The Microbial Communities of Leaves and Roots Associated with Turtle Grass (Thalassia testudinum) and Manatee Grass (Syringodium filliforme) are Distinct from Seawater and Sediment Communities, but Are Similar between Species and Sampling Sites
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Alpha Diversity
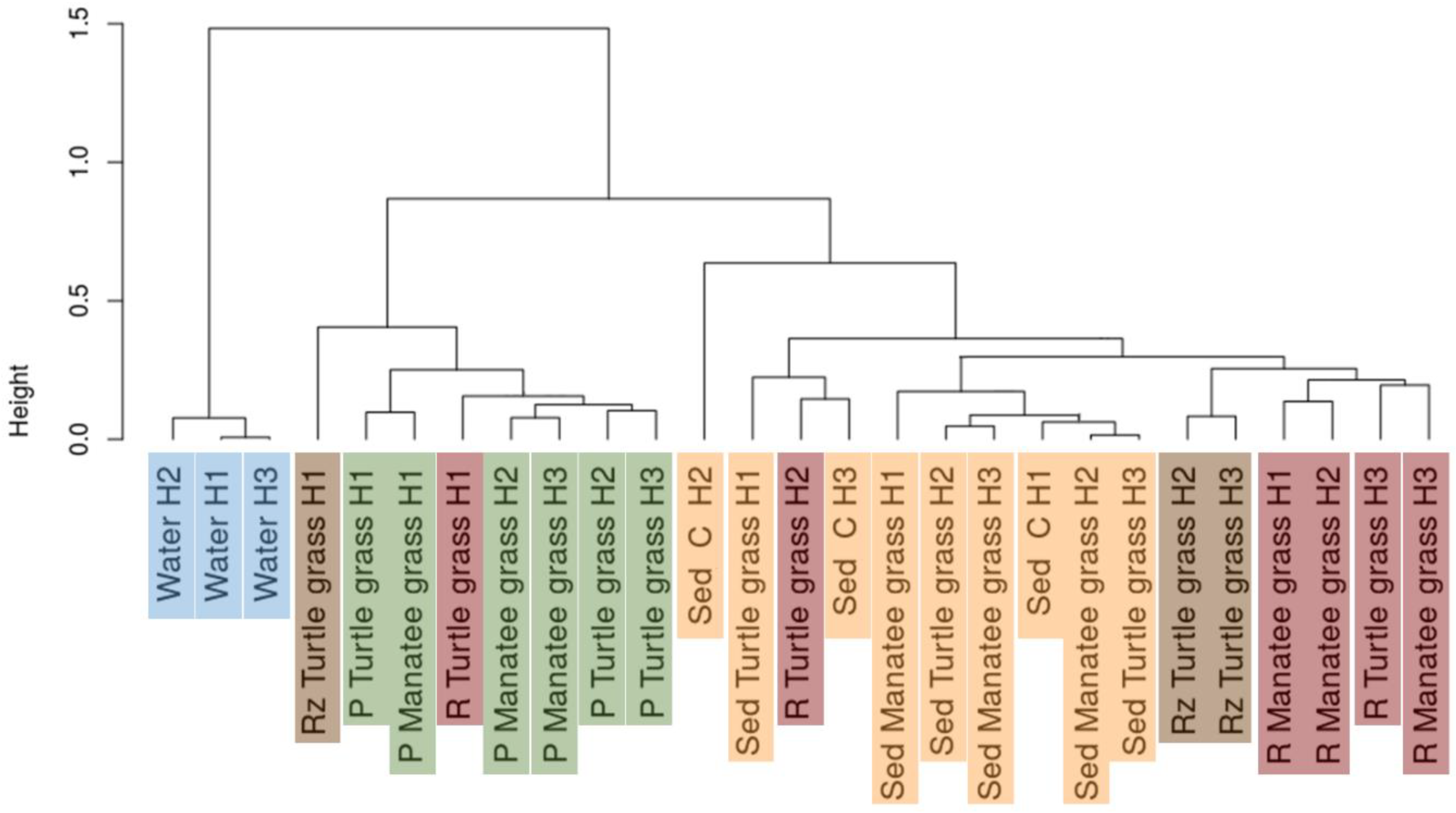
3.2. Beta Diversity
3.3. Sequence Variants and Abundant Genera
3.3.1. Aboveground
3.3.2. Belowground
3.3.3. Leaves vs. Roots and Rhizomes
3.3.4. Turtle Grass Sample-Types
3.3.5. Manatee Grass Sample-Types
3.3.6. Differences in Genera-Abundance by Seagrass Species
3.3.7. Seawater by Site
3.3.8. Phyllosphere by Site
3.3.9. Rhizosphere by Site
3.3.10. Differences in Abundance of Taxa by Site
3.4. Predictive Metagenome Analysis
4. Discussion
4.1. Microbial Communities Across Seagrass Sample-Types
4.2. Site and Species Comparison
4.3. Water and Phyllosphere
4.4. Roots, Sediments, and Rhizomes
4.5. Core Microbiome
4.6. Predictive Metabolism
4.7. Processing Methods Comparison
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schubert, N.; Demes, K.W. Phenotypic Plasticity in the Marine Angiosperm Halophila Decipiens (Hydrocharitaceae, Streptophyta). Mar. Ecol. Progress Ser. 2017, 575, 81–93. [Google Scholar] [CrossRef]
- Spalding, M.; Taylor, M.; Ravilious, C.; Short, F.; Green, E. The distribution and status of seagrasses. In World Atlas of Seagrasses; Green, E.P., Short, F.T., Eds.; University of California Press: Berkeley, CA, USA, 2003; pp. 5–26. [Google Scholar]
- Duarte, C.M.; Middelburg, J.J.; Caraco, N. Major Role of Marine Vegetation on the Oceanic Carbon Cycle. Biogeosciences 2005, 2, 1–8. [Google Scholar] [CrossRef]
- Charpy-Roubaud, C.; Sournia, A. The Comparative Estimation of Phytoplanktonic, Microphytobenthic and Macrophytobenthic Primary Production in the Oceans. Mar. Microb. Food Webs 1990, 4, 31–57. [Google Scholar]
- Ugarelli, K.; Chakrabarti, S.; Laas, P.; Stingl, U. The Seagrass Holobiont and Its Microbiome. Microorganisms 2017, 5, 81. [Google Scholar] [CrossRef]
- Mazarrasa, I.; Marba, N.; Lovelock, C.E.; Serrano, O.; Lavery, P.S.; Fourqurean, J.W.; Kennedy, H.; Mateo, M.A.; Krause-Jensen, D.; Steven, A.D.L.; et al. Seagrass Meadows as a Globally Significant Carbonate Reservoir. Biogeosciences 2015, 12, 4993–5003. [Google Scholar] [CrossRef]
- Reynolds, P.L. Seagrass and Seagrass Beds. Available online: http://ocean.si.edu/seagrass-and-seagrass-beds (accessed on 1 November 2018).
