoa beans | conventional nucleic acid extraction techniques | ITS1-5.8S-ITS2 region (ITS1-ITS4) ACT1 gene (CA1-CA5r; CA21-CA22r; Act1-f-CA22r) D1/D2 domain (NL1-NL4) | scs | Basic Local Alignment Search Tool (wnm) Ribosomal Database Project (RDP) | np | [286] | |
| ganjang | conventional nucleic acid extraction techniques | whole-genome sequencing | PacBio Sequel SMRT sequencer | Pairwise sequence alignment tool LAST (lastal, ver. 1060) | np | [246] | |
| olive | conventional nucleic acid extraction techniques | D1/D2 domain (ns-ref) | ns | NCBI database | Pre-grouping: RAPD-PCR (primer: M13) | [287] | |
| mix bases for ice cream | ns | D1/D2 domain (NL1-NL4) | scs | NCBI database | Pre-grouping: DGGE analysis of D1 domain (NL1-LS2) | [288] | |
| coffee fermentation | ns-ref | ITS1-5.8S-ITS2 region (ITS1-ITS4) | scs | NCBI database | np | [232] | |
| 2021 | mould-matured Turkish cheese | UltraClean™ microbial DNA isolation kit (Mo Bio Laboratories) | ITS1-5.8S-ITS2 region (ITS1-ITS4) | scs | NCBI database | Typing: rep-PCR (ns) | [244] |
| mango | ns | ITS1-5.8S-ITS2 region (ITS1-ITS4) | ns | NCBI database | np | [289] | |
| olives | conventional nucleic acid extraction techniques | D1/D2 domain (ns-ref) | ns | ns | np | [290] | |
| fermented honey by-products | InstaGene™ Matrix (Bio-Rad Laboratories) | D1/D2 domain (ns-ref) | ns | NCBI database | Pre-grouping: ITS-RFLP Typing: RAPD-PCR (primers: M13, P80); Tandem Repeat tRNA (TRtRNA) PCR | [259] | |
| honey | conventional nucleic acid extraction techniques | D1/D2 domain (NL1-NL4) | Genetic Analyzer (3500 series) | NCBI database | np | [291] | |
| cocoa | conventional nucleic acid extraction techniques | D1/D2 domain (NL1-LS2) | scs | NCBI database | Typing: ITS-RFLP | [260] | |
| Vino cotto | ns-ref | ITS-RFLP; D1/D2 (NL1-NL4) | scs | NCBI database, Ribosomal Database Project (RDP) | Typing: RAPD-PCR (primer: M13) | [252] | |
| red wines ageing in oak barrels | conventional nucleic acid extraction techniques | ITS-RFLP; ITS1-5.8S-ITS2 region (ITS1-ITS4) | ns (B. bruxellensis) | yeast-id.org database | Typing: Intron Splice Site PCR (ISS-PCR); Microsatellite analysis (ns-ref) | [253] | |
| cheese | ns | D1/D2 domain (NL1-NL4) | ns | NCBI database, SILVA database | np | [231] | |
| Tibetan kefir grains | TIANamp Yeast DNA Kit (Tiangen Biotechnology Co., Ltd.) | D1/D2 domain (NL1-NL4) | Sanger | Basic Local Alignment Search Tool (wnm) | np | [292] | |
| 2020 | chicha | conventional nucleic acid extraction techniques | ITS-RFLP | non-sequencing based method | yeast-id.org database | Typing: mtDNA-RFLP; Interdelta analysis (δ-PCR) (S. cerevisiae); Fingerprint analysis (primer: TdPIR3) (T. delbrueckii) | [254] |
| D1/D2 region (NL1-NL4) | Sanger | NCBI database | |||||
| tequila (fermentation) | conventional nucleic acid extraction techniques | ITS-RFLP | non-sequencing based method | yeast-id.org database, Ribosomal Database Project (RDP) | np | [255] | |
| D1/D2 region (NL1-NL4) | ns | NCBI database | |||||
| Sherry wines | conventional nucleic acid extraction techniques | ITS-RFLP | non-sequencing based method | yeast-id.org database | Typing: Microsatellite multiplex PCR (analysed loci: SC8132X, YOR267C, SCPTSY7) (flor genotypes belonging to S. cerevisiae) | [256] | |
| ITS1-5.8S-ITS2 region (ITS1-ITS4) | EZ-sequencing | EMBL database, CBS database | |||||
| grape must fermentations | ns-ref | ITS1-5.8S-ITS2 region (ITS1-ITS4) | scs | NCBI database | np | [293] | |
