Abstract
Staphylococcus xylosus belongs to the vast group of coagulase-negative staphylococci. It is frequently isolated from meat products, either fermented or salted and dried, and is commonly used as starter cultures in sausage manufacturing. Analysis of the S. xylosus genome together with expression in situ in a meat model revealed that this bacterium is well adapted to meat substrates, being able to use diverse substrates as sources of carbon and energy and different sources of nitrogen. It is well-equipped with genes involved in osmotic, oxidative/nitrosative, and acidic stress responses. It is responsible for the development of the typical colour of cured meat products via its nitrate reductase activity. It contributes to sensorial properties, mainly by the the catabolism of pyruvate and amino acids resulting in odorous compounds and by the limiting of the oxidation of fatty acids, thereby avoiding rancidity.
1. Occurrence of Staphylococcus xylosus in Meat Products
Staphylococcus xylosus is one of the 41 species belonging to the coagulase-negative group (CNS) of Staphylococcus genus affiliated to the phylum of Firmicutes. S. xylosus belongs to the Staphylococcus saprophyticus cluster group, which includes nine species (www.bacterio.net).
S. xylosus is a commensal species of the epithelium and mucous membranes of animals and more particularly of mammals. It was first isolated from human skin, but its occurrence is uncommon in humans compared with small mammals and farm animals [1,2]. The high presence of S. xylosus on the skin of livestock could explain its presence in meat.
In fresh sausages, S. xylosus accounted for 48% of the isolates followed by S. equorum (24%), S. pasteuri (13%) and S. warneri (9%) [3,4]. A high-throughput approach of 16S DNA pyrosequencing has revealed the presence of CNS including S. xylosus in samples of fresh meat, minced steaks of beef or veal, and poultry sausages [5].
Processes such as fermentation, salting and drying select CNS, which reach concentrations of 5 to 7 log CFU/g and constitute, with lactic acid bacteria (LAB), the main populations in this kind of product [6,7]. The ecology of S. xylosus in these products has been particularly studied. In traditional Mediterranean fermented sausages, S. xylosus often constitutes the dominant staphylococcal species [8,9,10,11,12,13,14,15,16,17]. In addition, species such as S. equorum, S. saprophyticus and S. succinus belong to this staphylococcal ecosystem. S. xylosus is also frequently isolated from natural casings used for sausage manufacturing [15,18]. In salted and dried products such as Iberian ham, S. xylosus was identified as the main species throughout the manufacturing process [19,20]. Brine was the main source of staphylococci, with S. xylosus accounting for 29% of the isolates [21]. Of the nine CNS species identified in Kitoza, a traditional Malagasy product made from dried or smoked pork or beef, S. xylosus represented 10 to 17% of the isolates [7].
S. xylosus is also one of the main starters inoculated in meat fermented products. In industrial sausage production where S. xylosus is used as starter, it remains dominant regardless of the fermentation stage, showing a good adaptation of the bacterium to the meat substrate and to the various manufacturing processes [22,23]. Complete genomes are available for three S. xylosus strains: C2a (our laboratory, LN554884), SMQ-121 [24], and HKUOPL8 [25]. Moreover, the draft genomes of two other strains are also available: NJ [26] and DMB3-Bh1 [27]. In this article, we essentially focus on the strain S. xylosus C2a.
2. Adaptation to Substrates
Meat contains proteins (15–22 g/100 g), lipids (1.5–4.0 g/100 g), minerals and traces of carbohydrates [28]. In sausage manufacturing, carbohydrates are added to favour fermentation. The metabolic potential of the strain S. xylosus C2a, the most characterised strain among the species sequenced, is discussed in relation to these substrates.
2.1. Carbon Substrates
Sugar transport and catabolism have been well characterised in S. xylosus C2a. Two types of transport have been identified: the phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS) and the PTS-independent system [29]. The PTS system consists of two nonspecific energy-coupling components, Enzyme I (EI, ptsI) and a heat stable phosphocarrier protein (HPr, pstH), and several sugar-specific multiprotein permeases known as enzymes II (EIIA, EIIB and EIIC) [30]. The PTS-independent system consists of a permease and a kinase [31].
Genome-based analyses of the metabolic pathways for energy production reveal that S. xylosus possesses the genetic potential for transport of 10 carbohydrates by the PTS system and eight by the non-PTS system (Table 1). We focused on the three main carbohydrates added to the meat batter, glucose, sucrose and lactose, and the lactate already present in meat.

Table 1.
Carbohydrate transport and catabolism in S. xylosus C2a.
Glucose can be transported by PST-dependent and independent systems in S. xylosus (Table 1) [29,30]. In S. xylosus, the genes ptsI and ptsH are co-transcribed forming the ptsHI operon (SXYL_01852-53) [32]. In the genome of S. xylosus C2a, three genes encoding specific glucose permeases, ptsG, SXYL_00253 and crr, are present. These genes are not organised in clusters, in contrast to the glcA and glcB described in S. carnosus [33]. In the PTS-independent system, glucose is transported through the membrane by GlcU permease (glcU) and then phosphorylated by a glucokinase (glkA) [31]. The gdh gene encoding a glucose dehydrogenase is downstream of glcU [31]. The Gdh enzyme allows the formation of gluconate from glucose and is co-expressed with GlcU. This co-expression suggests that S. xylosus uses two metabolic pathways in parallel for energy yield from glucose [31]. The entry of glucose into the cell depends on its concentration in the extracellular medium [29]. GlcU permease is functional at high concentration, whereas the glucose-specific PTS system transport is active at low concentration. The two main routes of catabolism of glucose in staphylococci are the Embden–Meyerhof–Parnas (EMP) pathway and the pentose phosphate (PP) pathway [30]. However, under growth conditions in a complex medium, about 85% of the glucose is catabolised by the EMP route. The main product of the anaerobic metabolism of glucose is lactate, although it is poorly produced in S. xylosus [30]. Acetate and CO2 are the main products of aerobic glucose metabolism [30,34,35,36]. Staphylococci can adapt to the oxygenation conditions via the EMP and PP pathways, the TCA cycle and their respiratory chain. The availability of carbon substrates leads to a regulatory process called catabolic carbon repression [29,37]. This results in the repression of genes or operons allowing the use of alternative carbon sources. In staphylococci, the catabolic repression of glucose is provided by a catabolite control protein A (CcpA) transcriptional regulator [30].
In a ground pork meat model containing 0.5% glucose and 2.77% of NaCl incubated at 22 °C in Petri dishes, monoculture of S. xylosus C2a consumed glucose and lactate simultaneously [38]. The entry of glucose was via the PTS-independent system, and it was catabolised through the EMP and the PP pathways [38]. Lactate was imported by a lactate permease and catabolised to pyruvate by a lactate–quinone oxidoreductase (Table 1). Acetyl-CoA generated by the degradation of glucose and lactate can feed the TCA cycle and, in part, was catabolised to acetate, which was excreted in meat. The genes encoding an ATP synthase (atp, SXYL_00824-31) were overexpressed, leading to an energy supply necessary for S. xylosus [38]. This simultaneous consumption of glucose and lactate has been observed for S. aureus in laboratory media [35].
In S. xylosus, four scr genes allow the use of sucrose (Table 1) [39]. The scrA gene encodes a sucrose PTS permease composed of EIIBC domains. Internalised sucrose-6-phosphate is cleaved by a sucrose phosphate hydrolase or sucrase encoded by the scrB gene to yield glucose-6-phosphate and fructose, which will rejoin the glycolysis [39]. The fructose is then phosphorylated by a fructokinase encoded by scrK [40]. The genes scrB and scrK are organised in an operon. The scrA gene is independent [40]. The expression of the two genes scrA and scrB is induced by the concentration of sucrose present in the medium. This regulation is under the control of a repressor encoded by the scrR gene, located upstream of the scrB gene [41].
In S. xylosus, lactose is assimilated, unphosphorylated, by a lactose permease, which transports galactosides and pentoses (Table 1) [42]. The system comprises lacP and lacH genes, which respectively encode a lactose permease and a β-galactosidase, which hydrolyses lactose to glucose and galactose. A lacR regulatory gene is positioned upstream of the operon and oriented opposite to the lacPH operon [30,43]. Galactose is further degraded along the Leloir pathway leading to glucose 1-P via the galKET cluster with galR upstream encoding a regulator.
2.2. Nitrogen Substrates, Peptides, Amino Acids
Proteins are the main components of meat. They are hydrolysed mainly into peptides by endogenous proteinases during sausage fermentation and ripening [28,44]. The proteolytic activity of staphylococci is low [45]. The peptides can be further catabolised into amino acids by endogenous and bacterial peptidases [44]. S. xylosus used as starter in association with a lactic acid bacterium contributed to the enrichment of free amino acids in sausages [44,46].
We identified, in the S. xylosus C2a genome, two clusters of genes encoding oligopeptide transport systems that belong to the ATP-binding cassette (ABC) family of transporters (Table 2). They are composed of five subunits: an extracellular oligopeptide-binding protein, which specifically captures the substrates, two transmembrane proteins forming the pore and two proteins in charge of ATP hydrolysis. At a genetic level, the five genes are always organised in an operon. Bacteria can have two operons that can be transcribed differently [47]. In S. xylosus, only the operon SXYL_00298-00302 was upregulated in a meat model [38]. S. xylosus C2a has a potentially high genetic capacity to degrade peptides, with 20 genes encoding peptidases (Table 2) and 14 encoding putative peptidases (LN554884). Four of these genes were overexpressed in a meat model and could contribute to the nutrition of S. xylosus C2a, map and amps encoding methionine and leucyl aminopeptidases and the genes encoding U32 family peptidases (SXYL_01247-48) [38].

Table 2.
Peptide transport, peptidases and amino acid catabolism in S. xylosus C2a.
The genome of S. xylosus contains 59 tRNAs for all amino acids. S. xylosus is prototrophic, it can grow on a medium containing ammonium sulphate as the sole nitrogen source [48]. In a meat model that contains variable levels of amino acids, 34 genes involved in synthesis of branched chain and aromatic amino acids, histidine and arginine were downregulated and genes involved in transport of alanine and lysine were upregulated [38]. Most of these genes are under the control of CodY, a global repressor of the transcription that controls the genes involved in the utilisation of nitrogen [49], while the genes involved in arginine synthesis are under the control of CcpA [50].
