Diagnostic Procedures to Detect Chlamydia trachomatis Infections
Abstract
:1. Introduction
2. Pathogenesis, Genotypes and Clinical Manifestation
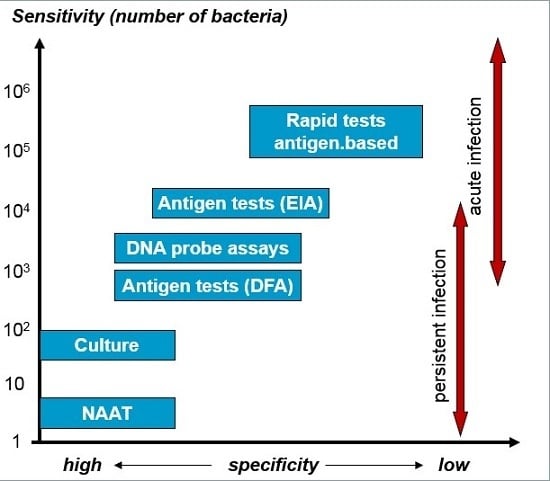
3. Diagnostic Procedures
4. Isolation of C. trachomatis in Cell Culture
5. Nucleic Acids Amplification Tests (NAATs)
6. Clinical Specimens for CT Testing
7. Rapid Diagnostic Tests (RDTs)
8. Serology
9. Conclusions
Conflicts of Interest
References
- Global Incidence and Prevalence of Selected Curable Sexually Transmitted Infections—2008. Available online: http://apps.who.int/iris/bitstream/10665/75181/1/9789241503839_eng.pdf (accessed on 15 June 2016).
- 2014 Sexually Transmitted Diseases Surveillance. Available online: http://www.cdc.gov/std/stats14/chlamydia.htm (accessed on 15 June 2016).
- European Centre for Disease Prevention and Control. Sexually Transmitted Infections in Europe 2013; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2015. [Google Scholar]
- Bastidas, R.J.; Elwell, C.A.; Engel, J.N.; Valdivia, R.H. Chlamydia intracellular survival strategies. Cold Spring Harb. Perspect. Med. 2013, 3, a010256. [Google Scholar] [CrossRef] [PubMed]
- Hybiske, K.; Stephens, R.S. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. USA 2007, 104, 11430–11435. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.R.; Burton, M.J.; Haddad, D.; West, S.; Wright, H. Trachoma. Lancet 2014, 384, 2142–2152. [Google Scholar] [CrossRef]
- Lanjouw, E.; Ouburg, S.; de Vries, H.J.; Stary, A.; Radcliffe, K.; Unemo, M. 2015 European guideline on the management of Chlamydia trachomatis infections. Int. J. STD AIDS 2016, 27, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Darville, T. Chlamydia trachomatis infections in neonates and young children. Semin. Pediatr. Infect. Dis 2005, 16, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Haggerty, C.L.; Gottlieb, S.L.; Taylor, B.D. Risk of sequelae after Chlamydia trachomatis genital infection in women. J. Infect. Dis. 2010, 201, S134–S155. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Chlamydia Control in Europe: Literature Review; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2014. [Google Scholar]
- White, J.A. Manifestations and management of lymphogranuloma venereum. Curr. Opin. Infect. Dis. 2009, 22, 57–66. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, N.H.; de Vries, H.J. Lymphogranuloma venereum among men who have sex with men. An epidemiological and clinical review. Expert Rev. Anti. Infect. Ther. 2014, 12, 697–704. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, N.H.; van Rooijen, M.; Schim van der Loeff, M.F.; de Vries, H.J. Anorectal and inguinal lymphogranuloma venereum among men who have sex with men in Amsterdam, The Netherlands: Trends over time, symptomatology and concurrent infections. Sex. Transm. Infect. 2013, 89, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Papp, J.R.; Schachter, J.; Gaydos, C.A.; van der Pol, B. Recommendations for the laboratory-based detection of Chlamydia trachomatis and Neisseria gonorrhoeae—2014. MMWR Recomm. Rep. 2014, 63, 1–19. [Google Scholar]
- Johnson, R.E.; Newhall, W.J.; Papp, J.R.; Knapp, J.S.; Black, C.M.; Gift, T.L.; Steece, R.; Markowitz, L.E.; Devine, O.J.; Walsh, C.M.; et al. Screening tests to detect Chlamydia trachomatis and Neisseria gonorrhoeae infections—2002. MMRW Recomm. Rep. 2002, 11, 1–38. [Google Scholar]