| laphet | conventional nucleic acid extraction techniques | ITS1-5.8S-ITS2 region (ITS1-ITS4) | ns | NCBI database, Ribosomal Database Project (RDP) | Pre-grouping: ITS-RFLP Typing: RAPD (primers: M13, P80); Tandem Repeat tRNA (TRtRNA) PCR | [261] | |
| coffee beans | conventional nucleic acid extraction techniques | ITS1-5.8S-ITS2 region (ITS1-ITS4) | scs | NCBI database | np | [294] | |
| sourdoughs | Wizard® Genomic DNA Purification Kit (Promega Corp.) | D1/D2 domain (NL1-NL4) | Genetic Analyzer (ABI 3730) | NCBI database | np | [295] |
| Year | Food | DNA Extraction | PCR: Amplified Region (Used Primer Pair: Forward-Reverse); Used Enzyme/Kit (If Specified) | Sequencing Technique | Identification Database | Reference |
|---|---|---|---|---|---|---|
| 2024 | fruit and vegetable biowastes | Combination of conventional nucleic acid extraction techniques and QIAamp DNA Stool Mini Kit (Qiagen) | ITS2 (ITS2f-ITS2r) (Modified primers possibly developed by the authors); Advantage Polymerase Mix (Clontech) (Modified primers. In this study?) | 454 Life Sciences | UNITE database | [248] |
| cheese | E.Z.N.A.® Soil DNA Kit (Omega Bio-tek) | D1/D2 domain (ns-ref a) | Illumina MiSeq | internal database for fungi [265]; Basic Local Alignment Search Tool | [296] | |
| irradiated ready-to-eat chicken feet | MagAttract PowerSoil Pro DNA Kit (Qiagen) | ITS1 region (ITS1-F-ITS2) (The primer names given in the article have been corrected based on the sequence provided.) | ns b | RDP classifier; Unite database | [274] | |
| daqu (indispensable starter) | E.Z.N.A.® Soil DNA Kit (Omega Bio-tek) | ITS1 region (ITS5-ITS1) (Both specified primers are forward primers. The sequence of primers is not given.) | Illumina NovaSeq | UNITE database | [297] | |
| Baijiu | E.Z.N.A.® Soil DNA Kit (Omega Bio-tek) | ITS1 region (ITS1-ITS2) | Illumina MiSeq | ns | [298] | |
| green table olive | PowerFood™ Microbial DNA Isolation Kit (MoBio) | ITS1 region (ITS1-F_KYO2-ITS2_KYO2); KAPA HiFi HotStart ReadyMix (Roche) | Illumina MiSeq | UNITE database | [299] | |
| natural whey starters (used in cheese making) | DNeasy PowerFood Microbial Kit (Qiagen) | ITS2 region (ITS3-ITS4_KYO1) (The forward primer name given in the article has been corrected based on the sequence provided.); Taq polymerase (Q Biogene) | Illumina MiSeq | UNITE database | [300] | |
| grape berry and juice | DNeasy PowerSoil Kit (Qiagen) | ITS1 (ns) | lllumina MiSeq | UNITE database | [301] | |
| table olives | MasterPure™ Complete DNA and RNA Purification Kit (Biosearch Technologies) | D1 domain (LS2-MF-NL4) (The reverse primer name given in the article has been corrected based on the sequence provided.) | lllumina MiSeq | NCBI database, SILVA database | [238] | |
| 2023 | black olives | ns-ref | D1/D2 domain (LS2 and NL4MS) (LS2 is a reverse primer. NL4 is a reverse primer too. In the referred article there is no NL4MS primer.) | Illumina MiSeq | NCBI database, SILVA database | [239] |
| coffee fermentation | DNeasy PowerLyzer PowerSoil Kit (Qiagen) | ITS2 region (ITS3F-ITS4R) (The primer sequences are not given, no reference is provided. Probably ITS3 and ITS4 primers.) | Illumina MiSeq | UNITE database | [302] | |
| kombucha | E.Z.N.A.® Soil DNA Kit (Omega Bio-tek) | ITS1 region (ITS5-ITS2); Fast Pfu DNA Polymerase | Illlumina NovaSeq | UNITE database | [303] | |
| baijiu | FastDNA™ SPIN Kit for Soil (MP Biomedical) | ITS region (ITS4-ITS9) (ITS9 primer sequence or reference is not provided. It is not clear which region was exactly amplified.); DNA polymerase (TransGen Biotech) | PacBio | UNITE database | [266] | |