Arginine is commonly found in meat and could be used as an alternative energy source via the arginine deiminase (ADI) pathway. The ADI pathway comprises three enzymatic reactions catalysed by arginine deiminase (ADI) encoded by arcA, ornithine transcarbamoylase (OTC) encoded by arcB and carbamate kinase encoded by arcC. In the three completed genomes of S. xylosus, the genes arcB and arcC are present, but arcA encoding the arginine deiminase is absent (Table 2). However, the arcA gene has been detected by a PCR approach in 3 out of 13 S. xylosus strains, and among these three strains, one had an arginine catabolic mobile element, ACME-associated arcA gene [51,52]. Thus, this gene seems to be found infrequently in S. xylosus species. Arginine could be also catabolised by arginase encoded by arg, a gene frequent in S. xylosus, present in the three strains sequenced and in 12 out of 13 strains [51]. Arginase activity leads to ornithine and urea, and urea can be further catabolised by a urease to carbonic acid and two molecules of ammonia serving as a nitrogen source. In a meat model, concentration of arginine decreased after 48 h of incubation and at this time glucose and lactate were exhausted, and genes related to urea pathway were upregulated [53].
Glutamate and glutamine are present in meat. Glutamate is a key component; it serves as an amino group donor and is a link between nitrogen and carbon metabolism. Glutamate can be imported by a glutamate symporter encoded by gltT and catabolised by glutamate dehydrogenases (gluD1, gluD2) that provide alpha-ketoglutarate, which will fuel the TCA cycle [38]. Furthermore, two clusters of genes gltBCD and SXYL_00105-108 linked respectively with glutamate and glutamine interconversion were highly overexpressed in meat (Table 2).
2.3. Nucleosides
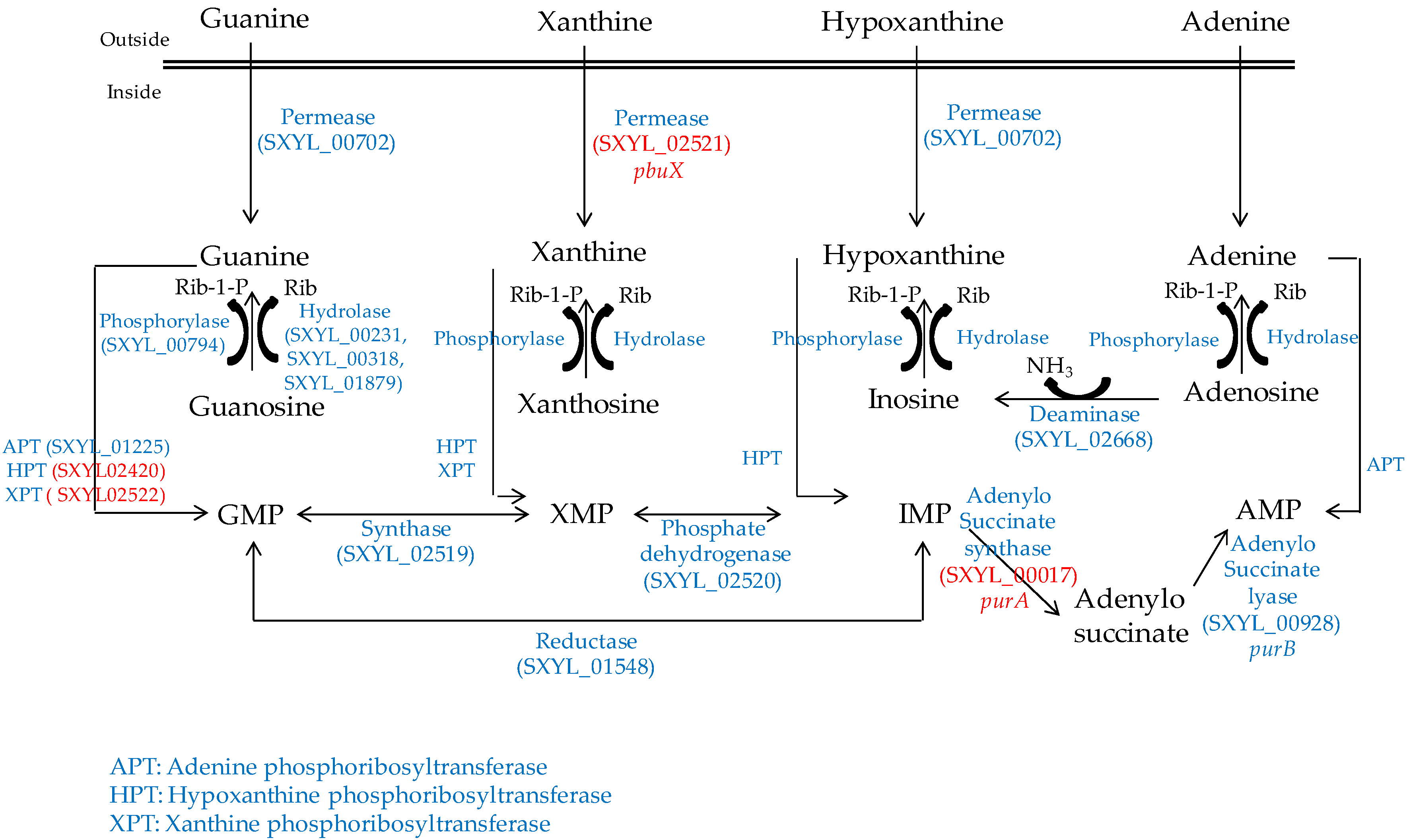
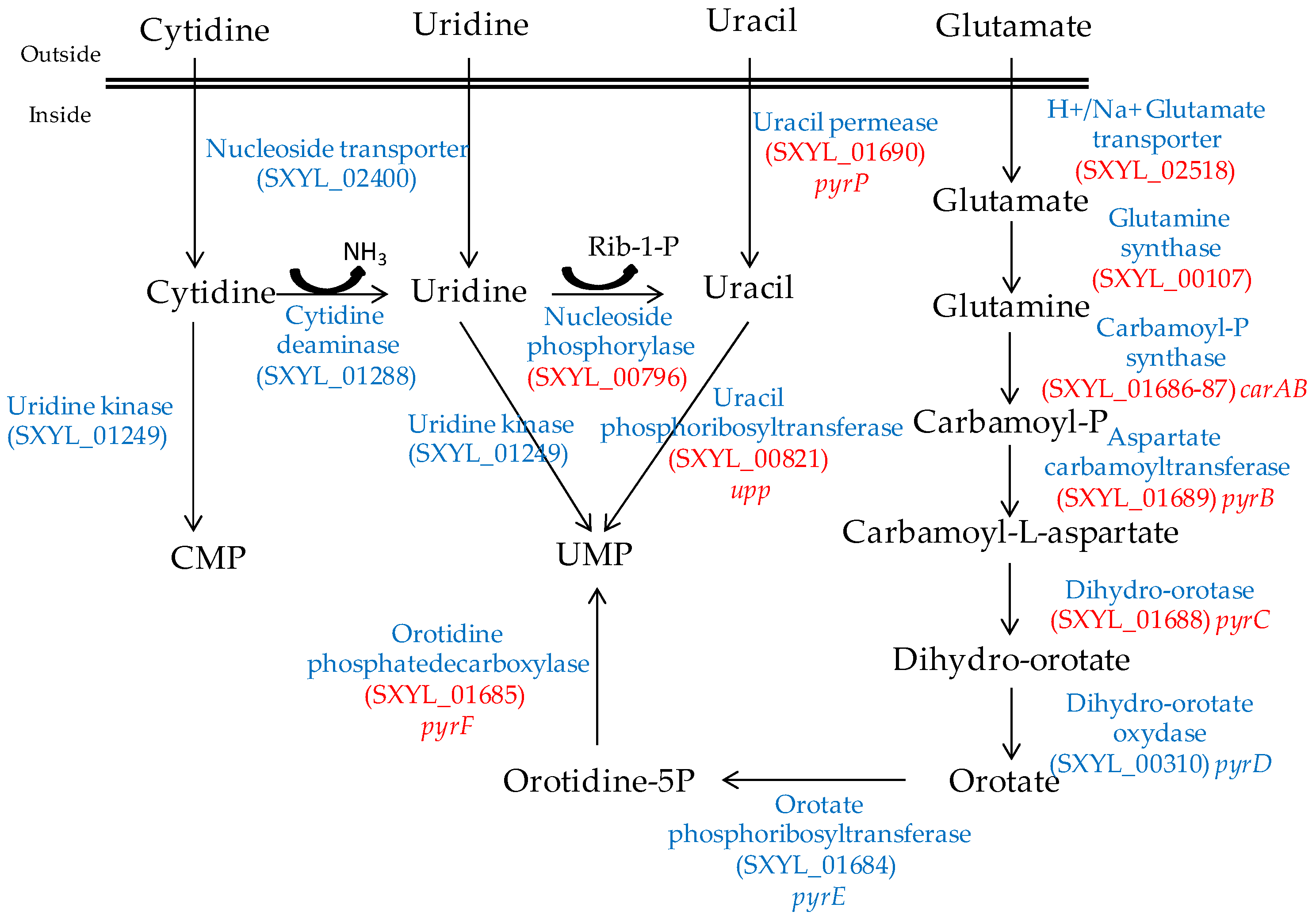
Nucleosides are released from the ATP hydrolysis that occurs during the maturation of the meat and during the fermentation process [54]. The S. xylosus genome contains the catabolic genes involved in purine and pyrimidine transport, salvage and interconversion (Figure 1 and Figure 2) as described for Bacillus subtilis [55]. Purine and pyrimidine bases and nucleosides are transported into the cells by specific permeases and four corresponding genes were identified in S. xylosus. The bases are converted to nucleoside monophosphates by phosphoribosyltransferases. Ribonucleosides are cleaved by phosphorylases into free bases and ribose-1 P or by hydrolases into free bases and ribose (Figure 1 and Figure 2). Then, ribose will fuel the PP pathway. The conversion of adenosine to inosine and that of cytidine to uridine are catalysed by specific deaminases releasing NH3 that may serve as nitrogen source. The catabolism of inosine and adenosine has been highlighted in coagulase-negative staphylococci [52] and in Lactobacillus sakei [56,57].