- Robinson, A.J.; Ridgway, G.L. Modern diagnosis and management of genital Chlamydia trachomatis infection. Br. J. Hosp. Med. 1996, 55, 388–393. [Google Scholar]
- Nwokolo, N.C.; Dragovic, B.; Patel, S.; Tong, C.Y.; Barker, G.; Radcliffe, K. 2015 UK national guideline for the management of infection with Chlamydia trachomatis. Int. J. STD AIDS 2016, 27, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.; Chernesky, M.; Jang, D.; Hook, E.W.; Cartwright, C.P.; Howell-Adams, B.; Ho, S.; Welk, J.; Lai-Zhang, J.; Brashear, J.; et al. Characteristics of the m2000 automated sample preparation and multiplex real-time PCR system for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 2007, 45, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Qian, Q.; Kirby, J.E. Evaluation of the Abbott RealTime CT/NG assay in comParison to the Roche Cobas AmplicorCT/NG assay. J. Clin. Microbiol. 2011, 49, 1294–1300. [Google Scholar] [CrossRef] [PubMed]
- Chernesky, M.A.; Jang, D.A.; Luinstra, K.; Chong, S.; Smieja, M.; Cai, W.J.; Hayhoe, B.; Portillo, E.; MacRitchie, C.; Main, C.; et al. High analytical sensitivity and low rates of inhibition may contribute to detection of Chlamydia trachomatis in significantly more women by the APTIMA Combo 2 assay. J. Clin. Microbiol. 2006, 44, 400–405. [Google Scholar] [PubMed]
- Ripa, T.; Nilsson, P.A. A Chlamydia trachomatis strain with a 377-bp deletion in the cryptic plasmid causing false negative nucleic acid amplification tests. Sex. Transm. Dis. 2007, 34, 255–256. [Google Scholar] [PubMed]
- Harris, S.R.; Clarke, I.N.; Seth-Smith, H.M.B. Whole genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat. Genet. 2012, 44, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Samboona, N.; Wan, R.; Ojcius, D.M.; Pettengill, M.A.; Joseph, S.J.; Chang, A.; Hsu, R.; Read, T.D.; Dean, D. Hypervirulent Chlamydia trachomatis clinical strain is a recombinant between lymphogranulkoma venereum (L2) and D lineages. MBio 2011, 2, e00045-11. [Google Scholar]
- Möller, J.K.; Pedersen, L.N.; Persson, K. ComParison of the Abbott RealTime CT new formulation assay with two other commercial assays for detection of wild-type and new variant strains of Chlamydia trchomatis. J. Clin. Microbiol. 2010, 48, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Levett, P.N.; Brandt, K.; Olenius, K.; Brown, C.; Montgomery, K.; Horsman, G.B. Evaluation of three automated nucleic acid amplification systems for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in first-void urine specimens. J. Clin. Microbiol. 2008, 46, 2109–2111. [Google Scholar] [CrossRef] [PubMed]
- Schachter, J.; Chow, J.M.; Howard, H.; Bolan, G.; Moncada, J. Detection of Chlamydia trachomatis by nucleic acid amplification testing: Our evaluation suggests that CDC-recommended approaches for confirmatory testing are ill-advised. J. Clin. Microbiol. 2006, 44, 2512–2517. [Google Scholar] [CrossRef] [PubMed]
- Black, C.M.; Driebe, E.M.; Howard, L.A.; Fajman, N.N.; Sawyer, M.K.; Girardet, R.G.; Sautter, R.L.; Greenwald, E.; Beck-Sague, C.M.; Unger, E.R.; et al. Multicenter study of nucleic acid amplification tests for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in children being evaluated for sexual abuse. Pediatr. Infect. Dis. J. 2009, 28, 608–613. [Google Scholar] [CrossRef] [PubMed]
- United Kingdom National Guideline on the Management of Sexually Transmitted Infections and Related Conditions in Children and Young People—2010. Available online: http://www.bashh.org/documents/2674.pdf (accessed on 15 June 2016).