- Guannel, G.; Arkema, K.; Ruggiero, P.; Verutes, G. The Power of Three: Coral Reefs, Seagrasses and Mangroves Protect Coastal Regions and Increase Their Resilience. PLoS ONE 2016, 11, e0158094. [Google Scholar] [CrossRef]
- Christianen, M.J.A.; van Belzen, J.; Herman, P.M.J.; van Katwijk, M.M.; Lamers, L.P.M.; van Leent, P.J.M.; Bouma, T.J. Low-Canopy Seagrass Beds Still Provide Important Coastal Protection Services. PLoS ONE 2013, 8, e62413. [Google Scholar] [CrossRef]
- Kannan, R.R.R.; Arumugam, R.; Anantharaman, P. Antibacterial Potential of Three Seagrasses against Human Pathogens. Asian Pac. J. Trop. Med. 2010, 3, 890–893. [Google Scholar] [CrossRef]
- Castelblanco-Martinez, D.N.; Barba, E.; Schmitter-Soto, J.J.; Hernandez-Arana, H.A.; Morales-Vela, B. The Trophic Role of the Endangered Caribbean Manatee Trichechus manatus in an Estuary with Low Abundance of Seagrass. Estuaries Coasts 2012, 35, 60–77. [Google Scholar] [CrossRef]
- Best, R.C. Foods and Feeding Habits of Wild and Captive Sirenia. Mamm. Rev. 1981, 11, 3–29. [Google Scholar] [CrossRef]
- Seagrasses. Available online: http://myfwc.com/research/habitat/seagrasses/%0A (accessed on 15 October 2018).
- Gullstrom, M.; de la Torre-Castro, M.; Bandeira, S.O.; Bjork, M.; Dahlberg, M.; Kautsky, N.; Ronnback, P.; Ohman, M.C. Seagrass Ecosystems in the Western Indian Ocean. Ambio 2002, 31, 588–596. [Google Scholar] [CrossRef]
- Bertelli, C.M.; Unsworth, R.K.F. Protecting the Hand That Feeds Us: Seagrass (Zostera marina) Serves as Commercial Juvenile Fish Habitat. Mar. Pollut. Bull. 2014, 83, 425–429. [Google Scholar] [CrossRef]
- Lilley, R.J.; Unsworth, R.K.F. Atlantic Cod (Gadus morhua) Benefits from the Availability of Seagrass (Zostera marina) Nursery Habitat. Glob. Ecol. Conserv. 2014, 2, 367–377. [Google Scholar] [CrossRef]
- Waycott, M.; Duarte, C.M.; Carruthers, T.J.B.; Orth, R.J.; Dennison, W.C.; Olyarnik, S.; Calladine, A.; Fourqurean, J.W.; Heck, K.L.; Hughes, A.R.; et al. Accelerating Loss of Seagrasses across the Globe Threatens Coastal Ecosystems. Proc. Natl. Acad. Sci. USA 2009, 106, 12377–12381. [Google Scholar] [CrossRef]
- Terrados, J.; Duarte, C.M.; Fortes, M.D.; Borum, J.; Agawin, N.S.R.; Bach, S.; Thampanya, U.; Kamp-Nielsen, L.; Kenworthy, W.J.; Geertz-Hansen, O.; et al. Changes in Community Structure and Biomass of Seagrass Communities along Gradients of Siltation in SE Asia. Estuar. Coast. Shelf Sci. 1998, 46, 757–768. [Google Scholar] [CrossRef]
- De Falco, G.; Baroli, M.; Murru, E.; Piergallini, G.; Cancemi, G. Sediment Analysis Evidences Two Different Depositional Phenomena Influencing Seagrass Distribution in the Gulf of Oristano (Sardinia, Western Mediterranean). J. Coast. Res. 2006, 22, 1043–1050. [Google Scholar] [CrossRef]
- Cavazza, W.; Immordino, F.; Moretti, L.; Peirano, A.; Pironi, A.; Ruggiero, F. Sedimentological Parameters and Seagrasses Distributions as Indicators of Anthropogenic Coastal Degradation at Monterosso Bay (Ligurian Sea, NW Italy). J. Coast. Res. 2000, 16, 295–305. [Google Scholar]
- Dahl, M.; Deyanova, D.; Lyimo, L.D.; Naslund, J.; Samuelsson, G.S.; Mtolera, M.S.P.; Bjork, M.; Gullstrom, M. Effects of Shading and Simulated Grazing on Carbon Sequestration in a Tropical Seagrass Meadow. J. Ecol. 2016, 104, 654–664. [Google Scholar] [CrossRef]
- Jones, B.L.; Unsworth, R.K.F. The Perilous State of Seagrass in the British Isles. R. Soc. Open Sci. 2016, 3, 150596. [Google Scholar] [CrossRef]
- Bergmann, N.; Winters, G.; Rauch, G.; Eizaguirre, C.; Gu, J.; Nelle, P.; Fricke, B.; Reusch, T.B.H. Population-Specificity of Heat Stress Gene Induction in Northern and Southern Eelgrass Zostera marina Populations under Simulated Global Warming. Mol. Ecol. 2010, 19, 2870–2883. [Google Scholar] [CrossRef]
- Burkholder, J.M.; Tomasko, D.A.; Touchette, B.W. Seagrasses and Eutrophication. J. Exp. Mar. Biol. Ecol. 2007, 350, 46–72. [Google Scholar] [CrossRef]
- Walker, D.I.; McComb, A.J. Seagrass Degradation in Australian Coastal Waters. Mar. Pollut. Bull. 1992, 25, 191–195. [Google Scholar] [CrossRef]
