Abstract
A strictly anaerobic bacterium denoted as strain NIT-TF6 of the genus Desulfitobacterium was isolated from a trichloroethene-dechlorinating culture with formate. Cells were straight rods of 1.6–6 µm long and 0.25–0.5 µm in diameter and used H2, lactate, pyruvate, and malate as electron donors and thiosulfate and Fe (III)-citrate as electron acceptors. The genome of strain NIT-TF6 was 4.8 Mbp in size and included nine 16S rRNA genes. Phylogenetic analysis based on 16S rRNA sequences showed that NIT-TF6 shared the highest sequence similarity (96.39%) with Desulfitobacterium hafniense DCB-2ᵀ, forming an independent clade in the phylogenetic tree. Digital DNA-DNA hybridization (dDDH) and average nucleotide identity (ANI) values between strain NIT-TF6 and other Desulfitobacterium species ranged from 15.9 to 16.9% and from 71.68 to 72.51%, respectively. These are well below the thresholds for species delineation. A distinguishing feature of strain NIT-TF6 was its possession of both L-lactate dehydrogenase (L-LDH) and D-lactate dehydrogenase (D-LDH), in contrast to other Desulfitobacterium strains that exclusively express D-LDH. Based on the dDDH and ANI results, combined with physiological, phylogenetic, morphological, biochemical, genomic, and metabolic iron-related characteristics, strain NIT-TF6 has been proposed as a novel species within the genus Desulfitobacterium. The name Desulfitobacterium elongatum sp. nov. has been proposed for this strain, with NIT-TF6ᵀ designated as the type strain.
1. Introduction
Desulfitobacterium belongs to the Bacillota and plays a key role in bioremediation by using organohalides such as chlorinated ethenes (CEs), chloroethanes, and chlorinated phenolic compounds. It consists of three validated species, that is, D. hafniense DCB-2T [], D. dehalogenans JW/IU-DC1T [], D. metallireducens 853-15T [], and the yet-to-be validated “D. dichloroelimans DCA1” []. The genus also previously included D. aromaticivorans. However, this species has been reclassified as Paradesulfitobacterium aromaticivorans []. In terms of their valid species, D. chlororespirans DSM 11544 is a scaffold genome, and D. frappieri ATCC 700357 has been merged under the first described name, namely, D. hafniense. Desulfitobacterium species have been isolated from municipal sludge, methanogenic lake sediment, compost soil, uranium-contaminated aquifer sediment, and the soil matrix of an anoxic water-saturated layer, respectively. They are strictly anaerobic, rod-shaped, and spore-forming bacteria that have versatile energy conversions, including fermentation and respiration using a wide range of electron donors and acceptors. These valid strains confirmed their heterotrophic growth by gaining energy through versatile respiration with Fe (III) and nitrate, coupling with hydrogen or organic acids.
Recently, the genus has been receiving considerable attention in supporting Dehalococcoides, a representative organohalide-respiring bacterium that can fully dechlorinate trichloroethene (TCE) to non-toxic ethene [,,]. In our previous study, a microcosm (FOR) of groundwater supplemented with formate was reported to dechlorinate TCE to ethene efficiently in the augmentation of Dehalococcoides. The FOR includes Dehalococcoides, Methanosphaerula, Rectinema, and Desulfitobacterium. It showed more rapid and efficient dechlorination compared to microcosms supplemented with other C2- and C3-organic acids, and absence of Desulfitobacterium []. Dehalococcoides use H2 and acetate as the sole electron donor and carbon source, respectively, and require cobalamin as a cofactor [,,]. These are generally supplied by heterotrophs fermenting organic matter in the environment. Desulfitobacterium possesses genes for formate hydrogenlyase, which converts formate into H2 and CO2 []. It also produces cobalamin for Dehalococcoides [] and carries the complete genome for the acetyl-CoA (Wood–Ljungdahl) pathway. This can convert CO2 into acetate via an autotrophic pathway through formate as an intermediate [,].
So far, the contributions of Desulfitobacterium, particularly in supporting TCE-to-ethene dechlorination via formate use, remain unclear. Therefore, further research is required. In this study, a novel strain NIT-TF6 of Desulfitobacterium was isolated from the FOR culture, and its potential role was clarified using physiological and whole-genomic analysis of the strain. It was compared with D. hafniense [], D. dehalogenans [], D. metallireducens [], and “D. dichloroeliminans” [].