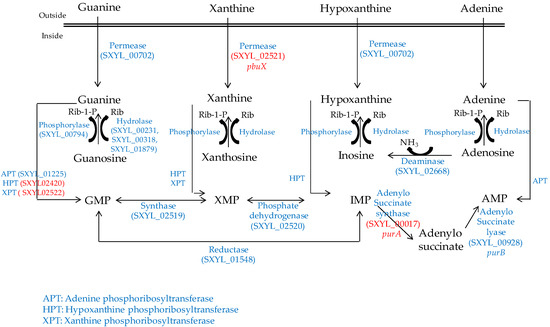
Figure 1.
Purine transport, salvage and interconversion in S. xylosus C2a. In red, genes overexpressed in a meat model.
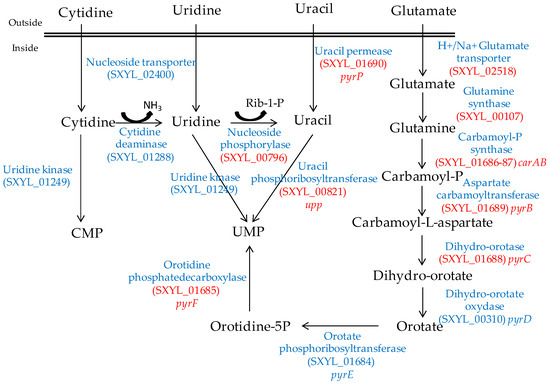
Figure 2.
Synthesis of UMP by S. xylosus C2a from pyrimidine and glutamate/glutamine. In red, genes overexpressed in a meat model.
In meat, intermediates from ATP such as xanthine and uracil are present [58]. S. xylosus C2a modulates thirteen genes involved in nucleotide transport and metabolism in a meat model [38]. Xanthine could be imported by the xanthine permease encoded by pbuX and catabolised into XMP by the xanthine and hypoxanthine phosphoribosyltransferases encoded by xpt and hpt (Figure 1). AMP could be synthesised by adenylosuccinate synthase (purA) from IMP (Figure 1). Uracil could be taken up via uracil permease (pyrP) and catabolised into UMP by uracil phosphoribosyltransferase (upp) (Figure 2). Remarkably, UMP could also be synthesised by the import of glutamate or glutamine, which are both present in meat, and then catabolised through six enzymatic reactions encoded by the cluster pyrEFcarABpyrCB and pyrD (Figure 2).
2.4. Iron Uptake
Meat is an iron-rich substrate including hemic (myoglobin and haemoglobin) and non-hemic (ferritin and transferrin) iron sources. S. xylosus C2a has developed six systems (sit, sfa, hts, fhu, sst, SXYL_00561-63) to acquire iron that can be separated into two general mechanisms. The first, represented by sitABC (SXYL_02216-18) and the cluster SXYL_00561-63, involves a direct contact between the bacteria and the exogenous sources of iron. The cluster sitABC, encoding an iron-regulated ABC transport involved in divalent metal uptake, was influenced in the presence of ferrous iron (FeSO4), while the cluster SXYL_00561-63 was highly upregulated in the presence of ferritin [59]. This cluster encodes an oxidoreductase, a monooxygenase and a transporter and was identified in other species belonging to the S. saprophyticus cluster group [59]. Remarkably, the Isd (iron responsive surface determinant) system responsible for iron acquisition from heme in S. aureus [60] was absent in the three S. xylosus sequenced strains; only isdG encoding the monooxygenase was present. The second mechanism relies on siderophores, which are small molecules that are secreted by bacteria and have an exceptionally high affinity for iron [61]. It has been clearly demonstrated that S. aureus produces two distinct siderophores: staphyloferrin A (SA) and staphyloferrin B [62]. In the S. xylosus C2a genome, only the clusters sfaABCD and htsABC coding for the synthesis and transport of SA were present. The C2a strain was able to produce siderophore in a staphylococcal siderophore detection medium [53]. Furthermore, S. xylosus possesses the Fhu system involved in the uptake of hydroxamate-type and the Sst system for catechol-type siderophores. This strain could be able to use exogenous siderophores to scavenge iron from various sources.
3. Adaptation to Stressful Manufacturing Processes
3.1. Osmotic Stress
S. xylosus is consistently isolated from fermented sausages and dry cured meat products. It is able to grow in the presence of curing salts. It is a remarkably osmotolerant bacterium, like other staphylococci. Osmosprotection appears to be crucial in this salted environment. S. xylosus has developed several mechanisms to cope with the osmotic stress. It possesses several osmoprotectant systems, such as solute uptakes for proline, serine/alanine/glycine and glycine betaine/carnitine/choline, and synthesis of glycine betaine (Table 3).

Table 3.
Potential of S. xylosus C2a to cope with osmotic stress.
The response of S. xylosus to the presence of NaCl was studied in the meat model [38]. It responded by under-expressing mscL (SXYL_01536), which encodes a mechanosensitive channel, which prevents the efflux of solute. It also involved different mechanisms of accumulation of osmoprotectants and Na+-dependent antiporters (Table 3). Thus, S. xylosus overexpressed the genes encoding two systems of uptake and synthesis of glycine betaine, a major osmoprotectant. The opuC cluster encodes a betaine/carnitine/choline type ABC carrier for the uptake of the carnitine present in meat, which can be catabolised to glycine betaine by an l-carnitine dehydrogenase encoded by lcdH. The cudTCA genes encode enzymes involved in the acquisition of choline and its dehydrogenation to glycine betaine by a choline dehydrogenase encoded by betA [63]. In parallel, the two mnh clusters encoding Na+/H+ antiporter systems were overexpressed by S. xylosus in the meat model [38].
3.2. Oxidative, Nitrosative Stress
Meat processing generates changes in oxygen levels and redox potential. This level varies during mincing, and an oxygen gradient is established during fermentation between the surface and the heart of the sausage. Furthermore, nitrite added during manufacturing undergoes chemical reactions that lead to reactive nitrogen species (RNS) including NO [64]. These RNS and reactive oxygen species (ROS) will generate nitrosative and oxidative stress, especially in meat in which iron contributes to Fenton chemistry generating highly reactive hydroxyl radicals. Staphylococci have several mechanisms to overcome the deleterious effects of this stress [65].
S. xylosus C2a possesses 14 genes coding for enzymes involved in the detoxification of ROS and RNS and, in particular, one superoxide dismutase, three catalases, four peroxiredoxins, four nitroreductases and a nitric oxide synthase (Table 4). Nine of them were overexpressed in response to nitrosative stress in a meat model containing curing salts, nitrate and nitrite (Table 4) [53]. S. xylosus possesses a single superoxide dismutase (SOD) encoded by sodA involved in protection against oxidative stress generated by hyperbaric oxygen and paraquat [66]. The expression of this gene was not modulated in the meat model with or without curing salts [38,53]. In S. xylosus C2a, the detoxification of H2O2 is accomplished by three catalases (KatA, KatB, KatC). The genes encoding these three catalases are also present in the genomes of the strains SMQ-121 and HKUOPL8. Most staphylococci have one catalase, as S. aureus [65], but the strain S. carnosus TM300 has two [67], as do some strains of S. equorum, S. saprophyticus and S. xylosus [68]. The transcription of katA of S. xylosus C2a was induced upon entry in the stationary phase, by oxygen and hydrogen peroxide, and was repressed by iron and manganese [69]. This gene was down-regulated, while katB and katC were upregulated in response to a nitrosative stress in a meat model [53]. In addition to catalases, ahpC encoding alkyl hydroperoxide reductase subunit C, bsaA encoding glutathione peroxidase and bcp encoding a bacterioferritin comigratory protein were upregulated. AhpC confers resistance to ROS and BsaA detoxifies H2O2 and also other peroxides (ROOR) [65]. Bacterioferritin comigratory protein functions as an iron chelator and is homologous to a thioreductase-peroxidase contributing to the reduction of thiol-dependent peroxides [65]. The genome of S. xylosus C2a contains four genes encoding nitroreductases and three were upregulated to cope with nitrosative stress in a meat model with curing salts [53]. These nitroreductases may help to maintain the thiol disulphide balance as shown for S. aureus [70].

Table 4.
Potential of S. xylosus C2a to cope with oxidative/nitrosative stress.
A gene encoding a nitric oxide synthase (NOS) is present in S. xylosus genomes as in all staphylococci. In S. xylosus C2a, this enzyme protects against peroxide stress [71]. Furthermore, the loss of NOS activity in this strain resulted in the modulation of the expression of genes encoding catalases, with upregulation of katA and downregulation of katB and katC [71]. In this study, a nos deficient mutant displayed higher colony pigmentation than the C2a wild-type strain. All these results are in agreement with those on S. aureus, attesting that NOS activity protects against oxidative stress [72,73]. S. xylosus C2a grown in a meat model overexpressed the cluster crtPQMN (SXYL_00051-54) involved in carotenoid pigment biosynthesis pathway [38]. In S. aureus, the pigment protects against oxidative stress by scavenging free radicals [74].
Proteins and amino acids can be oxidised or modified by ROS and RNS. Staphylococci have developed mechanisms to repair protein damage [65]. Thioredoxin and glutaredoxin are essential to maintain protein thiols in their reduced forms. They are major contributors to oxidative stress resistance by facilitating the reduction of H2O2, scavenging hydroxyl radicals and donating reducing equivalents to peroxiredoxins [65]. The S. xylosus C2a genome has four genes encoding thioredoxins and one encoding glutaredoxin (Table 3). The transcription of trxB encoding a thioredoxin reductase was increased in the presence of RNS in a meat model [53]. Similarly, stressors such as hydroperoxide and disulphide induce transcription of trxA and trxB in S. aureus [75]. S. xylosus C2a, as S. aureus, has three msrA genes and one msrB encoding methionine sulphoxide reductase involved in the repair of oxidised methionine (Table 4). In S. aureus, MsrA1 is the major contributor against H2O2 stress [76]. This gene was also overexpressed in S. xylosus to counter nitrosative stress in a meat model [53].