- Unsworth, R.K.F.; Jones, B.L.; Cullen-Unsworth, L.C. Seagrass Meadows Are Threatened by Expected Loss of Peatlands in Indonesia. Glob. Chang. Biol. 2016, 22, 2957–2958. [Google Scholar] [CrossRef]
- Connell, S.D.; Fernandes, M.; Burnell, O.W.; Doubleday, Z.A.; Griffin, K.J.; Irving, A.D.; Leung, J.Y.S.; Owen, S.; Russell, B.D.; Falkenberg, L.J. Testing for Thresholds of Ecosystem Collapse in Seagrass Meadows. Conserv. Biol. 2017, 31, 1196–1201. [Google Scholar] [CrossRef]
- Duarte, C.M. Submerged Aquatic Vegetation in Relation to Different Nutrient Regimes. Ophelia 1995, 41, 87–112. [Google Scholar] [CrossRef]
- Mabrouk, L.; Hamza, A.; Ben Brahim, M.; Bradai, M.N. Variability in the Structure of Epiphyte Assemblages on Leaves and Rhizomes of Posidonia Oceanica in Relation to Human Disturbances in a Seagrass Meadow off Tunisia. Aquat. Bot. 2013, 108, 33–40. [Google Scholar] [CrossRef]
- Short, F.; Green, E.P. World Atlas of Seagrasses; University of California Press: Berkeley, CA, USA, 2003. [Google Scholar]
- Larkum, A.W.D.D.; Orth, R.J.; Duarte, C.M. Seagrasses: Biology, Ecology, and Conservation; Springer: Cham, The Netherlands, 2006. [Google Scholar]
- Borum, J.; Pedersen, O.; Greve, T.M.; Frankovich, T.A.; Zieman, J.C.; Fourqurean, J.W.; Madden, C.J. The Potential Role of Plant Oxygen and Sulphide Dynamics in Die-off Events of the Tropical Seagrass, Thalassia testudinum. J. Ecol. 2005, 93, 148–158. [Google Scholar] [CrossRef]
- Carlson, P.R.; Yarbro, L.A.; Barber, T.R. Relationship of Sediment Sulfide to Mortality of Thalassia-testudinum in Florida Bay. Bull. Mar. Sci. 1994, 54, 733–746. [Google Scholar]
- Brodersen, K.E.; Nielsen, D.A.; Ralph, P.J.; Kuhl, M. Oxic Microshield and Local PH Enhancement Protects Zostera muelleri from Sediment Derived Hydrogen Sulphide. New Phytol. 2015, 205, 1264–1276. [Google Scholar] [CrossRef]
- Holmer, M.; Pedersen, O.; Krause-Jensen, D.; Olesen, B.; Hedegård Petersen, M.; Schopmeyer, S.; Koch, M.; Lomstein, B.A.; Jensen, H.S. Sulfide Intrusion in the Tropical Seagrasses Thalassia testudinum and Syringodium filiforme. Estuar. Coast. Shelf Sci. 2009, 85, 319–326. [Google Scholar] [CrossRef]
- Hasler-Sheetal, H.; Holmer, M. Sulfide Intrusion and Detoxification in the Seagrass Zostera marina. PLoS ONE 2015, 10, e0129136. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The Importance of the Microbiome of the Plant Holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Moriarty, D.J.W.; Iverson, R.L.; Pollard, P.C. Exudation of Organic Carbon by the Seagrass Halodule wrightii Aschers. And Its Effect on Bacterial Growth in the Sediment. J. Exp. Mar. Biol. Ecol. 1986, 96, 115–126. [Google Scholar] [CrossRef]
- Penhale, P.A.; Smith, W.O. Excretion of Dissolved Organic Carbon by Eelgrass (Zostera marina) and Its Epiphytes. Limnol. Oceanogr. 1977, 22, 400–407. [Google Scholar] [CrossRef]
- Smith, G.W.; Hayasaka, S.S.; Thayer, G.W. Ammonification of Amino Acids by the Rhizoplane Microflora of Zostera marina L. and Halodule wrightii Aschers. Bot. Mar. 1984, 27, 23–28. [Google Scholar] [CrossRef]
- Wetzel, R.G.; Penhale, P.A. Transport of Carbon and Excretion of Dissolved Organic Carbon by Leaves and Roots/Rhizomes in Seagrasses and Their Epiphytes. Aquat. Bot. 1979, 6, 149–158. [Google Scholar] [CrossRef]
- Patriquin, D.; Knowles, R. Nitrogen Fixation in the Rhizosphere of Marine Angiosperms. Mar. Biol. 1972, 16, 49–58. [Google Scholar] [CrossRef]
- Sun, F.; Zhang, X.; Zhang, Q.; Liu, F.; Zhang, J.; Gong, J. Seagrass (Zostera marina) Colonization Promotes the Accumulation of Diazotrophic Bacteria and Alters the Relative Abundances of Specific Bacterial Lineages Involved in Benthic Carbon and Sulfur Cycling. Appl. Environ. Microbiol. 2015, 81, 6901–6914. [Google Scholar] [CrossRef]
- O’Donohue, M.J.; Moriarty, D.J.; Rae, I.C. Nitrogen Fixation in Sediments and the Rhizosphere of the Seagrass Zostera capricorni. Microb. Ecol. 1991, 22, 53–64. [Google Scholar] [CrossRef]
- Welsh, D. Nitrogen Fixation in Seagrass Meadows: Regulation, Plant ± Bacteria Interactions and Significance to Primary Productivity. Ecol. Lett. 2000, 3, 58–71. [Google Scholar] [CrossRef]