| lambic beer | Combination of conventional nucleic acid extraction techniques and DNeasy Blood and Tissue Kit (Qiagen) | ns-ref | Ion Torrent | in-house database containing a representative genome sequence | [233] | |
| hongqu rice wines | FastDNA™ SPIN Kit for Soil (MP Biomedical) | ITS-5.8S rRNA gene (ITS-F-ITS-R) (The primer sequence is given, but no reference. Not clear whether their own primer was developed or not.) | PacBio | ns | [267] | |
| caciofiore cheese | E.Z.N.A.® Soil DNA Kit (Omega Bio-tek) | D1/D2 region (ns-ref) | Illumina MiSeq | SILVA database, Basic Local Alignment Search Tool | [304] | |
| ciauscolo PGI salami | Quick-RNA Miniprep Kit (Zymo Research) | D1 domain (ns-ref) | Illumina MiSeq | internal database for fungi [265]; NCBI database | [305] | |
| 2022 | kombucha | DNeasy PowerSoil Kit (Qiagen) | D1/D2 domain (NL1-LS2); PCR-DGGE (separation, extraction reamplification without GC clamp) | scm | NCBI database | [154] |
| different cheeses | Combination of conventional nucleic acid extraction techniques and DNeasy Blood and Tissue Kit (Qiagen) | D1/D2 domain (NL1-NL4) | Sanger | NCBI database | [306] | |
| grape; must | MasterPure™ Complete DNA and RNA Purification Kit (Biosearch Technologies) | D1 domain (ns-ref); Kapa HiFi HotStart ReadyMix (Roche) | Illumina MiSeq | internal database for fungi [265] | [243] | |
| soy sauce | ns-ref | ITS region (ns) | Illumina HiSeq | ns | [285] | |
| olive | DNeasy PowerSoil Kit (Qiagen) | ITS2 region (ITS3-ITS4); AccuPrime™ Taq DNA polymerase system (Invitrogen) | Illumina MiSeq | ns | [287] | |
| coffee fermentation | QIAamp DNA Mini Kit (Qiagen) | ITS1 region (ITS1-ITS2) | Illumina MiSeq | ns | [232] | |
| table olive | PowerFood™ Microbial DNA Isolation Kit (MoBio) | ITS1 region (ITS1-F_KYO2-ITS2_KYO2); KAPA HiFi HotStart ReadyMix (Roche) | Illumina MiSeq | UNITE database | [307] | |
| huangshui (a byproduct of baijiu fermentation) | E.Z.N.A.® Soil DNA Kit (Omega Bio-tek) | ITS1 region (ITS5-ITS2) (The primer names given in the article have been corrected based on the sequence provided.) | Illumina NovaSeq | UNITE database | [308] | |
| grape | E.Z.N.A.® Soil DNA Kit (Omega Bio-tek) | D1/D2 domain (LS2-MF-NL4) (The reverse primer name given in the article has been corrected based on the sequence provided.) | Illumina MiSeq | SILVA database, internal 26S database for fungi [265], Basic Local Alignment Search Tool | [309] | |
| 2021 | grape, must | FastDNA™ SPIN Kit for Soil (MP Biomedical) | ITS1 region (ITS1-F-ITS2); FastStart Master Mix (Roche) | Illumina MiSeq | UNITE database; Ribosomal Database Project (RDP), NCBI database | [310] |
| beer, cider | DNeasy PowerLyzer Microbial Kit (Qiagen) | ITS1 region (BITS-B58S3) | Illumina MiSeq | NCBI database | [153] | |
| cocoa | DNeasy PowerLyzer PowerSoil (Qiagen) | ITS2 region (ITS3-ITS4) (The primer names given in the article have been corrected based on the sequence provided.) | Illumina MiSeq | UNITE database | [311] | |
| cheese | DNeasy Blood and Tissue Kit (Qiagen) with a supplementary initial enzymatic lysis | ITS2 region (ITS3f-ITS4_Kyo1) (The primer sequences are not given. In the referred article there is no ITS3f primer, only ITS3.) | Illumina MiSeq | UNITE database | [231] | |
| ready-to-eat pineapple | conventional nucleic acid extraction techniques | ITS2 region (ITS3- ITS4); AccuPrime™ Taq DNA polymerase system (Invitrogen) | Illumina MiSeq | UNITE database, Basic Local Alignment Search Tool | [312] | |