2. Materials and Methods
2.1. Isolation and Growth Conditions
Desulfitobacterium elongatum NIT-TF6 was isolated from a FOR culture containing Dehalococcoides mccartyi NIT01. This was originally obtained from an enriched culture designated YN3 []. The inoculum used to prepare the YN3 culture was sediment from the Arako River, Nagoya, Japan []. The DS-basal medium used for isolating and cultivating the strain consisted of 20 g/L NaCl, 0.3 g/L KCl, 0.5 g/L NH4Cl, 0.1 g/L CaCl2·2H2O, 4 g/L MgCl2·6H2O, 0.6 g/L KH2PO4, 2.5 g/L NaHCO3, 1 mL/L SL-10, 1 mL/L Se/W solution, vitamin solution (per L consisted of 20 mg D-biotin, 20 mg folic acid, 100 mg pyridoxine-HCl, 50 mg vitamin B1 hydrochloride, 50 mg riboflavin, 50 mg nicotinic acid, 50 mg D-panthothenic acid calcium salt, 75 mg vitamin B12, 50 mg p-aminobenzoic acid, 50 mg nicotinamide, 50 mg lipoic acid, 50 mg hemin and 50 mg 1, 4-naphthoquinone), and 0.2 mg/L resazurin (all purchased from and manufactured in Japan). The basal medium was prepared anaerobically by flushing with a N2:CO2 gas mixture (80:20, v/v) []. NIT-TF6 was isolated using an anaerobic DS-FT agar-shake medium that comprises DS-basal medium supplemented with 10 mM lactate and 10 mM thiosulfate, and 1.5% agarose. After incubating for 7–14 d at 28 °C, the colony was purified through repeated agar-shake cultivation and re-cultivated in liquid DHB-CO3-LT medium. This comprises DHB-CO3 medium supplemented with 10 mM lactate and 10 mM thiosulfate. The purified culture was phylogenetically identified based on the 16S rRNA gene sequence amplified from cell lysates [] and designated as strain NIT-TF6. Routine cultivation of NIT-TF6 was performed in liquid DHB-CO3-LT medium, with 7–14 d of anaerobic incubation at 28 °C required for complete growth. The source of the reference strains D. dehalogenans ATCC 51507T, “D. dichloroeliminans” DCA1, D. hafniense DCB-2T, D. metallireducens DSM 15288T were methanogenic lake sediment, the soil matrix of a 1 m deep, anoxic, water-saturated layer that had been continuously contaminated with 1,2-dichloroethane for 30 years beneath an industrial storage tank, municipal sludge, and uranium-contaminated aquifer sediment.
2.2. Physiological, Morphological, and Biochemical Analyses
Acetate, fumarate, pyruvate, lactate, propionate, butyrate, methanol, ethanol, isopropanol, benzoate (all at 10 mM), and H2+acetate (6% v/v and 10 mM) were tested as potential electron donors for thiosulfate reduction. The suitability of various electron acceptors was evaluated by monitoring growth and detecting the oxidation of 10 mM pyruvate in the presence of thiosulfate, nitrate, nitrite, Fe (III) citrate, and air (1%) []. The morphology of Desulfitobacterium elongatum was examined using scanning electron microscopy (JSM-7800F; JEOL Ltd., Tokyo, Japan) at an operating voltage of 1.0 kV []. Gram-staining data were procured from Rapid Annotation using Subsystem Technology (RAST) server (https://rast.nmpdr.org/, accessed on 9 February 2025).
2.3. Sanger Sequencing of 16S rRNA Gene and Genome Pyrosequencing
Genomic DNA was extracted from Desulfitobacterium elongatum, and sanger sequencing was performed []. The sequencing was conducted using the Illumina MiSeq and Nanopore MinION platforms. A total of 1.8 million Illumina paired-end reads (1.02 Gbp, 150 bp × 2) and 0.37 million Nanopore reads (1.76 Gbp) were processed for error correction using Short Read Manager and assembled with Unicycler (v0.4.7, https://github.com/rrwick/Unicycler/releases/tag/v0.4.7, accessed on 9 February 2025). The complete genome of NIT-TF6 was successfully assembled, and gene prediction and annotation were performed using DFAST (https://dfast.ddbj.nig.ac.jp/, accessed on 9 February 2025) []. Comparative genomic analysis between NIT-TF6 and other Desulfitobacterium species was conducted using bidirectional best hits with a threshold of 40% identity and 80% query coverage, RAST (Rapid Annotation using Subsystem Technology) (http://rast.nmpdr.org/rast.cgi, accessed on 9 February 2025) [], and the BLAST 2.17.0 (http://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 9 February 2025) tool from the NCBI database. The complete genome sequence of strain NIT-TF6 was deposited in DDBJ/GenBank under the BioProject number PRJDB19082 with accession number AP038772 (BioSample identifier: SAMD00830004).