- Cole, L.W.; McGlathery, K.J. Nitrogen Fixation in Restored Eelgrass Meadows. Mar. Ecol. Prog. Ser. 2012, 448, 235–246. [Google Scholar] [CrossRef]
- Caffrey, J.; Kemp, W. Nitrogen Cycling in Sediments with Estuarine Populations of Potamogeton perfoliatus and Zostera marina. Mar. Ecol. Prog. Ser. 1990, 66, 147–160. [Google Scholar] [CrossRef]
- Smith, R.D.; Dennison, W.C.; Alberte, R.S. Role of Seagrass Photosynthesis in Root Aerobic Processes. Plant Physiol. 1984, 74, 1055–1058. [Google Scholar] [CrossRef]
- Agawin, N.S.R.; Ferriol, P.; Cryer, C.; Alcon, E.; Busquets, A.; Sintes, E.; Vidal, C.; Moyà, G. Significant Nitrogen Fixation Activity Associated with the Phyllosphere of Mediterranean Seagrass Posidonia oceanica: First Report. Mar. Ecol. Prog. Ser. 2016, 551, 53–62. [Google Scholar] [CrossRef]
- Smith, A.C.; Kostka, J.E.; Devereux, R.; Yates, D.F. Seasonal Composition and Activity of Sulfate-Reducing Prokaryotic Communities in Seagrass Bed Sediments. Aquat. Microb. Ecol. 2004, 37, 183–195. [Google Scholar] [CrossRef]
- Isaksen, M.F.; Finster, K. Sulphate Reduction in the Root Zone of the Seagrass Zostera noltii on the Intertidal Flats of a Coastal Lagoon (Arcachon, France). Mar. Ecol. Prog. Ser. 1996, 137, 187–194. [Google Scholar] [CrossRef]
- Lee, K.S.; Dunton, K.H. Diurnal Changes in Pore Water Sulfide Concentrations in the Seagrass Thalassia testudinum Beds: The Effects of Seagrasses on Sulfide Dynamics. J. Exp. Mar. Biol. Ecol. 2000, 255, 201–214. [Google Scholar] [CrossRef]
- Blaabjerg, V.; Finster, K. Sulphate Reduction Associated with Roots and Rhizomes of the Marine Macrophyte Zostera marina. Aquat. Microb. Ecol. 1998, 15, 311–314. [Google Scholar] [CrossRef]
- Martin, B.C.; Bougoure, J.; Ryan, M.H.; Bennett, W.W.; Colmer, T.D.; Joyce, N.K.; Olsen, Y.S.; Kendrick, G.A. Oxygen Loss from Seagrass Roots Coincides with Colonisation of Sulphide-Oxidising Cable Bacteria and Reduces Sulphide Stress. ISME J. 2018. [Google Scholar] [CrossRef]
- Dineen, J. Thalassia testudinum. Available online: http://www.sms.si.edu/irlspec/thalas_testud.htm (accessed on 15 October 2018).
- Dawes, C. The Dynamic Seagrasses of the Gulf of Mexico and Florida Coasts. In Proceedings of the Symposium on Subtropicaltropical Seagrasses of the Southeastern United States; Durako, M.J., Phillips, R.C., Lewis, R.I., Eds.; Florida Department of Natural Resources Bureau of Marine Research: St. Petersburg, Russia, 1987; pp. 25–38. [Google Scholar]
- Moore, D.R. Distribution of the Sea Grass, Thalassia, in the United States. Bull. Mar. Sci. 1963, 13, 329–342. [Google Scholar]
- Short, F.T.; Carruthers, T.J.R.; van Tussenbroek, B.; Zieman, J. Syringodium filiforme. Available online: http://dx.doi.org/10.2305/IUCN.UK.2010-3.RLTS.T173378A7003203.en (accessed on 15 October 2018).
- Weidner, S.; Arnold, W.; Pühler, A. Diversity of Uncultured Microorganisms Associated with the Seagrass Halophila stipulacea Estimated by Restriction Fragment Length Polymorphism Analysis of PCR-Amplified 16s RRNA Genes. Appl. Environ. Microbiol. 1996, 62, 766–771. [Google Scholar]
- White, L.J.; Brözel, V.S.; Subramanian, S. Isolation of Rhizosphere Bacterial Communities from Soil. Bio-Protocol 2015, 5. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S RRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4516–4522. [Google Scholar] [CrossRef]
- Available online: QIIME 2 https://qiime2.org/ (accessed on 15 October 2018).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-Species Living Tree Project (LTP)” Taxonomic Frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Charif, D.; Lobry, J.R. Seqin{R} 1.0-2: A Contributed Package to the {R} Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis. In Structural Approaches to Sequence Evolution: Molecules, Networks, Populations; Bastolla, U., Port, M., Roman, H.E., Vendruscolo, M., Eds.; Springer: New York, NY, USA, 2007; pp. 207–232. [Google Scholar]
- Walesiak, M.; Dudek, A. ClusterSim: Searching for Optimal Clustering Procedure for a Data Set. 2017. Available online: https://rdrr.io/cran/clusterSim/ (accessed on 15 August 2018).