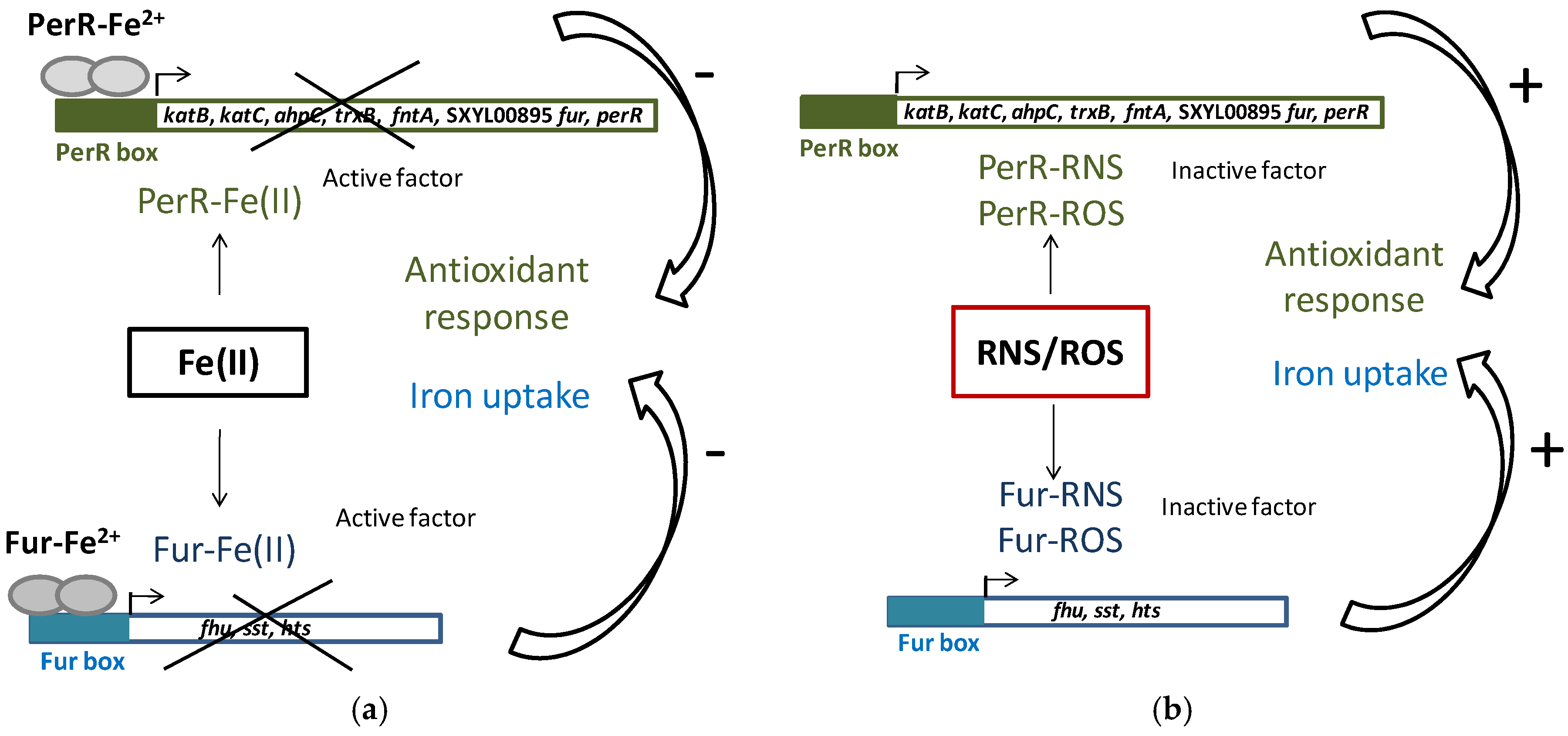
Iron homeostasis is essential because of its involvement in the generation of ROS. To maintain iron homeostasis, S. xylosus C2a, in addition to the six iron-acquiring systems described above, can store iron in ferritin (ftnA) and bacterioferritin comigratory protein (bcp) (Table 4). The clusters (fhu, sst, sfa, hts) and the genes ftnA and bcp were upregulated in a meat model containing nitrate and nitrite (Table 4) [53]. Some of these genes (hts, fhu, sst) belong to the Fur (ferric uptake regulator) regulon. Fur can be inactivated by NO from nitrite, thus derepressing the regulon (Figure 3) [53]. In addition, perR encoding the transcriptional regulator PerR, a member of the Fur family and identified as a peroxide-sensing protein, was upregulated. The genes katB, katC, ahpC, trxB and fntA, all of which are upregulated in the presence of RNS, are under the control of PerR [53]. PerR, as Fur, can be inactivated by NO from nitrite (Figure 3).
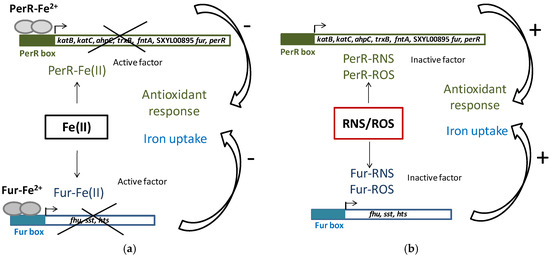
Figure 3.
PerR and Fur regulation in S. xylosus C2a. (a) Classic model of PerR and Fur repression of antioxidant response and iron uptake; (b) Impact of reactive nitrogen species (RNS) and reactive oxygen species (ROS), which compete with Fe(II) for PerR and Fur binding, cause antioxidant response and iron uptake.
3.3. Acid Stress
During sausage fermentation, pH decreases from 5.8 to the range 4.5 to 5.3, depending on the carbohydrate added and the starters inoculated [77]. Bacteria have developed several mechanisms to cope with acidity [78]. One of them relies on proton extrusion by F1F0-ATPase, which plays a key role in maintaining the internal pH near neutral. A link between ATPase and acid tolerance has been established for several lactic acid bacteria [78]. The cluster atp encoding F1F0-ATPase and atpI (SXYL_00823) encoding a putative ATP synthase protein I were highly overexpressed in S. xylosus C2a, which has to adapt to the pH 5.9 of a meat model as the inoculum was grown in chemical defined medium at pH 7.0 [38].
S. xylosus C2a also highly overexpressed the cluster dlt (SXYL_01987-90) involved in d-alanylation of teichoic acids. The degree of d-alanylation varies depending on environmental conditions such as pH, temperature or salt [79]. Inactivation of dltC in Streptococcus mutans resulted in the generation of an acid-sensitive strain that could not grow below pH 6.5 [80].
In addition, S. xylosus C2a can generate ammonia to neutralise acids, in particular via arginase and urease; the cluster ureDGFECBA encoding urease was overexpressed in a meat model [53]. Similarly, increased urease activity appeared to be an important factor in the acid defence in S. aureus [81]. The gene vraS (SXYL_00951) involved in a two-component system was upregulated in S. xylosus in a meat model [38]; this gene was also upregulated in S. aureus after an acid shock and is involved in the cell wall stimulon response [81]. This production of ammonia could contribute to the pH increase during Mediterranean sausage ripening and to the flavour [6].
4. Functional Properties
S. xylosus used as starter culture in sausage manufacturing contributes to the development of sensorial quality, in particular the typical cured colour via its nitrate reductase activity, and to flavour, by producing odorous metabolites from pyruvate and amino acid catabolism and limiting oxidation of free fatty acids [6,82,83,84].
4.1. Colour Development
The typical cured colour pigment, nitrosomyoglobin, results from a series of reactions involving the formation of nitrogen oxide (NO), which interacts with the iron Fe2+ of the cofactor heme of the myoglobin [64]. The substrate to produce NO could be nitrate or nitrite. Nitrite undergoes chemical reactions that lead to NO in the sausage. These reactions are favoured by the acidification caused by lactic acid bacteria. Addition of nitrate leads to its reduction to nitrite by nitrate reductase of staphylococci. S. xylosus strains exhibit variable nitrate reductase activity. Sanchez Mainar and Leroy [85] showed in 13 strains of S. xylosus that about 1/3 strains have high activity, 1/3 moderate activity and the remaining have little or no nitrate reductase activity. Similarly, among 23 strains of S. xylosus, Mauriello and colleagues noted that 57% have a high activity and 17% an intermediate activity [86]. The nitrate reductase of S. carnosus is encoded by the nar operon [67,87]. This species also has a nitrite reductase, which reduces nitrite to ammonia and is encoded by the nir operon [67,88]. These operons are similar in S. xylosus. The operon narGHJI (SXYL_00539-42) encodes the subunits α, β, δ and γ of the nitrate reductase and the gene narT (SXYL_00547) is involved in the transport of nitrate. Upstream, the operon nir is composed of five genes nirR, sirA, nirB, nirD and sirB (SXYL_00531-36) with nirR encoding a regulator, nirBD encoding the nitrite reductase and sirA and sirB are necessary for biosynthesis of the siroheme prosthetic group. Downstream from the nar operon, the operon nreABC (SXYL_00543-45) is involved in the regulation of the operon nar and nir in anaerobiosis and in the presence of nitrate [89]. Transcription of the operons nar, nir and nre was enhanced in a meat model without added nitrite or nitrate, probably due to anaerobic conditions [38]. When nitrate and nitrite were added to a meat model, reduction of nitrate by S. xylosus was mostly achieved after 24 h of incubation [53].
Safety considerations about nitrite and its potential to form carcinogenic nitrosamines have led to the development of alternatives to this additive [64]. The formation of nitrosomyoglobin by S. xylosus has been evidenced in laboratory media and meat but the mechanisms remained to be demonstrated [90,91]. This formation could rely on nitric oxide synthase (NOS), which produces NO from arginine. NOS activity was evidenced in S. xylosus C2a through nitrosomyoglobin formation [71]. In parallel, this strain forms oxymyoglobin, probably by its capacity to reduce the Fe3+ of metmyoglobin to Fe2+.