2.4. Phylogenetic Analysis Based on 16S rRNA Gene and Core Genome
The full-length 16S rRNA gene sequence of Desulfitobacterium elongatum was extracted from the complete genome data. Those for all valid strains of Desulfitobacterium spp. were downloaded from the RAST database. Phylogenetic analyses were performed using MEGA X (v8, Max Planck Institute of Biochemistry (MPIB), Planegg, Germany) (https://www.megasoftware.net/, accessed on 9 February 2025) based on the neighbor-joining method and maximum composite likelihood model []. Statistical support for the branches of the phylogenetic trees was determined using bootstrap analysis based on 1000 resamplings []. Digital DNA-DNA hybridization (dDDH) and ANI have replaced traditional DDH as the gold standard in prokaryote taxonomy for defining new species [,,]. The dDDH and ANI values were calculated using Genome-to-Genome Distance version 3.0 [] and the ANI calculator on the EzBioCloud platform [].
2.5. TB Tool II for Genome Analysis
TBtools-II (v1.106) from the Kyoto Encyclopedia of Genes and Genomes (KEGG) framework was used to construct a circular genome of Desulfitobacterium elongatum (v1.106, available at https://github.com/CJ-Chen/TBtools-Manual, accessed on 9 February 2025).
2.6. GC-FID Analysis for Dechlorination Experiments
Volatile compounds such as TCE, cis-DCE, VC, ethene, and methane were analyzed by injecting the headspace gas into a gas chromatograph (model GC-2014, Shimadzu, Kyoto, Japan) equipped with a Porapak Q column (GL Sciences, Tokyo, Japan) and a flame ionization detector (FID) [,].
2.7. GC-MS for Detection of 13C-Labeled Acetate
13C-labeled acetate was detected using Gas Chromatography–Mass Spectrometry (GC-MS). For this, a 1 mL sample solution was mixed with 4 mL ethyl acetate, and the separate solvent was recovered by the addition of anhydrous sodium sulfate. Helium was considered a carrier gas with splitless injection. The column oven and injection temperature were maintained at 50 °C and 280 °C, respectively. The column oven temperature increased to 80 °C [].
3. Results and Discussion
3.1. Isolation and Morphology of Strain NIT-TF6
Desulfitobacterium elongatum NIT-TF6 was isolated using deep agar cultivation supplemented with 10 mM lactate and 10 mM thiosulfate from the FOR microcosm. The isolated strain includes long, straight rod-shaped cells measuring 1.6–6 µm in length and 0.25–0.5 µm in diameter (Figure 1).
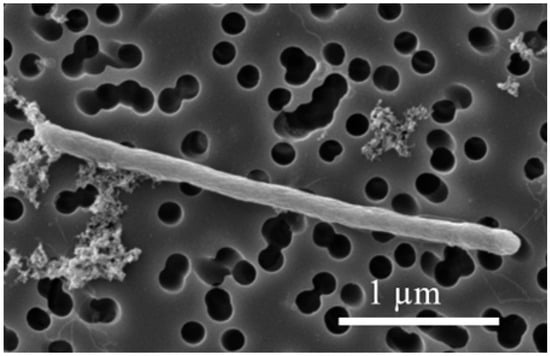
Figure 1.
Scanning electron microscopic images of spore-forming strain NIT-TF6 in the exponential phase.
3.2. Physiological Characteristics of Strain NIT-TF6
Desulfitobacterium elongatum NIT-TF6 can use various substrates, such as pyruvate, lactate, and H2+acetate, with thiosulfate as the electron acceptor (Table 1). Strain NIT-TF6 did not grow on thiosulfate respiration with a combination of acetate, fumarate, propionate, butyrate, methanol, ethanol, isopropanol, and benzoate. The substrates were chosen to represent a range of potential carbon sources and electron donors/acceptors that are relevant to the strain’s ecological niche. The selection was based on prior studies of related Desulfitobacterium species. Therefore, these substrates can cover the representative substrates that the strain can utilize.

Table 1.
Useful characteristics for differentiating strain NIT-TF6 from closely related members of the genus Desulfitobacterium.
3.3. Phylogenetic Analysis
Desulfitobacterium elongatum NIT-TF6 shared the highest sequence similarity with D. hafniense DCB-2T (96%) and formed a cluster with the closest relative and D. metallireducens DSM 15288T, D. dehalogenans ATCC 51507T, and “D. dichloroeliminans” DCA1. Except for the mentioned genera, strain NIT-TF6 had the highest sequence similarity with Desulfosporosinus orientis DSM 765T (94%). Based on these findings, strain NIT-TF6 likely represents a novel species within the family Desulfitobacteriaceae (Figure 2). A phylogenetic tree based on the whole genome of the strain NIT-TF6 and its close relatives, type strains within the Desulfitobacteriaceae family was considered based on TYGS (https://tygs.dsmz.de/, accessed on 7 January 2025) results (Figure S1—Supplementary Materials). However, the constructed phylogenetic tree is interchangeable within the species and subspecies level and does not necessarily represent similar data analyzed by 16S rRNA. This was clarified using dDDH and ANI analysis to identify species delineation.