- Gaydos, C.A.; Cartwright, C.P.; Colaninno, P.; Welsch, J.; Holden, J.; Ho, S.Y.; Webb, E.M.; Anderson, C.; Bertuzis, R.; Zhang, L.; et al. Performance of the Abbott RealTime CT/NG for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 2010, 48, 3236–3243. [Google Scholar] [CrossRef] [PubMed]
- Van der Pol, B.; Ferrero, D.; Buck-Barrington, L.; Hook, E.W., III; Lenderman, C.; Quinn, T.; Gaydos, C.A.; Lovchik, J.; Schachter, J.; Moncada, J.; et al. Multicenter evaluation of the BD ProbeTec ET system for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine specimens, female endocervical swabs, and male urethral swabs. J. Clin. Microbiol. 2001, 39, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, C.A.; White, J.A.; Michel, C.E.; Mahilum-Tapay, L.; Magbanua, J.P.V.; Nadala, E.C.B., Jr.; Barber, P.J.; Goh, B.T.; Lee, H.H. Optimal method of collection of first-void urine for diagnosis of Chlamydia trachomatis infection in men. J. Clin. Microbiol. 2008, 46, 1466–1469. [Google Scholar] [CrossRef] [PubMed]
- Michel, C.E.; Sonnex, C.; Carne, C.A.; White, J.A.; Magbanua, J.P.; Nadala, E.C., Jr.; Lee, H.H. Chlamydia trachomatis load at matched anatomic sites: Implications for screening strategies. J. Clin. Microbiol. 2007, 45, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Schachter, J.; McCormack, W.M.; Chernesky, M.A.; Martin, D.H.; van der Pol, B.; Rice, P.A.; Hook, E.W., III; Stamm, W.E.; Quinn, T.C.; Chow, J.M. Vaginal swabs are appropriate specimens for diagnosis of genital tract infections with Chlamydia trachomatis. J. Clin. Microbiol. 2003, 41, 3784–3789. [Google Scholar] [PubMed]
- Dudareva Vizule, S.; Haar, K.; Sailer, A.; Flores, J.A.; Silva-Santisteban, A.; Galea, J.T.; Coates, T.J.; Klausner, J.D.; Caceres, C.F. Prevalence of pharyngeal and rectal Chlamydia trachomatis and Neisseria gonorrhoeae infections among men who have sex with men in Germany. Sex. Transm. Infect. 2014, 90, 46–51. [Google Scholar]
- Kent, C.K.; Chaw, J.K.; Wong, W.; Liska, S.; Gibson, S.; Hubbard, G.; Klausner, J.D. Prevalence of rectal, urethral, and pharyngeal chlamydia and gonorrhea detected in 2 clinical settings among men who have sex with men: San Francisco, California, 2003. Clin. Infect. Dis. 2005, 41, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Schachter, J.; Moncada, J.; Liska, S.; Shayevich, C.; Klausner, J.D. Nucleic acid amplifications tests in the diagnosis of chlamydial and gonococcal infections of the oropharynx and rectum of men who have sex with men. Sex. Transm. Dis. 2008, 35, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.V.; Tamari, I.E.; Smieja, M.; Jamieson, F.; Jones, K.E.; Towns, L.; Juzkiw, J.; Richardson, S.E. Detection of Neisseria gonorrhoeae and Chlamydia trachomatis in pharyngeal and rectal specimens using the BD ProbeTec ET system, the Gen-Probe Aptima Combo 2 assay and culture. Sex. Transm. Infect. 2009, 85, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Hammerschlag, M.R.; Roblin, P.M.; Gelling, M.; Tsumura, N.; Jule, J.E.; Kutlin, A. Use of polymerase chain reaction for the detection of Chlamydia trachomatis in ocular and nasopharyngeal specimens from infants with conjunctivitis. Pediatr. Infect. Dis. J. 1997, 16, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Hong, K.C.; Schachter, J.; Moncada, J.; Lekew, T.; House, J.I.; Zhou, Z.; Neuwelt, M.D.; Rutar, T.; Halfpenny, C.; et al. Detection of Chlamydia trachomatis ocular infection in trachoma-endemic communities by rRNA amplification. Investig. Ophthalmol. Vis. Sci. 2009, 50, 90–94. [Google Scholar] [CrossRef] [PubMed]