4.2. Flavour Development
4.2.1. Pyruvate Catabolism
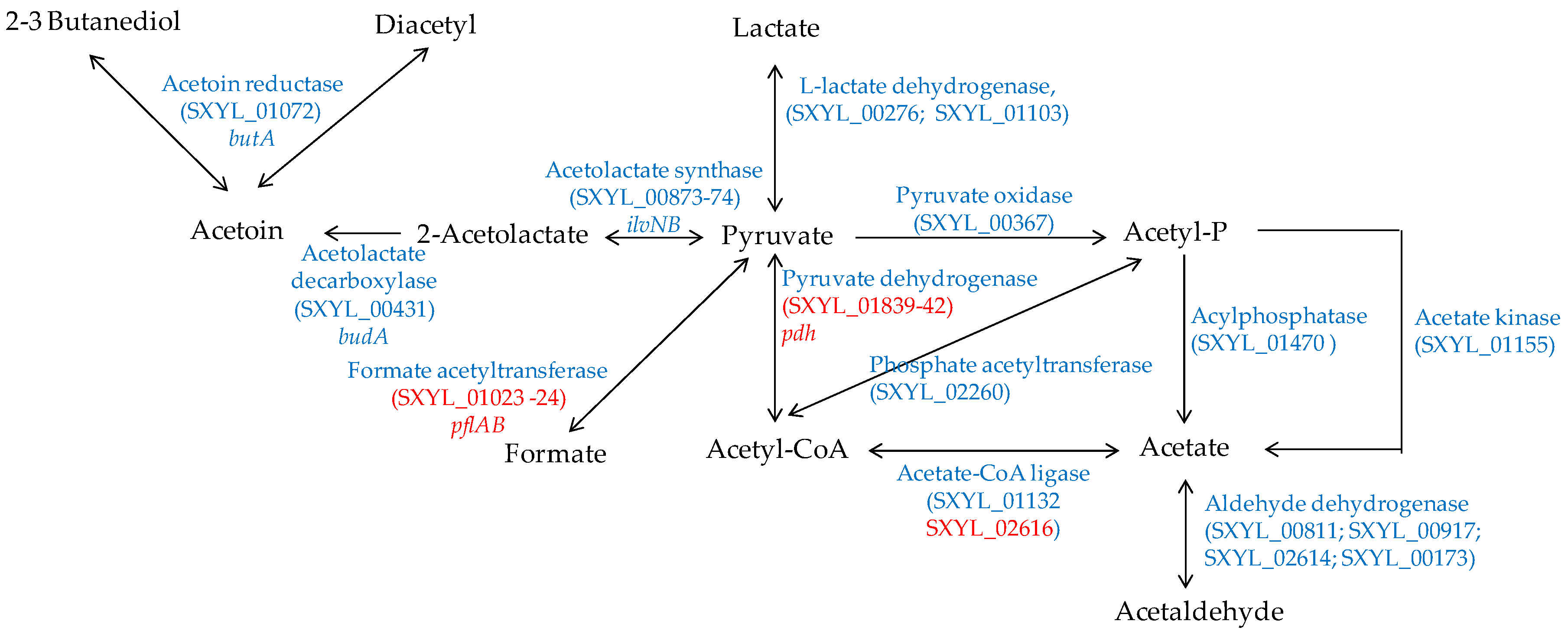
In a meat model, S. xylosus C2a produced mainly acetate from glucose and lactate, which were catabolised simultaneously [38]. Similarly, acetate was found in sausages containing glucose and inoculated by different strains of S. xylosus [92,93]. Acetate contributes to the acidic taste and to the aroma in sausage by providing a hint of vinegar [94]. In a meat model, S. xylosus C2a catabolised pyruvate to acetyl-CoA by the pyruvate dehydrogenase encoded by the pdh cluster, and then to acetate by acetate CoA-ligase (Figure 4) [38]. In addition, S. xylosus C2a has the genetic potential to synthesise acetate from acetyl-phosphate originating either from pyruvate or from acetyl-CoA (Figure 4). The formate acetyltransferase encoded by pflAB was overexpressed in a meat model, but formate was not measured [38].
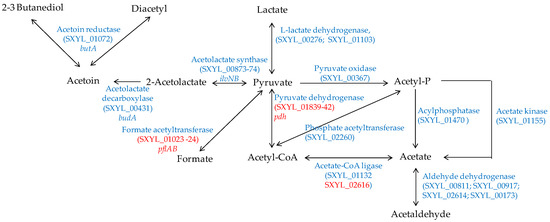
Figure 4.
Pyruvate catabolism in S. xylosus C2a. In red, genes overexpressed in a meat model.
S. xylosus C2a can synthesise acetoin, diacetyl and butanediol from pyruvate (Figure 4). These compounds with a buttery odour were found in sausages inoculated by S. xylosus [92,93]. Acetoin was produced with intraspecies variability by several strains of S. xylosus in a meat simulation medium [95]. One of these strains produced acetoin in a northern European sausage and acetoin and diacetyl in a southern European sausage [95].
4.2.2. Amino Acid Catabolism
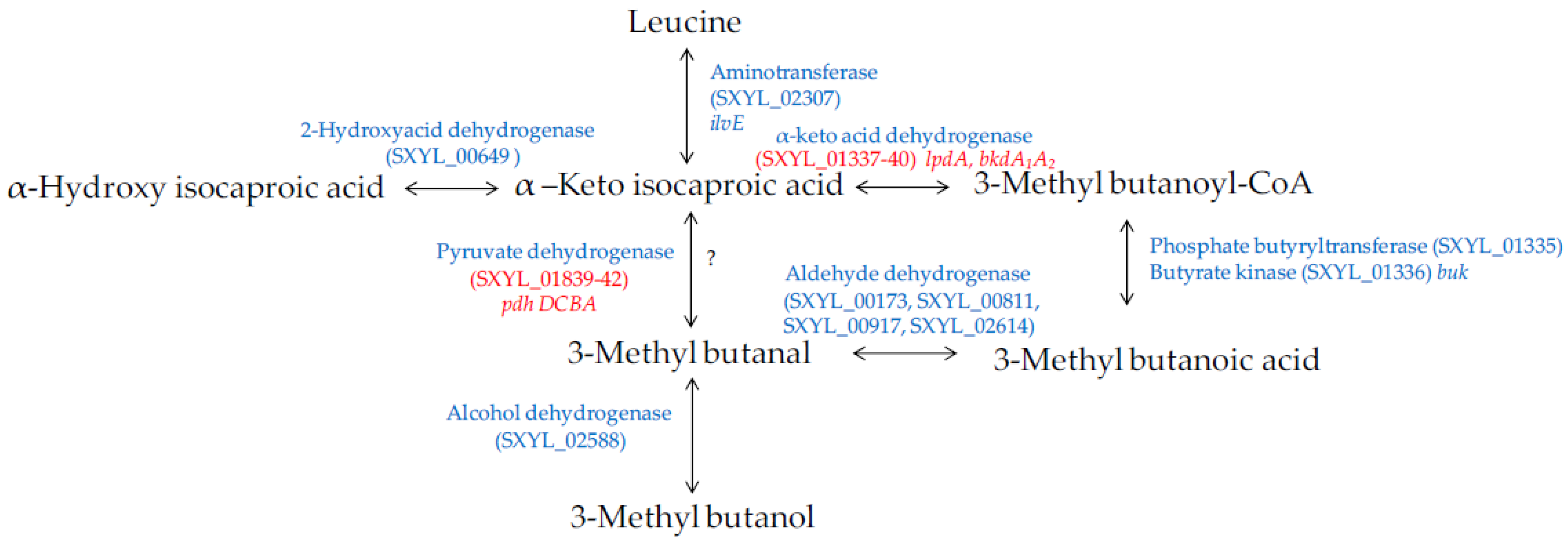
Amino acids, in particular branched-chain amino acids (leucine, isoleucine, and valine), are catabolised into aldehydes, alcohols and acids during sausage manufacturing [94,96]. Methyl-aldehydes with malty odours, methyl-alcohols with fruity odours and methyl-acids with cheesy odours contribute to the aroma of sausages. The catabolism of these amino acids is modulated by S. xylosus [92,95,97]. The major metabolite identified was 3-methyl butanol arising from leucine catabolism in minced meat or in sausage [92,95] and in laboratory media [98,99]. The production of this metabolite was substantial during the growth of S. xylosus and was observed at different pHs (5.0 to 6.0) and temperatures (20 to 30 °C), parameters relevant for sausage manufacturing [98]. The other metabolites, methyl-aldehyde and methyl-acid, were often identified as minor compounds and depending on the temperature, pH and salt [100,101]. The studies of Beck et al. [102,103] showed that methyl-aldehydes were oxidised into methyl-acids becoming the major metabolites. S. xylosus C2a catabolises leucine to alpha-ketoisocaproic acid by a transaminase encoded by ilvE (Figure 5). Then, a cluster of 6 genes (SXYL_01335-40) could be involved in the formation of 3-methyl butanoic acid. First, the branched-chain alpha-keto acid dehydrogenase complex leads to the formation of 3-methyl butanoyl-CoA, and phosphate butyryltransferase and butyrate kinase lead to 3-methylbutanoic acid. In S. xylosus C2a, the cluster SXYL_01337-40 was overexpressed in a meat model [38]. From 3-methylbutanoic acid, 3-methylbutanal can be formed by aldehyde dehydrogenase and then 3-methylbutanol by alcohol dehydrogenase (Figure 5). We did not find any gene encoding a branched-chain keto acid decarboxylase, thus it seemed that the formation of 3-methyl butanal from alpha-ketoisocaproic acid is not possible by S. xylosus C2a, and this does not confirm the hypothesis of Beck et al. [102]. Eventually, the pyruvate dehydrogenase could catalyse the decarboxylation of alpha-ketoisocaproic as described for Bacillus subtilis [104].
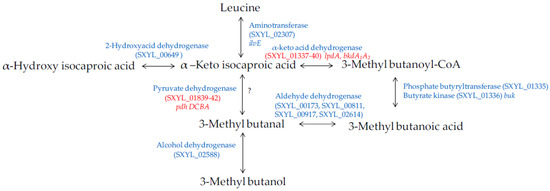
Figure 5.
Leucine catabolism in S. xylosus C2a. In red, genes differentially expressed in a meat model.
4.2.3. Lipolysis and Fatty Acid Oxidation
Lipolysis occurs during sausage manufacturing and releases mostly long-chain fatty acids and is to a great extent due to endogenous triglyceride lipases and phospholipases [6,105]. S. xylosus can contribute to lipolysis as most strains were able to hydrolyse pork fat [86]. For S. xylosus, two lipases have been described: SXL and GehM, which share 53% identity [106,107]. Extracellular lipase SXL alone is well characterised. It is a monomeric protein (43 kDa), which is almost identical to the lipases of Staphylococcus aureus and Staphylococcus simulans. Its peak activity is at pH 8.2 and 45 °C, it is able to hydrolyse triacylglycerols without chain length specificity, and it is stable between pH 5 and 8.5 and thermostable [107]. A second lipase SXL named SXL2 shares 98.7% identity with SXL [108]. It is also an alkaline lipase (pH 8.5) which acts at high temperature (55 °C) and hydrolyses preferentially short-chain substrates. The structural stability of SXL lipase is modulated by Zn2+ ions [109]. The SXL genes are not present in the three completed genomes of S. xylosus strains. The lipase GehM is thermostable with a peak activity at pH 9 and 42 °C [110]. The expression of gehM was downregulated by the presence of triglycerides in the culture medium [111]. gehM is present in the genome of SMQ-121 and HKUOPL8 strains, but is a pseudogene in the strain C2a. In the genome of the strain S. xylosus C2a, five genes encoding putative lipases, one encoding a putative phospholipase and three encoding putative lysophospholipases are present (LN554884).