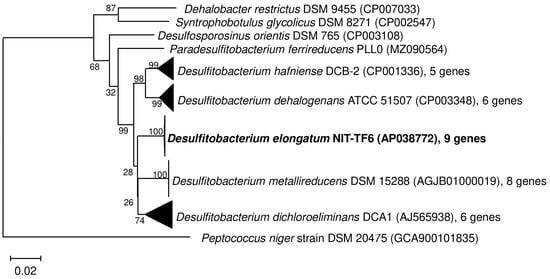
Figure 2.
16S rRNA gene sequence-based neighbor-joining phylogenetic tree of the strain NIT-TF6 and its close relatives, namely type strains within the Desulfitobacteriaceae family. Bar indicates 0.02 substitutions per nucleotide position.
3.4. General Genomic Feature
Desulfitobacterium elongatum NIT-TF6 contains a single circular chromosome consisting of 4,807,969 base pairs, 4655 protein-coding sequences (CDS), and 119 RNA sequences. Its G+C content is approximately 43.2 mol% and in the range observed in other Desulfitobacterium species (41.9–47.5%). Other genomic characteristics, such as genome size and CDSs, were consistent in the range of 3.1–5.2 Mb and 3138–5247, respectively, with previously reported values. However, strain NIT-TF6 contains nine copies of the 16S rRNA gene, which is higher than the typical range (5–8) reported in other Desulfitobacterium species (Table 2).

Table 2.
Genomic features of strain NIT-TF6 with other Desulfitobacterium valid strains. Column headers 1, 2, 3, 4, and 5 indicate the following: 1: Desulfitobacterium elongatum NIT-TF6; 2: D. dehalogenans ATCC 51507T []; 3: “D. dichloroeliminans” DCA1 []; 4: D. hafniense DCB-2T []; 5: D. metallireducens DSM 15288T [].
The recommended thresholds for classification of prokaryotic novel species are 60–70% for dDDH and 95–96% for ANI []. In this study, dDDH and ANI values between the strain NIT-TF6 and other Desulfitobacterium species were 15.9–16.9% and 71.68–72.51%, respectively. These are substantially below the widely accepted species threshold for identification. To further examine the functional annotation of genes associated with specific categories, a circular genome map of NIT-TF6 was constructed using TBtools-II (Figure 3). The highest gene numbers were associated with the metabolism of amino acids and their derivatives (148 genes). The amino acid metabolism dominance implicates that strain NIT-TF6 could sustain in an environment where amino acid is the primary energy source. This was followed by genes involved in carbohydrate metabolism (130 genes) that could support the growth phase of strain NIT-TF6 during nutrient-limiting conditions. Strain NIT-TF6 possesses genes related to the metabolism of cofactors and vitamins (80 genes) and energy metabolism (74 genes).
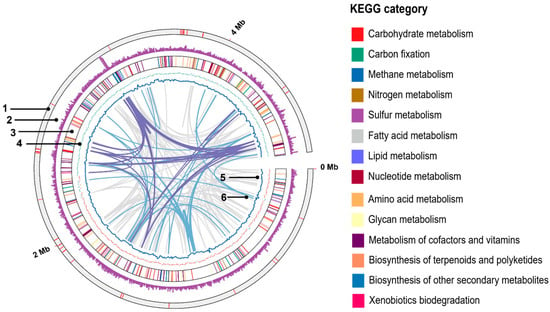
Figure 3.
Circular genome map of Desulfitobacterium elongatum NIT-TF6 illustrated with a circular representation generated using TBtools-II. The rings are numbered sequentially from the outside to the inside as follows: 1, location of c-type cytochromes; 2, gene density; 3, protein-coding sequences colored based on KEGG category; 4, G+C skew (red, positive; green, negative); 5, G+C content; 6, links showing repetitive sequence ≥95% identity (pink, >500 bp; purple, >2 kbp). KEGG, Kyoto Encyclopedia of Genes and Genomes.
3.5. Metabolic Potential and Unique Characteristics Based on the Genome
The genome of Desulfitobacterium elongatum NIT-TF6 contains genes encoding dehydrogenases for hydrogen (EC: 1.12.1.2), pyruvate (EC 2.7.1.40), malate (EC 1.1.1.38 and 1.1.1.40), and lactate. Fermentation of sugar and organic acids does not occur due to the absence of glucose transporter and acylphosphatase (EC: 3.6.1.7) for the substrate-level phosphorylation, respectively. The uniqueness in the genome for strain NIT-TF6 is the presence of genes coding L-lactate dehydrogenase (L-LDH, EC 1.1.1.27) in addition to another different type of L-lactate dehydrogenase complex (LldEFG, no EC number) []. Meanwhile, all other Desulfitobacterium species have only the gene of LldEFG (Figure 4). D-lactate dehydrogenase (D-LDH, EC 1.1.1.28) is not only present in D. metalllireducens DSM 15288T among Desulfitobacterium species. Various bacterial species can produce D-lactate or both D- and L-lactates. However, major enzymes transforming lactate, such as LldEFG and LDH, have isometric specificity with some exceptions []. The presence of two types of L-lactate dehydrogenase and D-LDH in strain NIT-TF6 increases the availability of lactate for energy conversion and assimilation.