- De Vries, H.J.C.; Zingoni, A.; Kreuter, A.; Moi, H.; White, J.A. 2013 European guideline on the management of lymphogranuloma venereum. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.; Arndt, R.; von Krosigk, A.; Plettenberg, A. Repeated detection of lymphogranuloma venereum caused by Chlamydia trachomatis L2 in homosexual men in Hamburg. Sex. Transm. Infect. 2005, 81, 91–92. [Google Scholar] [CrossRef] [PubMed]
- Morre, S.A.; Spaargaren, J.; Fennema, J.S.; de Vries, H.J.; Coutinho, R.A.; Peña, A.S. Real-time polymerase chain reaction to diagnose lymphogranuloma venereum. Emerg. Infect. Dis. 2005, 11, 1311–1312. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Chi, K.H.; Alexander, S.; Martin, I.M.; Liu, H.; Ison, C.A.; Ballard, R.C. The molecular diagnosis of lymphogranuloma venereum : Evaluation of a real time multiplex polymerase chain reaction test using rectal and urethral specimens. Sex. Transm. Dis. 2007, 34, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Quint, K.D.; Bom, R.J.; Bruisten, S.M.; van Doorn, L.J.; Nassir-Hajipour, N.; Melchers, W.J.; de Vries, H.J.; Morre, S.A.; Quint, W.G. ComParison of three genotyping methods to identify Chlamydia trachomatis genotypes in positive men and women. Mol. Cell. Probes. 2010, 24, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Van Dommelen, L.; van Tiel, F.H.; Ouburg, S.; Brouwers, E.E.; Terporten, P.H.; Savelkoul, P.H.; Morré, S.A.; Bruggeman, C.A.; Hoebe, C.J. Alarmingly poor performance in Chlamydia trachomatis point-of-care testing. Sex. Transm. Infect. 2010, 86, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Nunez-Foreno, L.; Moyano-Ariza, L.; Gaitan-Duarte, H.; Ángel-Müller, E.; Ruiz-Parra, A.; González, P.; Rodríguez, A.; Tolosa, J.E. Diagnostic accuracy of rapid tests for sexually transmitted infections in symptomatic women. Sex. Transm. Infect. 2016, 92, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Hislop, J.; Quayyum, Z.; Flett, G.; Boachie, C.; Fraser, C.; Mowatt, G. Systematic review of the clinical effectiveness and cost-effectiveness of rapid point-of-care tests for the detection of genital chlamydia infection in women and men. Health Technol. Assess. 2010, 14. [Google Scholar] [CrossRef] [PubMed]
- Van der Helm, J.J.; Sabajo, L.O.A.; Grunberg, A.W.; Morré, S.A.; Speksnijder, A.G.; de Vries, H.J. Point-of-care test for detection of urogenital Chlamydia in women show low sensitivity. A performance evaluation study in two clinics in Surinam. PLoS ONE 2012, 7, e32122. [Google Scholar]
- Hurly, D.S.; Buhrer-Skinner, M.; Badman, S.G.; Bulu, S.; Tabrizi, S.N.; Tarivonda, L.; Muller, R. Field evaluation of the CRT and ACON chlamydia point-of-care tests in a tropical, low-resource setting. Sex. Transm. Infect. 2014, 90, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Abbai Shaik, N.S.; Reddy, T.; Govender, S.; Ramjee, G. Poor performance of the Chlamydia Rapid Test device for the detection of asymptomatic infections in South African men: A pilot study. J. Sex. Transm. Dis. 2016. [Google Scholar] [CrossRef] [PubMed]