The oxidation of unsaturated fatty acids released by lipolysis results in numerous volatile compounds, including aldehydes, alcohols and ketones, which contribute to the aroma of the sausages [6,96]. This oxidation is essentially a chemical peroxidation via ROS. As underlined above, S. xylosus is well equipped to detoxify ROS and thus to limit this fatty acid oxidation. Indeed, it was able to inhibit the oxidation of linoleic and linolenic unsaturated fatty acids in laboratory media [112]. Furthermore, mutants deficient in SOD or catalase activity of S. xylosus C2a were less efficient than the wild type in limiting oxidation of unsaturated fatty acids [113].
5. Conclusions
The genome analysis of S. xylosus used as a meat starter culture together with transcriptomic approaches in situ in meat have highlighted that this bacterium has all functions necessary for its adaptation to meat substrates and to technological stress, and has the potential to contribute to the sensorial quality of sausages. Nevertheless, these functional properties vary according to the strains. Furthermore, the selection of strains for starter cultures should include safety criteria, such as the lack of production of biogenic amines [82,83,114] and enterotoxins and of transferable antibiotic resistance genes [82,114].
Acknowledgments
The authors are grateful to David Marsh for correcting the English.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kloos, W.E.; Zimmerman, R.J.; Smith, R.F. Preliminary studies on the characterization and distribution of Staphylococcus and Micrococcus species on animal skin. Appl. Environ. Microbiol. 1976, 31, 53–59. [Google Scholar] [PubMed]
- Nagase, N.; Sasaki, A.; Yamashita, K.; Shimizu, A.; Wakita, Y.; Kitai, S.; Kawano, J. Isolation and species distribution of staphylococci from animal and human skin. J. Vet. Med. Sci. 2002, 64, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Rantsiou, K.; Iacumin, L.; Urso, R.; Cantoni, C.; Comi, G. Study of the ecology of fresh sausages and characterization of populations of lactic acid bacteria by molecular methods. Appl. Environ. Microbiol. 2004, 70, 1883–1894. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Iacumin, L.; Cantoni, C.; Comi, G.; Cocolin, L. Ecology and characterization by molecular methods of Staphylococcus species isolated from fresh sausages. Int. J. Food Microbiol. 2005, 97, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Chaillou, S.; Chaulot-Talmon, A.; Caekebeke, H.; Cardinal, M.; Christieans, S.; Denis, C.; Desmonts, M.H.; Dousset, X.; Feurer, C.; Hamon, E.; et al. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J. 2015, 9, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Talon, R.; Leroy, S. Fermented meat products and the role of starter cultures. In Encyclopedia of Food Microbiology; Batt, C.A., Tortorello, M.L., Eds.; Elsevier; Academic Press: London, UK, 2014; Volume 1, pp. 870–874. [Google Scholar]
- Ratsimba, A.; Leroy, S.; Chacornac, J.P.; Rakoto, D.; Arnaud, E.; Jeannoda, V.; Talon, R. Staphylococcal ecosystem of kitoza, a traditional malagasy meat product. Int. J. Food Microbiol. 2017, 246, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Manzano, M.; Aggio, D.; Cantoni, C.; Comi, G. A novel polymerase chain reaction (PCR)—Denaturing gradient gel electrophoresis (DGGE) for the identification of micrococcaceae strains involved in meat fermentations. Its application to naturally fermented italian sausages. Meat Sci. 2001, 58, 59–64. [Google Scholar] [CrossRef]
- Coppola, S.; Mauriello, G.; Aponte, M.; Moschetti, G.; Villani, F. Microbial succession during ripening of naples-type salami, a southern italian fermented sausage. Meat Sci. 2000, 56, 321–329. [Google Scholar] [CrossRef]
- Rossi, F.; Tofalo, R.; Torriani, S.; Suzzi, G. Identification by 16S-23S rDNA intergenic region amplification, genotypic and phenotypic clustering of Staphylococcus xylosus strains from dry sausages. J. Appl. Microbiol. 2001, 90, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Drosinos, E.H.; Mataragas, M.; Xiraphi, N.; Moschonas, G.; Gaitis, F.; Metaxopoulos, J. Characterization of the microbial flora from a traditional greek fermented sausage. Meat Sci. 2005, 69, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Garcίa-Varona, M.; Santos, E.M.; Jaime, I.; Rovira, J. Characterisation of micrococcaceae isolated from different varieties of chorizo. Int. J. Food Microbiol. 2000, 54, 189–195. [Google Scholar] [CrossRef]
- Greppi, A.; Ferrocino, I.; La Storia, A.; Rantsiou, K.; Ercolini, D.; Cocolin, L. Monitoring of the microbiota of fermented sausages by culture independent rRNA-based approaches. Int. J. Food Microbiol. 2015, 212, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Aymerich, T.; Martίn, B.; Garriga, M.; Hugas, M. Microbial quality and direct PCR identification of lactic acid bacteria and nonpathogenic staphylococci from artisanal low-acid sausages. Appl. Environ. Microbiol. 2003, 69, 4583–4594. [Google Scholar] [CrossRef] [PubMed]
- Leroy, S.; Giammarinaro, P.; Chacornac, J.P.; Lebert, I.; Talon, R. Biodiversity of indigenous staphylococci of naturally fermented dry sausages and manufacturing environments of small-scale processing units. Food Microbiol. 2010, 27, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Połka, J.; Rebecchi, A.; Pisacane, V.; Morelli, L.; Puglisi, E. Bacterial diversity in typical italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 2015, 46, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Aquilanti, L.; Garofalo, C.; Osimani, A.; Clementi, F. Ecology of lactic acid bacteria and coagulase-negative cocci in fermented dry sausages manufactured in italy and other mediterranean countries: An overview. Int. Food Res. J. 2016, 23, 429–445. [Google Scholar]
- Rebecchi, A.; Pisacane, V.; Miragoli, F.; Połka, J.; Falasconi, I.; Morelli, L.; Puglisi, E. High-throughput assessment of bacterial ecology in hog, cow and ovine casings used in sausages production. Int. J. Food Microbiol. 2015, 212, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Rodrίguez, M.; Núñez, F.; Córdoba, J.J.; Sanabria, C.; Bermúdez, E.; Asensio, M.A. Characterization of Staphylococcus spp. and Micrococcus spp. isolated from iberian ham throughout the ripening process. Int. J. Food Microbiol. 1994, 24, 329–335. [Google Scholar] [CrossRef]
- Vilar, I.; García Fontán, M.C.; Prieto, B.; Tornadijo, M.E.; Carballo, J. A survey on the microbiological changes during the manufacture of dry-cured lacon, a spanish traditional meat product. J. Appl. Microbiol. 2000, 89, 1018–1026. [Google Scholar] [CrossRef] [PubMed]
- Cordero, M.R.; Zumalacárregui, J.M. Characterization of micrococcaceae isolated from salt used for spanish dry-cured ham. Lett. Appl. Microbiol. 2000, 31, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Blaiotta, G.; Pennacchia, C.; Villani, F.; Ricciardi, A.; Tofalo, R.; Parente, E. Diversity and dynamics of communities of coagulase-negative staphylococci in traditional fermented sausages. J. Appl. Microbiol. 2004, 97, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Corbière Morot-Bizot, S.; Leroy, S.; Talon, R. Monitoring of staphylococcal starters in two french processing plants manufacturing dry fermented sausages. J. Appl. Microbiol. 2007, 102, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; El Haddad, L.; Tremblay, D.M.; Plante, P.L.; Wasserscheid, J.; Dumaresq, J.; Dewar, K.; Corbeil, J.; Moineau, S. First complete genome sequence of Staphylococcus xylosus, a meat starter culture and a host to propagate Staphylococcus aureus phages. Genome Announc. 2014, 2, e00671-14. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.P.; Jiang, J.; Tun, H.M.; Mauroo, N.F.; Yuen, C.S.; Leung, F.C. Complete genome sequence of Staphylococcus xylosus HKUOPL8, a potential opportunistic pathogen of mammals. Genome Announc. 2014, 2, e00653-14. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Liu, L.; Liu, S.; Yang, D.; Liu, Y.; Yang, S.; Jia, A.; Qin, N. Genome of Staphylococcus xylosus and comparison with S. aureus and S. epidermidis. J. Genet. Genom. 2014, 41, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Arora, A.; Sathyabama, S.; Mubin, N.; Verma, S.; Mayilraj, S.; Agrewala, J.N. Genome sequencing, assembly, annotation and analysis of Staphylococcus xylosus strain DMB3-Bh1 reveals genes responsible for pathogenicity. Gut Pathog. 2016, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Toldrá, F.; Reig, M. The biochemistry of meat and fat. In Handbook of Fermented Meat and Poultry, 2nd ed.; Toldrá, F., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2015; pp. 49–54. [Google Scholar]
- Götz, F.; Bannerman, T.; Schleifer, K.-H. The genera Staphylococcus and Macrococcus. Prokaryotes 2006, 4, 5–75. [Google Scholar] [CrossRef]
- Brückner, R.; Rosenstein, R. Carbohydrate catabolism: Pathways and regulation—Chapter 34. In Gram-Positive Pathogens, 2nd ed.; Fischetti, V., Novick, R., Ferretti, J., Portnoy, D., Rood, J., Eds.; American Society of Microbiology Press: Washington, DC, USA, 2006; pp. 427–433. [Google Scholar]
- Fiegler, H.; Bassias, J.; Jankovic, I.; Brückner, R. Identification of a gene in Staphylococcus xylosus encoding a novel glucose uptake protein. J. Bacteriol. 1999, 181, 4929–4936. [Google Scholar] [PubMed]
- Jankovic, I.; Brückner, R. Carbon catabolite repression by the catabolite control protein CcpA in Staphylococcus xylosus. J. Mol. Microbiol. Biotechnol. 2002, 4, 309–314. [Google Scholar] [PubMed]