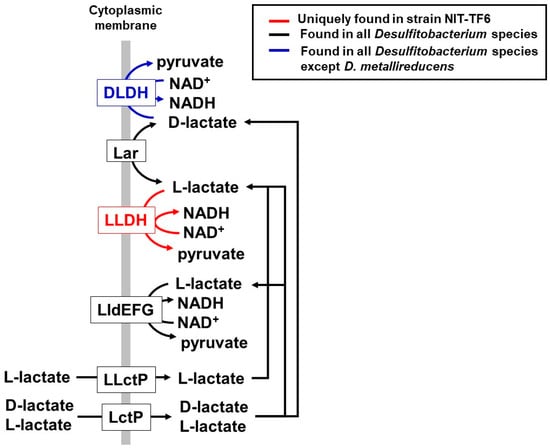
Figure 4.
Lactate metabolism-related genes found in the genome of NIT-TF6 and other Desulfitobacterium species. L-lactate dehydrogenase (L-LDH, EC: 1.1.1.27) is only found in Desulfitobacterium elongatum NIT-TF6; L-lactate permease (L-LctP, T.C.: 2.A.14.1.1), lactate permease (LctP, T.C.: 2.A.14), lactate racemase (Lar, EC: 5.1.2.1), and L-lactate dehydrogenase complex (LldEFG, no EC number) are found in all Desulfitobacterium species. D-LDH: D-lactate dehydrogenase (EC: 1.1.1.28) was found in all Desulfitobacterium species except D. metallireducens DSM 15288T.
Due to the strain NIT-TF6 not having fermentative growth, oxidative phosphorylation is the only energy conservation mechanism in the strain. The NAD(P)Hs released from e−donors initiate the electron transport chain, which results in proton motive force for ATP synthesis. The electron transport can be terminated via thiosulfate reduction to sulfite and sulfide, coupling the oxidization of menaquinol by thiosulfate reductase (EC 1.8.5.5). A set of genes for dissimilative sulfite reduction (dsrABC) is present in the genome of strain NIT-TF6 despite its inability to grow on sulfite as well as on all Desulfitobacterium that grow on sulfite. The reason why there is no sulfite reduction in the strain has not been clarified in this study. The gene coding ferredoxin-nitrite reductase (EC 1.7.7.1) was present, suggesting that nitrite can be transformed into ammonia. Regarding assimilation, carbon dioxide can be fixed with formate to acetate via the reductive acetyl-coA pathway. This pathway is commonly conserved in all Desulfitobacterium spp. and D. restrictus DSM 9455T.
3.6. Unique Menaquinone Synthesis Pathway Based on the Genome
The uniqueness in the genome contents and particularly in strain NIT-TF6 among Desulfitobacterium spp. in the assimilation was the menaquinone synthesis pathway. It was only the strain NIT-TF6 that had the most essential genes for the futalosine pathway (mqnABCDE) for menaquinone synthesis. However, all the other Desulfitobacterium mainly had the o-succinylbenzoate (OSB) pathway for synthesis (Figure 5). Besides Desulfitobacterium, Dehalobacter restrictus has OSB pathway-related genes. Meanwhile, Syntrophobotulus glycolicus, Desulfosporosinus orientis, and Paradesulfitobacterium ferrireducens had futalosine pathway-related genes. D. orientis also had partial genes for the OSB pathway. The absence of menF and mqnF in all strains was likely underestimated. This is because the identification system did not identify the gene as the target when the gene was most relevant to other genes, including showing sufficient e-values with the target. The differentiation of menaquinone biosynthesis has been reported to be an adaptation to the great oxygenation event. The futalosine pathway is the more ancient route, mostly found in aerobes and facultative anaerobes. Meanwhile, the OSB pathway is only found in aerobic, facultative anaerobic, and anaerobic prokaryotes. Differentiation in menaquinone synthesis among the Desulfitobacteriaceae family likely happened because of differentiation in genera concomitantly within the family.
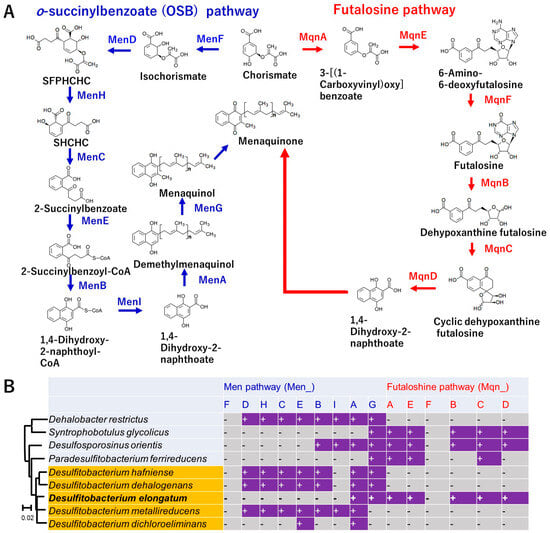
Figure 5.