| Tibetan kefir grains | PowerFood™ Microbial DNA Isolation Kit (MoBio) | ITS1 region (ITS1-ITS2) (The primer names given in the article have been corrected based on the sequence provided.) | Illumina MiSeq | RDP classifier, UNITE database | [292] | |
| cheese | E.Z.N.A.® Soil DNA Kit (Omega Bio-tek) | D1 domain (ns-ref) | Illumina MiSeq | internal database for fungi [265]; Basic Local Alignment Search Tool | [313] | |
| beer | ns-ref | ITS1 region (BITS-B58S3); Titanium® Taq DNA polymerase (Takara Bio) | Illumina MiSeq | UNITE database | [152] | |
| cheese | ZymoBIOMICS DNA Miniprep Kit (Zymo Research) | ITS1 region (BITS-B58S3) | Illumina MiSeq | UNITE database | [314] | |
| 2020 | tequila (fermentation) | PowerFood™ Microbial DNA Isolation Kit (MoBio) | D1/D2 domain (NL1-LS2); TaKaRa Ex Taq® DNA Polymerase (Takara Bio); PCR-DGGE (separation, extraction reamplification without GC clamp) | scm | NCBI database | [255] |
| ITS-5.8S region (ITS1-ITS4) | Illumina MiSeq | Ribosomal Database Project (RPD) | ||||
| laphet | DNeasy PowerFood Microbial Kit (Qiagen) | D1/D2 domain (NL1-LS2); TaKaRa Ex Taq® DNA Polymerase (Takara Bio); PCR-DGGE (separation, extraction reamplification without GC clamp) | scm | ns | [261] | |
| ITS1 region (ITS1-F-ITS2) | Illumina MiSeq | UNITE database | ||||
| coffee beans | Combination of conventional nucleic acid extraction techniques and DNeasy Blood and Tissue Kit (Qiagen) | ITS1 region (ITS1-F -ITS2) (The forward primer name given in the article has been corrected based on the sequence provided.); AmpliTaq Gold™ 360 DNA Polymerase (Applied Biosystems) | Illumina MiSeq | UNITE database | [294] | |
| bean paste | ZymoBIOMICS DNA Miniprep Kit (Zymo Research) | ITS2 region (ITS3_KYO2-ITS4) | Illumina HiSeq | UNITE database | [315] | |
| wine | E.Z.N.A™ Mag-Bind Soil DNA Kit (Omega Bio-tek) | ITS1-ITS2 fragment (ITS1F -ITS2R) (The primer sequences are not given, no reference is provided. The primers used cannot be identified.) | Illumina MiSeq | UNITE database | [316] | |
| honey | DNeasy PowerSoil DNA Isolation Kit (Qiagen) | ITS2 region (ITS-u3-ITS-u4); Phusion™ High-Fidelity DNA Polymerase (NEB) | Illumina (platform not specified) | UNITE database | [317] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kovács, M.; Pomázi, A.; Taczman-Brückner, A.; Kiskó, G.; Dobó, V.; Kocsis, T.; Mohácsi-Farkas, C.; Belák, Á. Detection and Identification of Food-Borne Yeasts: An Overview of the Relevant Methods and Their Evolution. Microorganisms 2025, 13, 981. https://doi.org/10.3390/microorganisms13050981
Kovács M, Pomázi A, Taczman-Brückner A, Kiskó G, Dobó V, Kocsis T, Mohácsi-Farkas C, Belák Á. Detection and Identification of Food-Borne Yeasts: An Overview of the Relevant Methods and Their Evolution. Microorganisms. 2025; 13(5):981. https://doi.org/10.3390/microorganisms13050981
Chicago/Turabian StyleKovács, Mónika, Andrea Pomázi, Andrea Taczman-Brückner, Gabriella Kiskó, Viktória Dobó, Tamás Kocsis, Csilla Mohácsi-Farkas, and Ágnes Belák. 2025. "Detection and Identification of Food-Borne Yeasts: An Overview of the Relevant Methods and Their Evolution" Microorganisms 13, no. 5: 981. https://doi.org/10.3390/microorganisms13050981
APA StyleKovács, M., Pomázi, A., Taczman-Brückner, A., Kiskó, G., Dobó, V., Kocsis, T., Mohácsi-Farkas, C., & Belák, Á. (2025). Detection and Identification of Food-Borne Yeasts: An Overview of the Relevant Methods and Their Evolution. Microorganisms, 13(5), 981. https://doi.org/10.3390/microorganisms13050981