- Christiansen, I.; Hengstenberg, W. Staphylococcal phosphoenolpyruvate-dependent phosphotransferase system—Two highly similar glucose permeases in Staphylococcus carnosus with different glucoside specificity: Protein engineering in vivo? Microbiology 1999, 145, 2881–2889. [Google Scholar] [CrossRef] [PubMed]
- Arkouledos, J.S.; Nychas, G.J. Comparative studies of the growth of Staphylococcus carnosus with or without glucose. Lett. Appl. Microbiol. 1995, 20, 19–24. [Google Scholar]
- Ferreira, M.T.; Manso, A.S.; Gaspar, P.; Pinho, M.G.; Neves, A.R. Effect of oxygen on glucose metabolism: Utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLoS ONE 2013, 8, e58277. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Mainar, M.; Matheuse, F.; De Vuyst, L.; Leroy, F. Effects of glucose and oxygen on arginine metabolism by coagulase-negative staphylococci. Food Microbiol. 2017, 65, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Seidl, K.; Muller, S.; Francois, P.; Kriebitzsch, C.; Schrenzel, J.; Engelmann, S.; Bischoff, M.; Berger-Bachi, B. Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 2009, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Vermassen, A.; Dordet-Frisoni, E.; de La Foye, A.; Micheau, P.; Laroute, V.; Leroy, S.; Talon, R. Adaptation of Staphylococcus xylosus to nutrients and osmotic stress in a salted meat model. Front. Microbiol. 2016, 7, 87. [Google Scholar] [CrossRef] [PubMed]
- Brückner, R.; Wagner, E.; Götz, F. Characterization of a sucrase gene from Staphylococcus xylosus. J. Bacteriol. 1993, 175, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, I.; Brückner, R. Carbon catabolite repression of sucrose utilization in Staphylococcus xylosus: Catabolite control protein CcpA ensures glucose preference and autoregulatory limitation of sucrose utilization. J. Mol. Microbiol. Biotechnol. 2007, 12, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Gering, M.; Brückner, R. Transcriptional regulation of the sucrase gene of Staphylococcus xylosus by the repressor ScrR. J. Bacteriol. 1996, 178, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Poolman, B.; Knol, J.; van der Does, C.; Henderson, P.J.; Liang, W.J.; Leblanc, G.; Pourcher, T.; Mus-Veteau, I. Cation and sugar selectivity determinants in a novel family of transport proteins. Mol. Microbiol. 1996, 19, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Bassias, J.; Brückner, R. Regulation of lactose utilization genes in Staphylococcus xylosus. J. Bacteriol. 1998, 180, 2273–2279. [Google Scholar] [PubMed]
- Hughes, M.C.; Kerry, J.P.; Arendt, E.K.; Kenneally, P.M.; McSweeney, P.L.; O’Neill, E.E. Characterization of proteolysis during the ripening of semi-dry fermented sausages. Meat Sci. 2002, 62, 205–216. [Google Scholar] [CrossRef]
- Miralles, M.C.; Flores, J.; Perez-Martinez, G. Biochemical tests for the selection of Staphylococcus strains as potential meat starter cultures. Food Microbiol. 1996, 13, 227–236. [Google Scholar] [CrossRef]
- Aro Aro, J.M.; Nyam-Osor, P.; Tsuji, K.; Shimada, K.I.; Fukushima, M.; Sekikawa, M. The effect of starter cultures on proteolytic changes and amino acid content in fermented sausages. Food Chem. 2010, 119, 279–285. [Google Scholar] [CrossRef]
- Monnet, V. Bacterial oligopeptide-binding proteins. Cell. Mol. Life Sci. 2003, 60, 2100–2114. [Google Scholar] [CrossRef] [PubMed]
- Fiegler, H.; Brückner, R. Identification of the serine acetyltransferase gene of Staphylococcus xylosus. FEMS Microbiol. Lett. 1997, 148, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Majerczyk, C.D.; Dunman, P.M.; Luong, T.T.; Lee, C.Y.; Sadykov, M.R.; Somerville, G.A.; Bodi, K.; Sonenshein, A.L. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 2010, 192, 2861–2877. [Google Scholar] [CrossRef] [PubMed]
- Nuxoll, A.S.; Halouska, S.M.; Sadykov, M.R.; Hanke, M.L.; Bayles, K.W.; Kielian, T.; Powers, R.; Fey, P.D. CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism. PLoS Pathog. 2012, 8, e1003033. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Mainar, M.; Weckx, S.; Leroy, F. Coagulase-negative staphylococci favor conversion of arginine into ornithine despite a widespread genetic potential for nitric oxide synthase activity. Appl. Environ. Microbiol. 2014, 80, 7741–7751. [Google Scholar] [CrossRef] [PubMed]
- Janssens, M.; Van der Mijnsbrugge, A.; Sanchez Mainar, M.; Balzarini, T.; De Vuyst, L.; Leroy, F. The use of nucleosides and arginine as alternative energy sources by coagulase-negative staphylococci in view of meat fermentation. Food Microbiol. 2014, 39, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Vermassen, A.; de la Foye, A.; Loux, V.; Talon, R.; Leroy, S. Transcriptomic analysis of Staphylococcus xylosus in the presence of nitrate and nitrite in meat reveals its response to nitrosative stress. Front. Microbiol. 2014, 5, 691. [Google Scholar] [CrossRef] [PubMed]
- Batlle, N.; Aristoy, M.C.; Toldrá, F. ATP metabolites during aging of exudative and nonexudative pork meats. J. Food Sci. 2001, 66, 68–71. [Google Scholar] [CrossRef]
- Switzer, R.L.; Zalkin, H.; Saxild, H.H. Purine, pyrimidine, and pyridine nucleotide metabolism. In Bacillus Subtilis and Its Closest Relatives; Sonenshein, A., Hoch, J.A., Losick, R., Eds.; American Society of Microbiology Press: Washington, DC, USA, 2002. [Google Scholar]
- Chaillou, S.; Champomier-Vergès, M.C.; Cornet, M.; Crutz-Le Coq, A.M.; Dudez, A.M.; Martin, V.; Beaufils, S.; Darbon-Rongère, E.; Bossy, R.; Loux, V.; et al. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 2005, 23, 1527–1533. [Google Scholar] [CrossRef] [PubMed]
- Rimaux, T.; Vrancken, G.; Vuylsteke, B.; De Vuyst, L.; Leroy, F. The pentose moiety of adenosine and inosine is an important energy source for the fermented-meat starter culture Lactobacillus sakei CTC 494. Appl. Environ. Microbiol. 2011, 77, 6539–6550. [Google Scholar] [CrossRef] [PubMed]
- Batlle, N.; Aristoy, M.C.; Toldrá, F. Early postmortem detection of exudative pork meat based on nucleotide content. J. Food Sci. 2000, 65, 413–416. [Google Scholar] [CrossRef]
- Vermassen, A.; Talon, R.; Leroy, S. Ferritin, an iron source in meat for Staphylococcus xylosus? Int. J. Food Microbiol. 2016, 225, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Skaar, E.P.; Schneewind, O. Iron-regulated surface determinants (isd) of Staphylococcus aureus: Stealing iron from heme. Microbes Infect. 2004, 6, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, J.R.; Heinrichs, D.E. The iron-regulated staphylococcal lipoproteins. Front. Cell. Infect. Microbiol. 2012, 2, 41. [Google Scholar] [CrossRef] [PubMed]
- Hammer, N.D.; Skaar, E.P. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 2011, 65, 129–147. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, R.; Futter-Bryniok, D.; Götz, F. The choline-converting pathway in Staphylococcus xylosus C2a: Genetic and physiological characterization. J. Bacteriol. 1999, 181, 2273–2278. [Google Scholar] [PubMed]
- Hammes, W.P. Metabolism of nitrate in fermented meats: The characteristic feature of a specific group of fermented foods. Food Microbiol. 2012, 29, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Gaupp, R.; Ledala, N.; Somerville, G.A. Staphylococcal response to oxidative stress. Front. Cell. Infect. Microbiol. 2012, 2, 33. [Google Scholar] [CrossRef] [PubMed]
- Barrière, C.; Brückner, R.; Talon, R. Characterization of the single superoxide dismutase of Staphylococcus xylosus. Appl. Environ. Microbiol. 2001, 67, 4096–4104. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, R.; Nerz, C.; Biswas, L.; Resch, A.; Raddatz, G.; Schuster, S.C.; Götz, F. Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 2009, 75, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Blaiotta, G.; Fusco, V.; Ercolini, D.; Pepe, O.; Coppola, S. Diversity of Staphylococcus species strains based on partial kat (catalase) gene sequences and design of a PCR-restriction fragment length polymorphism assay for identification and differentiation of coagulase-positive species (S. aureus, S. delphini, S. hyicus, S. intermedius, S. pseudintermedius, and S. schleiferi subsp. coagulans). J. Clin. Microbiol. 2010, 48, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Barrière, C.; Brückner, R.; Centeno, D.; Talon, R. Characterisation of the katA gene encoding a catalase and evidence for at least a second catalase activity in Staphylococcus xylosus, bacteria used in food fermentation. FEMS Microbiol. Lett. 2002, 216, 277–283. [Google Scholar] [CrossRef]
- Tavares, A.F.; Nobre, L.S.; Melo, A.M.; Saraiva, L.M. A novel nitroreductase of Staphylococcus aureus with S-nitrosoglutathione reductase activity. J. Bacteriol. 2009, 191, 3403–3406. [Google Scholar] [CrossRef] [PubMed]
- Ras, G.; Zuliani, V.; Derkx, P.; Seibert, T.M.; Leroy, S.; Talon, R. Evidence for nitric oxide synthase activity in Staphylococcus xylosus mediating nitrosoheme formation. Front. Microbiol. 2017, 8, 598. [Google Scholar] [CrossRef] [PubMed]
- Van Sorge, N.M.; Beasley, F.C.; Gusarov, I.; Gonzalez, D.J.; von Köckritz-Blickwede, M.; Anik, S.; Borkowski, A.W.; Dorrestein, P.C.; Nudler, E.; Nizet, V. Methicillin-resistant Staphylococcus aureus bacterial nitric-oxide synthase affects antibiotic sensitivity and skin abscess development. J. Biol. Chem. 2013, 288, 6417–6426. [Google Scholar] [CrossRef] [PubMed]
- Sapp, A.M.; Mogen, A.B.; Almand, E.A.; Rivera, F.E.; Shaw, L.N.; Richardson, A.R.; Rice, K.C. Contribution of the nos-pdt operon to virulence phenotypes in methicillin-sensitive Staphylococcus aureus. PLoS ONE 2014, 9, e108868. [Google Scholar] [CrossRef] [PubMed]