Comparison of genes coding enzymes involved in the menaquinone synthesis among Desulfitobacteriaceae. Panel (A) indicates the names of genes with coding enzymes involved in menaquinone synthesis. Panel (B) indicates the list of genes present in strains of Desulfitobacteriaceae. The symbols “+” and “-” indicate the presence and absence of the genes in their genomes, respectively.
3.7. The RdhA Gene Cluster and Mechanism for the Inability to Dechlorinate Chloroethene
Genomic analysis showed the presence of the catalytic subunit of reductive dehalogenase (RdhA)—coding gene (rdhA)—in Desulfitobacterium elongatum NIT-TF6, which is closely related to pceA from Desulfitobacterium elongatum and shares 94–95% nucleotide identity with D. restrictus strains DSM 9455 and TeCB1, which have previously demonstrated the ability to reductively dechlorinate tetrachloroethene (PCE) to 1,2-cis-DCE (dichloroethane) and 1, 2, 4, 5-tetrachlorobenzene to 1,3- and 1,4-dichlorobenzene [,]. Besides rdhA, the membrane-bound subunit of reductive dehalogenase (rdhB) in NIT-TF6 had 98% identity with that of “D. dichloroeliminans DCA1”, and another membrane-bound subunit (rdhC) sharing 98% identity with D. restrictus and 96–97% identity with D. hafniense and “D. dichloroeliminans”. These reductive dehalogenase-related genes are present near a transposase-coding gene and are likely acquired through horizontal gene transfer as reported in other organohalide-respiring Desulfitobacterium.
The presence of full sets of Rdh-related genes in the genome suggested the organohalide respiration capability of the strain NIT-TF6. However, the strain NIT-TF6 did not dechlorinate tetrachloroethene, trichloroethene, 1,2-dichloroethene, and 1,2-cis-dichloroethene. Comparing amino acid sequences among strain NIT-TF6, Desulfitobacterium hafniense DCB-2, and D. restrictus DSM 9455 showed that the most important amino acid residues of the active site were identical among the three strains, except for one replacement of V333 from a non-polar amino acid to thiamine of polar amino acid. The replacement potentially inactivated the RdhA to dechlorinate CEs. Figure 6 illustrates amino acid sequences of NIT-TF6 and its closest relatives.
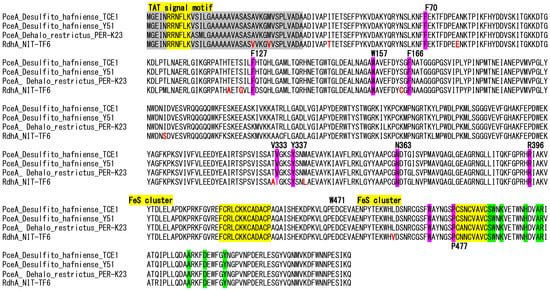
Figure 6.
Amino acid sequences between strain NIT-TF6, D. hafniense DCB-2T, and D. restrictus DSM 9455T. Red color indicates the difference in amino acid found only in strain NIT-TF6. The yellow region indicates the conserved domain of the TAT signal and Fe-S clusters. Pink-colored residues are important residues of the active site, while green indicates those for menaquinol binding sites.
3.8. Role of Strain NIT-TF6 in the TCE-Dechlorinating Culture
The genome suggests two potential roles for the Desulfitobacterium elongatum NIT-TF6 in supporting TCE dechlorination by Dehalococcoides in the microcosm: (1) participating in the dechlorination of chloroethenes in conjunction with Dehalococcoides, and (2) producing acetate through acetogenesis using formate and carbon dioxide. However, strain NIT-TF6 did not dechlorinate any chloroethenes or chloroethanes. Attempts to confirm the acetate formation from 13C-formate and 13C-carbonate were unsuccessful, as no significant increase in 13C-acetate was observed in cultures repeatedly transferred with 13C-labeled substrates. One potential role of strain NIT-TF6 could be the provision of H2 to Dehalococcoides. The strain was found to produce H2 from formate. However, this capability is not unique to Desulfitobacterium and is not a specialized trait of this genus. Therefore, the exact reason why Desulfitobacterium coexists with Dehalococcoides in the microcosm remains unclear based on the findings of this study.