- Wickham, H. Reshaping Data with the {reshape} Package. J. Stat. Softw. 2007, 21, 1–20. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. J Stat Softw 2010, 35, 65–88. [Google Scholar]
- Chen, H. VennDiagram: Generate High-Resolution Venn and Euler Plots. 2018. Available online: https://rdrr.io/cran/VennDiagram/ (accessed on 15 October 2018).
- Oksanen, J.; Blanchet, F.; Guillaume Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2018. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 15 October 2018).
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A Web-Based Tool for Comprehensive Statistical, Visual and Meta-Analysis of Microbiome Data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- ArcGIS and ArcMap, version 10.5.1, Esri: Redlands, CA, USA, 2017.
- Crump, B.C.; Wojahn, J.M.; Tomas, F.; Mueller, R.S. Metatranscriptomics and Amplicon Sequencing Reveal Mutualisms in Seagrass Microbiomes. Front. Microbiol. 2018, 9, 388. [Google Scholar] [CrossRef]
- Cúcio, C.; Engelen, A.H.; Costa, R.; Muyzer, G. Rhizosphere Microbiomes of European + Seagrasses Are Selected by the Plant, but Are Not Species Specific. Front. Microbiol. 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Kirchman, D.; Mazzella, L.; Alberte, R.; Mitchell, R. Epiphytic Bacterial Production on Zostera marina. Mar. Ecol. Prog. Ser. 1984, 15, 117–123. [Google Scholar] [CrossRef]
- Törnblom, E.; Søndergaard, M. Seasonal Dynamics of Bacterial Biomass and Production on Eelgrass Zostera marina Leaves. Mar. Ecol. Prog. Ser. 1999, 179, 231–240. [Google Scholar] [CrossRef]
- Williams, C.J.; Jaffé, R.; Anderson, W.T.; Jochem, F.J. Importance of Seagrass as a Carbon Source for Heterotrophic Bacteria in a Subtropical Estuary (Florida Bay). Estuar. Coast. Shelf Sci. 2009, 85, 507–514. [Google Scholar] [CrossRef]
- Nielsen, L.B.; Finster, K.; Welsh, D.T.; Donelly, A.; Herbert, R.A.; De Wit, R.; Lomstein, B.A.A. Sulphate Reduction and Nitrogen Fixation Rates Associated with Roots, Rhizomes and Sediments from Zostera noltii and Spartina maritima Meadows. Environ.Microbiol. 2001, 3, 63–71. [Google Scholar] [CrossRef]
- Kim, B.; Choi, A.; Lee, K.S.; Kang, C.K.; Hyun, J.H. Sulfate Reduction and Sulfur Cycles at Two Seagrass Beds Inhabited by Cold Affinity Zostera marina and Warm Affinity Halophila nipponica in Temperate Coastal Waters. Estuar. Coasts 2017, 40, 1346–1357. [Google Scholar] [CrossRef]
- Holmer, M.; Andersen, F.O.; Nielsen, S.L.; Boschker, H.T.S. The Importance of Mineralization Based on Sulfate Reduction for Nutrient Regeneration in Tropical Seagrass Sediments. Aquat. Bot. 2001, 71, 1–17. [Google Scholar] [CrossRef]
- Holmer, M.; Duarte, C.M.; Boschker, H.T.S.; Barrón, C. Carbon Cycling and Bacterial Carbon Sources in Pristine and Impacted Mediterranean Seagrass Sediments. Aquat. Microb. Ecol. 2004, 36, 227–237. [Google Scholar] [CrossRef]
- Jones, W.B.; Cifuentes, L.A.; Kaldy, J.E. Stable Carbon Isotope Evidence for Coupling between Sedimentary Bacteria and Seagrasses in a Sub-Tropical Lagoon. Mar. Ecol. Prog. Ser. 2003, 255, 15–25. [Google Scholar] [CrossRef]
- Mejia, A.Y.; Rotini, A.; Lacasella, F.; Bookman, R.; Thaller, M.C.; Shem-Tov, R.; Winters, G.; Migliore, L. Assessing the Ecological Status of Seagrasses Using Morphology, Biochemical Descriptors and Microbial Community Analyses. A Study in Halophila stipulacea (Forsk.) Aschers Meadows in the Northern Red Sea. Ecol. Indic. 2016, 60, 1150–1153. [Google Scholar] [CrossRef]