- Clauditz, A.; Resch, A.; Wieland, K.P.; Peschel, A.; Götz, F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 2006, 74, 4950–4953. [Google Scholar] [CrossRef] [PubMed]
- Uziel, O.; Borovok, I.; Schreiber, R.; Cohen, G.; Aharonowitz, Y. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 2004, 186, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Moskovitz, J. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: Expression of activity androles in tolerance of oxidative stress. Microbiology 2003, 149, 2739–2747. [Google Scholar] [CrossRef] [PubMed]
- Demeyer, D.; Raemaekers, M.; Rizzo, A.; Holck, A.; De Smedt, A.; Ten Brink, B.; Hagen, B.; Montel, C.; Zanardi, E.; Murbrekk, E.; et al. Control of bioflavour and safety in fermented sausages: First results of a european project. Food Res. Int. 2000, 33, 171–180. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C. Surviving the acid test: Responses of Gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 2003, 67, 429–453. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, F.C.; Baddiley, J. A continuum of anionic charge: Structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 686–723. [Google Scholar] [CrossRef] [PubMed]
- Boyd, D.A.; Cvitkovitch, D.G.; Bleiweis, A.S.; Kiriukhin, M.Y.; Debabov, D.V.; Neuhaus, F.C.; Hamilton, I.R. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 2000, 182, 6055–6065. [Google Scholar] [CrossRef] [PubMed]
- Bore, E.; Langsrud, S.; Langsrud, O.; Rode, T.M.; Holck, A. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 2007, 153, 2289–2303. [Google Scholar] [CrossRef] [PubMed]
- Leroy, S.; Vermassen, A.; Talon, R. Staphylococcus: Occurrence and properties. In Encyclopedia of Food and Health; Cabalerro, B., Finglas, P.M., Toldrá, F., Eds.; Elsevier Limited Publisher: Oxford, UK, 2016; Volume 5, pp. 104–145. [Google Scholar]
- Sánchez Mainar, M.; Stavropoulou, D.A.; Leroy, F. Exploring the metabolic heterogeneity of coagulase-negative staphylococci to improve the quality and safety of fermented meats: A review. Int. J. Food Microbiol. 2016, 247, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Cocconcelli, P.S.; Fontana, C. Bacteria. In Handbook of Fermented Meat and Poultry; Toldrá, F., Hui, H., Astiasarán, I., Sebranek, J.G., Talon, R., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2014; pp. 117–128. [Google Scholar]
- Sánchez Mainar, M.; Leroy, F. Process-driven bacterial community dynamics are key to cured meat colour formation by coagulase-negative staphylococci via nitrate reductase or nitric oxide synthase activities. Int. J. Food Microbiol. 2015, 212, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Mauriello, G.; Casaburi, A.; Blaiotta, G.; Villani, F. Isolation and technological properties of coagulase negative staphylococci from fermented sausages of southern italy. Meat Sci. 2004, 67, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Pantel, I.; Lindgren, P.E.; Neubauer, H.; Götz, F. Identification and characterization of the Staphylococcus carnosus nitrate reductase operon. Mol. Gen. Genet. 1998, 259, 105–114. [Google Scholar] [PubMed]
- Neubauer, H.; Pantel, I.; Götz, F. Molecular characterization of the nitrite-reducing system of Staphylococcus carnosus. J. Bacteriol. 1999, 181, 1481–1488. [Google Scholar] [PubMed]
- Schlag, S.; Fuchs, S.; Nerz, C.; Gaupp, R.; Engelmann, S.; Liebeke, M.; Lalk, M.; Hecker, M.; Götz, F. Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J. Bacteriol. 2008, 190, 7847–7858. [Google Scholar] [CrossRef] [PubMed]
- Morita, H.; Sakata, R.; Nagata, Y. Nitric oxide complex of iron(ii) myoglobin converted from metmyoglobin by Staphylococcus xylosus. J. Food Sci. 1998, 63, 352–355. [Google Scholar] [CrossRef]
- Li, P.; Kong, B.; Chen, Q.; Zheng, D.; Liu, N. Formation and identification of nitrosylmyoglobin by Staphylococcus xylosus in raw meat batters: A potential solution for nitrite substitution in meat products. Meat Sci. 2013, 93, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Sondergaard, A.K.; Stahnke, L.H. Growth and aroma production by Staphylococcus xylosus, S. carnosus and S. equorum—A comparative study in model systems. Int. J. Food Microbiol. 2002, 75, 99–109. [Google Scholar] [CrossRef]
- Stahnke, L.H. Volatiles produced by Staphylococcus xylosus and Staphylococcus carnosus during growth in sausage minces—Part II. The influence of growth parameters. LWT Food Sci. Technol. 1999, 32, 365–371. [Google Scholar] [CrossRef]
- Talon, R.; Leroy-Setrin, S.; Fadda, S. Dry fermented sausages. In Handbook of Food and Beverage Fermentation Technology; Hui, Y.H., Meunier-Goddick, L., Solvejg Hansen, A., Josephsen, J., Nip, W.K., Stanfield, P.S., Toldrá, F., Eds.; Marcel Dekker, Inc.: New York, NY, USA, 2004; pp. 397–416. [Google Scholar]
- Ravyts, F.; Steen, L.; Goemaere, O.; Paelinck, H.; De Vuyst, L.; Leroy, F. The application of staphylococci with flavour-generating potential is affected by acidification in fermented dry sausages. Food Microbiol. 2010, 27, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.; Olivares, A. Flavor. In Handbook of Fermented Meat and Poultry, 2nd ed.; Toldrá, F., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2015; pp. 217–226. [Google Scholar]
- Møller, J.K.S.; Hinrichsen, L.L.; Andersen, H.J. Formation of amino acid (l-leucine, l-phenylalanine) derived volatile flavour compounds by Moraxella phenylpyruvica and Staphylococcus xylosus in cured meat model systems. Int. J. Food Microbiol. 1998, 42, 101–117. [Google Scholar] [CrossRef]
- Ravyts, F.; Vrancken, G.; D’Hondt, K.; Vasilopoulos, C.; De Vuyst, L.; Leroy, F. Kinetics of growth and 3-methyl-1-butanol production by meat-borne, coagulase-negative staphylococci in view of sausage fermentation. Int. J. Food Microbiol. 2009, 134, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulou, D.A.; Borremans, W.; De Vuyst, L.; De Smet, S.; Leroy, F. Amino acid conversions by coagulase-negative staphylococci in a rich medium: Assessment of inter- and intraspecies heterogeneity. Int. J. Food Microbiol. 2015, 212, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Tjener, K.; Stahnke, L.H.; Andersen, L.; Martinussen, J. The pH-unrelated influence of salt, temperature and manganese on aroma formation by Staphylococcus xylosus and Staphylococcus carnosus in a fermented meat model system. Int. J. Food Microbiol. 2004, 97, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Olesen, P.T.; Meyer, A.S.; Stahnke, L.H. Generation of flavour compounds in fermented sausages-the influence of curing ingredients, Staphylococcus starter culture and ripening time. Meat Sci. 2004, 66, 675–687. [Google Scholar] [CrossRef]
- Beck, H.C.; Hansen, A.M.; Lauritsen, F.R. Catabolism of leucine to branched-chain fatty acids in Staphylococcus xylosus. J. Appl. Microbiol. 2004, 96, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Beck, H.C.; Hansen, A.M.; Lauritsen, F.R. Metabolite production and kinetics of branched-chain aldehyde oxidation in Staphylococcus xylosus. Enzym. Microb. Technol. 2002, 31, 94–101. [Google Scholar] [CrossRef]
- Yvon, M.; Rijnen, L. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 2001, 11, 185–201. [Google Scholar] [CrossRef]
- Talon, R.; Leroy-Sétrin, S.; Fadda, S. Bacterial starters involved in the quality of fermented meat products—Chapter 10. In Research Advances in Quality of Meat and Meat Products; Toldrá, F., Ed.; Research Signpost: Trivandrum, Kerala, India, 2002; pp. 175–191. [Google Scholar]
- Rosenstein, R.; Götz, F. Staphylococcal lipases: Biochemical and molecular characterization. Biochimie 2000, 82, 1005–1014. [Google Scholar] [CrossRef]
- Mosbah, H.; Sayari, A.; Mejdoub, H.; Dhouib, H.; Gargouri, Y. Biochemical and molecular characterization of Staphylococcus xylosus lipase. Biochim. Biophys. Acta 2005, 1723, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Bouaziz, A.; Horchani, H.; Ben Salem, N.; Gargouri, Y.; Sayari, A. Expression, purification of a novel alkaline Staphylococcus xylosus lipase acting at high temperature. Biochem. Eng. J. 2011, 54, 93–102. [Google Scholar] [CrossRef]
- Bertoldo, J.B.; Razzera, G.; Vernal, J.; Brod, F.C.; Arisi, A.C.; Terenzi, H. Structural stability of Staphylococcus xylosus lipase is modulated by Zn(2+) ions. Biochim. Biophys. Acta 2011, 1814, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Brod, F.C.; Pelisser, M.R.; Bertoldo, J.B.; Vernal, J.; Bloch, C., Jr.; Terenzi, H.; Arisi, A.C. Heterologous expression and purification of a heat-tolerant Staphylococcus xylosus lipase. Mol. Biotechnol. 2010, 44, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Iacumin, L.; Cocolin, L.; Cantoni, C.; Comi, G. Preliminary analysis of the lipase gene (gehM) expression of Staphylococcus xylosus in vitro and during fermentation of naturally fermented sausages (in situ). J. Food Prot. 2007, 70, 2665–2669. [Google Scholar] [CrossRef] [PubMed]
- Talon, R.; Walter, D.; Montel, M.C. Growth and effect of staphylococci and lactic acid bacteria on unsaturated free fatty acids. Meat Sci. 2000, 54, 41–47. [Google Scholar] [CrossRef]
- Barrière, C.; Centeno, D.; Lebert, A.; Leroy-Sétrin, S.; Berdague, J.L.; Talon, R. Roles of superoxide dismutase and catalase of Staphylococcus xylosus in the inhibition of linoleic acid oxidation. FEMS Microbiol. Lett. 2001, 201, 181–185. [Google Scholar] [CrossRef]
- Talon, R.; Leroy, S. Diversity and safety hasards of bacteria involved in meat fermentations. Meat Sci. 2011, 89, 303–309. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).