4. Conclusions
Strain NIT-TF6, belonging to the genus Desulfitobacterium, was isolated from a TCE-dechlorinating microcosm using formate as an energy and carbon source. The exact reason for the coexistence of Desulfitobacterium with Dehalococcoides in the culture remains unclear based on physiological and genomic characterization. This is because of the apparent uniqueness in the genome of strain NIT-TF6, namely the presence of genes for L-LDH and the futalosine pathway of menaquinone synthesis. The strain exhibited 91.31–96.39% sequence similarity in the 16S rRNA gene to D. hafniense DCB-2T and shared 15.9% dDDH and 71.88% ANI with its closest relative. As the unique genomic characteristics and these values are below the thresholds for species delineation, strain NIT-TF6 is proposed as the type strain of a novel species, Desulfitobacterium elongatum sp. nov., with the type strain NIT-TF6T.
Description of Desulfitobacterium elongatum sp. nov.
Desulfitobacterium elongatum (e.lon.ga’tum. L. neut. part. adj. elongatum, elongated) is gram-positive, spore-forming, straight rod-shaped, and strictly anaerobic. Thiosulfate and Fe (III) citrate were used as electron acceptors when pyruvate was used as electron donor. H2+acetate, pyruvate, lactate, and malate were used as electron donors when thiosulfate was used as electron acceptors. The strain did not grow on formate, acetate, citrate, fumarate, propionate, butyrate, methanol, ethanol, propanol, and benzoate in thiosulfate respiration. Sulfate, sulfite, nitrate, nitrite, and air were not reduced. The type strain, NIT-TF6T was isolated from Arako river sediment in Nagoya, Japan. The DNA G+C content of the type strain is 43.2 mol%, which was achieved based on the complete genome sequence (AP038772).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms13081863/s1, Table S1. Organic pollutants dechlorination activity for the strain NIT-TF6; Figure S1: Phylogenomic tree based on TYGS results showing relationship between strain NIT-TF6 and its related cluster of (a) Desulfitobacteriacea family (restricted query to the genomes). Leaf labels are annotated by affiliation to species and subspecies clusters, genomic G+C content, δ values, overall genome sequence length, number of proteins and kind of strain.
Author Contributions
U.B.: Data curation, formal analysis, investigation, visualization, and writing—original draft; R.T.: Methodology and data curation; L.X.: Formal analysis; N.Y.: Writing—original draft, conceptualization, supervision, funding acquisition, resources, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study received funding from the Joint Research Program (JRP) with the National Natural Science Foundation of China (NSFC).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Acknowledgments
We would like to thank Kyo Ikeru, Asuka Akita, and Tomomi Suzuki for their valuable technical assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Christiansen, N.; Ahring, B.K. Desulfitobacterium hafniense sp. nov., an anaerobic, reductively dechlorinating bacterium. Int. J. Syst. Bacteriol. 1996, 46, 442–448. [Google Scholar] [CrossRef]
- Utkin, I.; Woese, C.; Wiegel, J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int. J. Syst. Bacteriol. 1994, 44, 612–619. [Google Scholar] [CrossRef]
- Finneran, K.T.; Forbush, H.M.; Gaw VanPraagh, C.V.; Lovley, D.R. Desulfitobacterium metallireducens sp. nov., an anaerobic bacterium that couples growth to the reduction of metals and humic acids as well as chlorinated compounds. Int. J. Syst. Evol. Microbiol. 2002, 52, 1929–1935. [Google Scholar] [CrossRef]
- De Wildeman, S.; Diekert, G.; Van Langenhove, H.; Verstraete, W. Stereoselective microbial dehalorespiration with vicinal dichlorinated alkanes. Appl. Environ. Microbiol. 2003, 69, 5643–5647. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, G.; Yao, S.; Zhuang, L. Paradesulfitobacterium ferrireducens gen. nov., sp. nov., a Fe(III)-reducing bacterium from petroleum-contaminated soil and reclassification of Desulfitobacterium aromaticivorans as Paradesulfitobacterium aromaticivorans comb. nov. Int. J. Syst. Evol. Microbiol. 2021, 71, 005025. [Google Scholar] [CrossRef]
- Niggemyer, A.; Spring, S.; Stackebrandt, E.; Rosenzweig, R.F. Isolation and Characterization of a Novel As(V)-Reducing Bacterium: Implications for Arsenic Mobilization and the Genus Desulfitobacterium. Appl. Environ. Microbiol. 2001, 67, 5568–5580. [Google Scholar] [CrossRef] [PubMed]