- Vann, L.E.; Holland-Moritz, H.; Ettinger, C.; Lang, J.; Eisen, J.A. Natural Variation in Plants: A Survey of Host and Microbe Variation. Proquest Diss. Theses 2017, 71–104. [Google Scholar] [CrossRef]
- Bengtsson, M.M.; Bühler, A.; Brauer, A.; Dahlke, S.; Schubert, H.; Blindow, I. Eelgrass Leaf Surface Microbiomes Are Locally Variable and Highly Correlated with Epibiotic Eukaryotes. Front. Microbiol. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Ettinger, C.L.; Voerman, S.E.; Lang, J.M.; Stachowicz, J.J.; Eisen, J.A. Microbial Communities in Sediment from Zostera marina Patches, but Not the Z. marina Leaf or Root Microbiomes, Vary in Relation to Distance from Patch Edge. PeerJ 2017, 5, e3246. [Google Scholar] [CrossRef]
- Fahimipour, A.K.; Kardish, M.R.; Lang, J.M.; Green, J.L.; Eisen, J.A.; Stachowicz, J.J. Global-Scale Structure of the Eelgrass Microbiome. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Jiang, Z.-F.; Xia, F.; Johnson, K.W.; Bartom, E.; Tuteja, J.H.; Stevens, R.; Grossman, R.L.; Brumin, M.; White, K.P.; Ghanim, M. Genome Sequences of the Primary Endosymbiont “Candidatus Portiera Aleyrodidarum” in the Whitefly Bemisia Tabaci B and Q Biotypes. J. Bacteriol. 2012, 194, 6678–6679. [Google Scholar] [CrossRef]
- Campbell, A.M.; Fleisher, J.; Sinigalliano, C.; White, J.R.; Lopez, J.V. Dynamics of Marine Bacterial Community Diversity of the Coastal Waters of the Reefs, Inlets, and Wastewater Outfalls of Southeast Florida. Microbiologyopen 2015, 4, 390–408. [Google Scholar] [CrossRef]
- Yu, S.-X.; Pang, Y.-L.; Wang, Y.-C.; Li, J.-L.; Qin, S. Distribution of Bacterial Communities along the Spatial and Environmental Gradients from Bohai Sea to Northern Yellow Sea. PeerJ 2018, 6, e4272. [Google Scholar] [CrossRef]
- Lee, J.; Lim, J.-H.; Park, J.; Youn, S.-H.; Oh, H.-J.; Kim, J.-H.; Kim, M.K.; Cho, H.; Yoon, J.-E.; Kim, S.; et al. First Investigation of Microbial Community Composition in the Bridge (Gadeok Channel) between the Jinhae-Masan Bay and the South Sea of Korea. Ocean Sci. J. 2018, 53, 251–260. [Google Scholar] [CrossRef]
- Sassoubre, L.M.; Yamahara, K.M.; Boehm, A.B. Temporal Stability of the Microbial Community in Sewage-Polluted Seawater Exposed to Natural Sunlight Cycles and Marine Microbiota. Appl. Environ. Microbiol. 2015, 81, 2107–2116. [Google Scholar] [CrossRef]
- Rina, R.Y.; Yehuda, B.; Lea, R.; Uri, G. The Gill Microbiota of Invasive and Indigenous Spondylus Oysters from the Mediterranean Sea and Northern Red Sea. Environ. Microbiol. Rep. 2015, 7, 860–867. [Google Scholar]
- Garrity, G.M.; Bell, J.A.; Lilburn, T. Oceanospirillales ord. nov. BT. In Bergey’s Manual® of Systematic Bacteriology. Volume Two: The Proteobacteria Part B The Gammaproteobacteria; Brenner, D.J., Krieg, N.R., Staley, J.T., Garrity, G.M., Boone, D.R., De Vos, P., Goodfellow, M., Rainey, F.A., Schleifer, K.-H., Eds.; Springer: Boston, MA, USA, 2005; pp. 270–323. [Google Scholar]
- Marba, N.; Duarte, C.; Alexandre, A.; Cabaço, S. How Do Seagrasses Grow and Spread? In European Seagrasses: An Introduction to Monitoring and Management; Borum, J., Duarte, C.M., Krause-Jensen, D., Greve, T.M., Eds.; EU Project Monitoring and Managing of European Seagrasses (M&MS), 2004; Available online: http://www.seagrasses.org/ (accessed on 15 October 2018).
- Raja, S.; Thangaradjou, T.; Sivakumar, K.; Kannan, L. Rhizobacterial Population Density and Nitrogen Fixation in Seagrass Community of Gulf of Mannar, India. J. Environ. Biol. 2012, 33, 1033–1037. [Google Scholar]