- Gaydos, C.A.; van der Pol, B.; Jett-Gohenn, M.; Barnes, M.; Quinn, N.; Clark, C.; Daniel, G.E.; Dixon, P.B.; Hook, E.W., III; CT/NG Study Group. Performance of the Cephaid CT/NG Xpert rapid test for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 2013, 51, 1666–1672. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Gaydos, C.A.; Barnes, M.R.; Jett-Goheen, M.; Blake, D.R. Comparative effectiveness of a rapid point-of-care test for detection of Chlamydia trachomatis among women in a clinical setting. Sex. Transm. Infect. 2013, 89, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Pearce, D.M.; Shenton, D.P.; Holden, J.; Gaydos, C.A. Evaluation of a novel electrochemical detection method for Chlamydia trachomatis: Application for point-of-care diagnostics. IEEE Trans. Biomed. Eng. 2011, 58, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Krölov, K.; Frolova, J.; Tudoran, O.; Suhorutsenko, J.; Lehto, T.; Sibul, H.; Mäger, I.; Laanpere, M.; Tulp, I.; Langel, Ü. Sensitive and rapid detection of Chlamydia trachomatis by recombinase polymerase amplification directly from urine samples. J. Mol. Diagn. 2014, 16, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Morre, S.A.; Munk, C.; Persson, K.; Krüger-Kjaer, S.; van Dijk, R.; Meijer, C.J.; van den Brule, A.J. ComParison of three commercially available peptide-based immunoglobulin G (IgG) and IgA assays to microimmunofluorescence assay for detection of Chlamydia trachomatis antibodies. J. Clin. Microbiol. 2002, 40, 584–587. [Google Scholar] [CrossRef] [PubMed]
- Forsbach-Birk, V.; Simnacher, U.; Pfrepper, K.I.; Soutschek, E.; Kiselev, A.O.; Lampe, M.F.; Meyer, T.; Straube, E.; Essig, A. Identification and evaluation of a combination of Chlamydial antigens to support diagnosis of severe and invasive Chlamydia trachomatis infections. Clin. Microbiol. Infect. 2009, 16, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.S.; Maple, P.A.C.; Andrews, N.J.; Paul, I.D.; Caul, E.O. Measurement of IgG antibodies to Chlamydia trachomatis by commercial enzyme immunoassays and immunofluorescence in sera from pregnant women and patients with infertility, pelvic inflammatory disease, ectopic pregnancy, and laboratory diagnosed Chlamydia psittaci/Chlamydia pneumoniae infection. J. Clin. Pathol. 2003, 56, 225–230. [Google Scholar] [PubMed]
- Haralambieva, I.; Iankov, I.; Petrov, D.; Ivanova, R.; Kamarinchev, B.; Mitov, I. Cross-reaction between the genus-specific lipopolysaccharide antigen of Chlamydia spp. and the lipopolysaccharides of Porphyromonas gingivalis, Escherichia coli O119 and Salmonella newington: Implications for diagnosis. Diagn. Microbiol. Infect. Dis. 2001, 41, 99–106. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Lu, C.; Lei, L.; Yu, P.; Zhong, G. A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J. Immunol. 2010, 185, 1670–1680. [Google Scholar] [CrossRef] [PubMed]
- Budrys, N.M.; Gong, S.; Rodgers, A.K.; Wang, J.; Louden, C.; Shain, R.; Schenken, R.S.; Zhong, G. Chlamydia trachomatis antigens recognized in women with tubal factor infertility, normal fertility, and acute infection. Obstet. Gynecol. 2012, 119, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meyer, T. Diagnostic Procedures to Detect Chlamydia trachomatis Infections. Microorganisms 2016, 4, 25. https://doi.org/10.3390/microorganisms4030025
Meyer T. Diagnostic Procedures to Detect Chlamydia trachomatis Infections. Microorganisms. 2016; 4(3):25. https://doi.org/10.3390/microorganisms4030025
Chicago/Turabian StyleMeyer, Thomas. 2016. "Diagnostic Procedures to Detect Chlamydia trachomatis Infections" Microorganisms 4, no. 3: 25. https://doi.org/10.3390/microorganisms4030025
APA StyleMeyer, T. (2016). Diagnostic Procedures to Detect Chlamydia trachomatis Infections. Microorganisms, 4(3), 25. https://doi.org/10.3390/microorganisms4030025