- Villemur, R.; Lanthier, M.; Beaudet, R.; Lepine, F. The Desulfitobacterium genus. FEMS Microbiol. Rev. 2006, 30, 706–733. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Ge, R.; Shi, C.; Wan, Z.; Zheng, D.; Huang, W.; Wu, Y.; Yang, K.; Li, G.; Zhang, F. Reductive dechlorination of trichloroethene at concentrations approaching saturation by a Desulfitobacterium-containing community. J. Hazard. Mater. 2025, 486, 137005. [Google Scholar] [CrossRef]
- Tomita, R.; Yoshida, N.; Meng, L. Formate: A promising electron donor to enhance trichloroethene-to-ethene dechlorination in Dehalococcoides-augmented groundwater ecosystems with minimal bacterial growth. Chemosphere 2022, 307, 136080. [Google Scholar] [CrossRef]
- Maymó-Gatell, X.; Chien, Y.T.; Gossett, J.M.; Zinder, S.H. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 1997, 276, 1568–1571. [Google Scholar] [CrossRef]
- Löffler, F.E.; Yan, J.; Ritalahti, K.M.; Adrian, L.; Edwards, E.A.; Konstantinidis, K.T.; Muller, J.A.; Fullerton, H.; Zinder, S.H.; Spormann, A.M. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 2, 625–635. [Google Scholar]
- Yan, J.; Wang, J.; Solis, V.; Jin, H.; Chourey, K.; Li, X.; Yang, Y.; Yin, Y.; Hettich, R.L.; Löffler, F.E. Respiratory vinyl chloride reductive dechlorination to ethene in TceA-expressing Dehalococcoides mccartyi. Environ. Sci. Technol. 2021, 55, 4831–4841. [Google Scholar] [CrossRef]
- Nonaka, H.; Keresztes, G.; Shinoda, Y.; Ikenaga, Y.; Abe, M.; Naito, K.; Inatomi, K.; Furukawa, K.; Inui, M.; Yukawa, H. Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J. Bacteriol. 2006, 188, 2262–2274. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wei, S.; Liu, N.; Du, Y.; Ding, G. Interspecies transfer of biosynthetic cobalamin for complete dechlorination of trichloroethene by Dehalococcoides mccartyi. Water Sci. Technol. 2022, 85, 1335–1350. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Harzman, C.; Davis, J.K.; Hutcheson, R.; Broderick, J.B.; Marsh, T.L.; Tiedje, J. Genome sequence of Desulfitobacterium hafniense DCB-2, a Gram-positive anaerobe capable of dehalogenation and metal reduction. BMC Microbiol. 2012, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Asai, M.; Yoshida, N.; Kusakabe, T.; Ismaeil, M.; Nishiuchi, T.; Katayama, A. Dehalococcoides mccartyi NIT01, a novel isolate, dechlorinates high concentrations of chloroethenes by expressing at least six different reductive dehalogenases. Environ. Res. 2022, 207, 112150. [Google Scholar] [CrossRef]
- Ismaeil, M.; Yoshida, N.; Katayama, A. Identification of Multiple Dehalogenase Genes Involved in Tetrachloroethene-to-Ethene Dechlorination in a Dehalococcoides-Dominated Enrichment Culture. Biomed Res. Int. 2017, 2017, 9191086. [Google Scholar] [CrossRef]
- Yoshida, N.; Asahi, K.; Sakakibara, Y.; Miyake, K.; Katayama, A. Isolation and quantitative detection of tetrachloroethene (PCE)-dechlorinating bacteria in unsaturated subsurface soils contaminated with chloroethenes. J. Biosci. Bioeng. 2007, 104, 91–97. [Google Scholar] [CrossRef]
- Xie, L.; Yoshida, N.; Ishii, S.; Meng, L. Isolation and polyphasic characterization of Desulfuromonas versatilis sp. Nov., an electrogenic bacteria capable of versatile metabolism isolated from a graphene oxide-reducing enrichment culture. Microorganisms 2021, 9, 1953. [Google Scholar] [CrossRef]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018, 34, 1037–1039. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.; Gerdes, S.; Parello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Yoshida, N.; Ye, L.; Baba, D.; Katayama, A. A novel Dehalobacter species is involved in extensive 4,5,6,7-Tetrachlorophthalide dichlorination. Appl. Envirom. Microbiol. 2009, 75, 2400–2405. [Google Scholar] [CrossRef]
- Rodriguez-R., L.M.; Konstantinidis, K.T. Bypassing Cultivation To Identify Bacterial Species Culture-independent genomic approaches identify credibly distinct clusters, avoid cultivation bias, and provide true insights into microbial species. Microbe 2014, 9, 111–118. [Google Scholar]
- Pohanka, M. D-lactic acid as a metabolite: Toxicology, diagnosis, and detection. Biomed Res. Int. 2020, 2020, 3419034. [Google Scholar] [CrossRef]
- Alfán-Guzmán, R.; Ertan, H.; Manefield, M.; Lee, M. Isolation and characterization of Dehalobacter sp. strain TeCB1 including identification of TcbA: A novel tetra-and trichlorobenzene reductive dehalogenase. Front. Microbiol. 2017, 8, 558. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).