- Kaldy, J.; Eldridge, P.M.; Cifuentes, L.; Brian Jones, W. Utilization of DOC from Seagrass Rhizomes by Sediment Bacteria: 13C-Tracer Experiments and Modeling. MEPS 2006, 317, 41–55. [Google Scholar] [CrossRef]
- Wahbeh, M.I.; Mahasneh, A.M. Heterotrophic Bacteria Attached to Leaves, Rhizomes and Roots of Three Seagrass Species from Aqaba (Jordan). Aquat. Bot. 1984, 20, 87–96. [Google Scholar] [CrossRef]
- Lehnen, N.; Marchant, H.K.; Schwedt, A.; Milucka, J.; Lott, C.; Weber, M.; Dekaezemacker, J.; Seah, B.K.B.; Hach, P.F.; Mohr, W.; et al. High Rates of Microbial Dinitrogen Fixation and Sulfate Reduction Associated with the Mediterranean Seagrass Posidonia Oceanica. Syst. Appl. Microbiol. 2016, 39, 476–483. [Google Scholar] [CrossRef]
- Capone, D.G.; Budin, J.M. Nitrogen Fixation Associated with Rinsed Roots and Rhizomes of the Eelgrass Zostera marina. Plant Physiol. 1982, 70, 1601–1604. [Google Scholar] [CrossRef]
- Pereg, L.L.; Lipkin, Y.; Sar, N. Different Niches of the Halophila Stipulacea Seagrass Bed Harbor Distinct Populations of Nitrogen Fixing Bacteria. Mar. Biol. 1994, 119, 327–333. [Google Scholar] [CrossRef]
- Santos-Medellín, C.; Edwards, J.; Liechty, Z.; Nguyen, B.; Sundaresan, V. Drought Stress Results in a Compartment-Specific Restructuring of the Rice Root-Associated Microbiomes. MBio 2017, 8, e00764-17. [Google Scholar] [CrossRef]
- Smith, R.D.; Pregnall, A.M.; Alberte, R.S. Effects of Anaerobiosis on Root Metabolism of Zostera marina (Eelgrass): Implications for Survival in Reducing Sediments. Mar. Biol. 1988, 98, 131–141. [Google Scholar] [CrossRef]
- Kuever, J.; Rainy, F.A.; Widdel, F. Genus II Desulfocapsa. In Bergey’s Manual of Systematic Bacteriology: The Proteobacteria, PartC (The Alpha-, Beta-, Delta-, and Epsilonproteobacteria); Staley, J.T., Brenner, D.J., Krieg, N.R., Eds.; Springer Science & Business Media: New York, NY, USA, 2005; pp. 992–994. [Google Scholar]
- Dörries, M.; Wöhlbrand, L.; Kube, M.; Reinhardt, R.; Rabus, R. Genome and Catabolic Subproteomes of the Marine, Nutritionally Versatile, Sulfate-Reducing Bacterium Desulfococcus Multivorans DSM 2059. BMC Genom. 2016, 17, 918. [Google Scholar] [CrossRef]
- Rizzo, L.; Fraschetti, S.; Alifano, P.; Tredici, M.S.; Stabili, L. Association of Vibrio Community with the Atlantic Mediterranean Invasive Alga Caulerpa Cylindracea. J. Exp. Mar. Biol. Ecol. 2016, 475, 129–136. [Google Scholar] [CrossRef]
- Nilsson, E. Methods to Measure Biomass and Production of Bacteria and Photosynthetic Microbiota and Their Application on Illuminated Lake Sediments. A Literature Study (SKB-R--01-23); Swedish Nuclear Fuel and Waste Management Co.: Stockholm, Sweden, 2001. [Google Scholar]
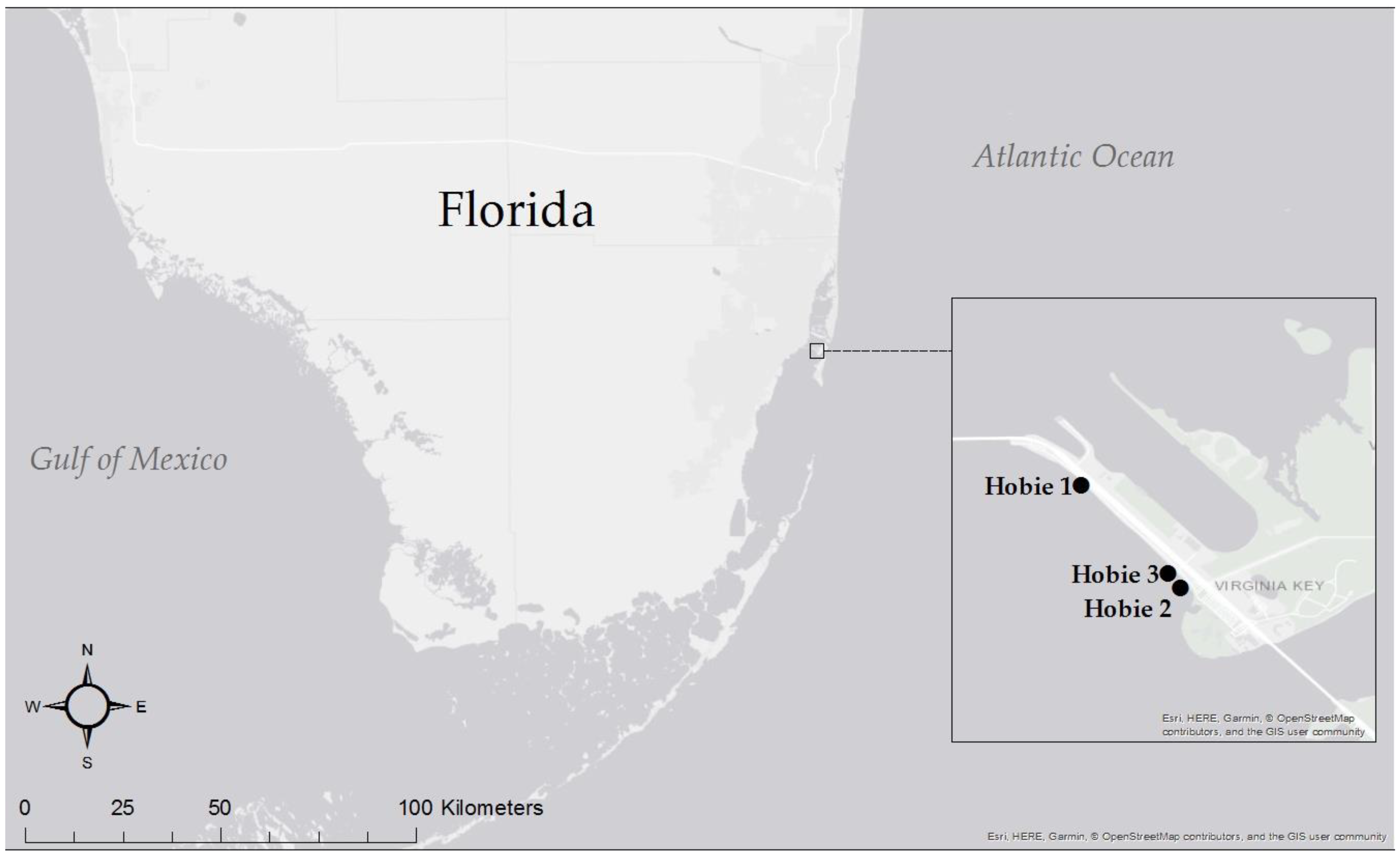



| Site | Latitude | Longitude | pH | Temperature (°C) | Salinity (ppt) | Conductivity (µS/cm) |
|---|---|---|---|---|---|---|
| Hobie 1 | 25.743298 | −80.173982 | 8.05 | 28.7 | 9.79 | 19.55 |
| Hobie 2 | 25.736188 | −80.167161 | 8.28 | 30.4 | 9.48 | 18.95 |
| Hobie 3 | 25.737209 | −80.167979 | 8.2 | 31 | 9.36 | 18.72 |
| Sample Type | Number of SV |
|---|---|
| Roots T | 3373 |
| Roots M | 2771 |
| Leaves T | 3347 |
| Leaves M | 4062 |
| Rhizome T | 3597 |
| Sediment C | 4650 |
| Sediment T | 4033 |
| Sediment M | 4033 |
| Seawater | 757 |
| All samples | 17686 |
| Taxa | Level | Higher Rank | This Study | Cúcio et al. 2016 [77] | Mejia et al. 2016 [86] | Ettinger et al. 2017 [89] | Fahimipour et al. 2017 [90] | Crump et al. 2018 [76] | Bengtsson et al. 2017 [88] |
|---|---|---|---|---|---|---|---|---|---|
| Acidimicrobiia | c | p Actinobacteria | + | + | + | + | + | + | |
| Acidobacteria | p | nma | + | + | |||||
| Alphaproteobacteria | c | p Proteobacteria | + | + | + | + | + | + | + |
| Alteromonadales OM60 | o/f | c Gammaproteobacteria | + | + | |||||
| Anaerolineae | c | p Chloroflexi | + | + | |||||
| Bacteroidia | c | p Bacteroidetes | + | + | + | + | |||
| Betaproteobacteria | c | p Proteobacteria | nma | + | + | + | + | ||
| Caldithrixae | c | p Bacteroidetes | + | + | |||||
| Campylobacterales | o | c Epsilonproteobacteria | nma | + | |||||
| Chloroflexi | p | + | + | + | + | ||||
| Clostridia | c | p Firmicutes | nma | + | + | + | + | + | |
| Cyanobacteria | p | nma (chloroplasts rfd) | + | + | |||||
| Cytophagia | c | p Bacteroidetes | + | + | + | ||||
| Deinococcus-Thermus | p | + | |||||||
| Deltaproteobacteria | c | + | + | + | + | + | + | ||
| Desulfobacterales | o | c Deltaproteobacteria | + | + | |||||
| Desulfovibrionaceae | f | o Desulfovibrionales, c Deltaproteobacteria | nma | + | |||||
| Desulfuromonadaceae | f | o Desulfuromonadales, c Deltaproteobacteria | nma | + | |||||
| Epsilonproteobacteria | c | p Proteobacteria | nma | + | + | + | |||
| Flavobacteriia | c | p Bacteroidetes | + | + | + | + | |||
| Gammaproteobacteria | c | p Proteobacteria | + | + | + | + | + | + | + |
| Gemmatimonadetes | p | nma | + | + | |||||
| Phycisphaerae | c | p Planctomycetes | nma | + | |||||
| Plantomycetia | c | p Planctomycetes | + | + | + | + | |||
| Plastid | plant | Chloroplasts | + (rfd) | + | |||||
| Saprospirae | f | c Sphingobacteria, o Sphingobacteriales | + | + | |||||
| Sphingobacteriia | c | p Bacteroidetes | nma | + | + | ||||
| Spirochaetes | p | + | + | ||||||
| Verrucomicrobia | p | nma | + | + | |||||
| WS3 | p | nma | + | + | |||||
| Seagrass Species | T. testudinum and S. filiforme | Z. marina, Z. noltii and C. nodosa | H. stipulacea | Z. marina | Z. marina | Z. marina and Z. japonica | Z. marina | ||
| Location | Key Biscayne, FL | Culatra Island, Portugal | Israel | Bodega bay, California | World wide | Netarts Bay or USA | Hiddensee, Germany |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ugarelli, K.; Laas, P.; Stingl, U. The Microbial Communities of Leaves and Roots Associated with Turtle Grass (Thalassia testudinum) and Manatee Grass (Syringodium filliforme) are Distinct from Seawater and Sediment Communities, but Are Similar between Species and Sampling Sites. Microorganisms 2019, 7, 4. https://doi.org/10.3390/microorganisms7010004
Ugarelli K, Laas P, Stingl U. The Microbial Communities of Leaves and Roots Associated with Turtle Grass (Thalassia testudinum) and Manatee Grass (Syringodium filliforme) are Distinct from Seawater and Sediment Communities, but Are Similar between Species and Sampling Sites. Microorganisms. 2019; 7(1):4. https://doi.org/10.3390/microorganisms7010004
Chicago/Turabian StyleUgarelli, Kelly, Peeter Laas, and Ulrich Stingl. 2019. "The Microbial Communities of Leaves and Roots Associated with Turtle Grass (Thalassia testudinum) and Manatee Grass (Syringodium filliforme) are Distinct from Seawater and Sediment Communities, but Are Similar between Species and Sampling Sites" Microorganisms 7, no. 1: 4. https://doi.org/10.3390/microorganisms7010004
APA StyleUgarelli, K., Laas, P., & Stingl, U. (2019). The Microbial Communities of Leaves and Roots Associated with Turtle Grass (Thalassia testudinum) and Manatee Grass (Syringodium filliforme) are Distinct from Seawater and Sediment Communities, but Are Similar between Species and Sampling Sites. Microorganisms, 7(1), 4. https://doi.org/10.3390/microorganisms7010004





