Identification of A Novel Arsenic Resistance Transposon Nested in A Mercury Resistance Transposon of Bacillus sp. MB24
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. PCR Amplification of Intact Transposon Regions
2.3. 5’ Rapid Amplification of cDNA ends (5′ RACE)
2.4. Real-Time Quantitative Reverse Transcription-PCR (qRT-PCR)
2.5. Genome Mining of Tn5084-like and TnARS1-like Transposon
3. Results and Discussion
3.1. Identification of TnMARS1 in Bacillus sp. MB24
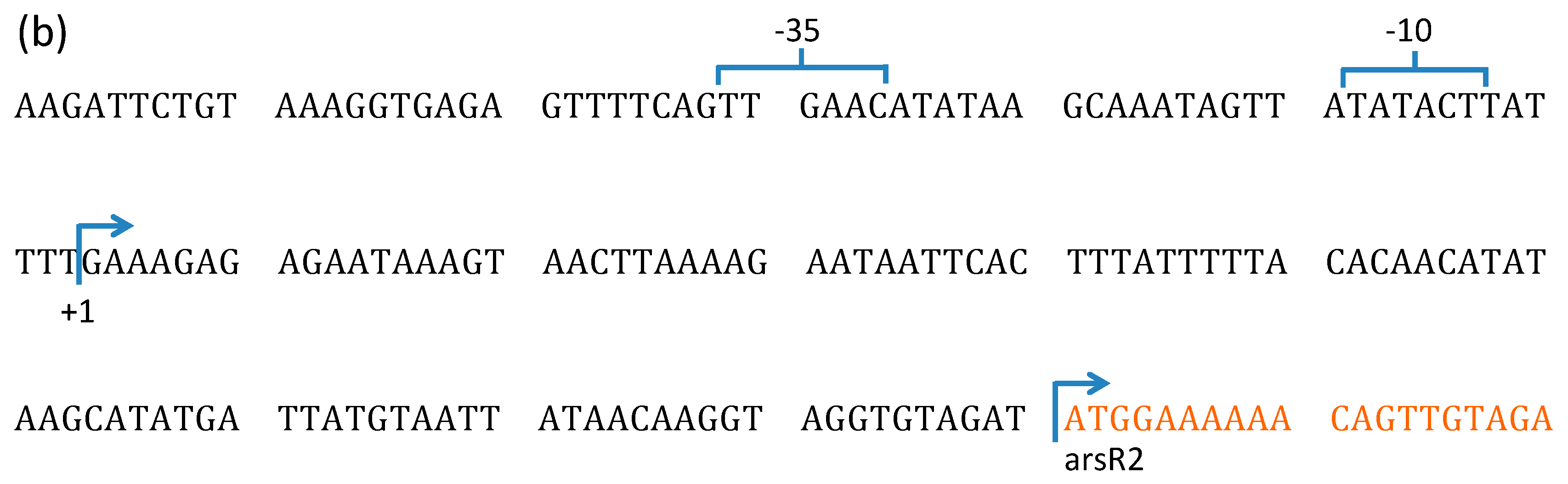
3.2. Characterization of TnARS1 in Bacillus sp. MB24.
3.2.1. Sequence Analysis of TnARS1
3.2.2. Transcription of ars Operons in Bacillus sp. MB24
3.2.3. Arsenic Resistance Conferred by ars Operon in Bacillus subtilis 168
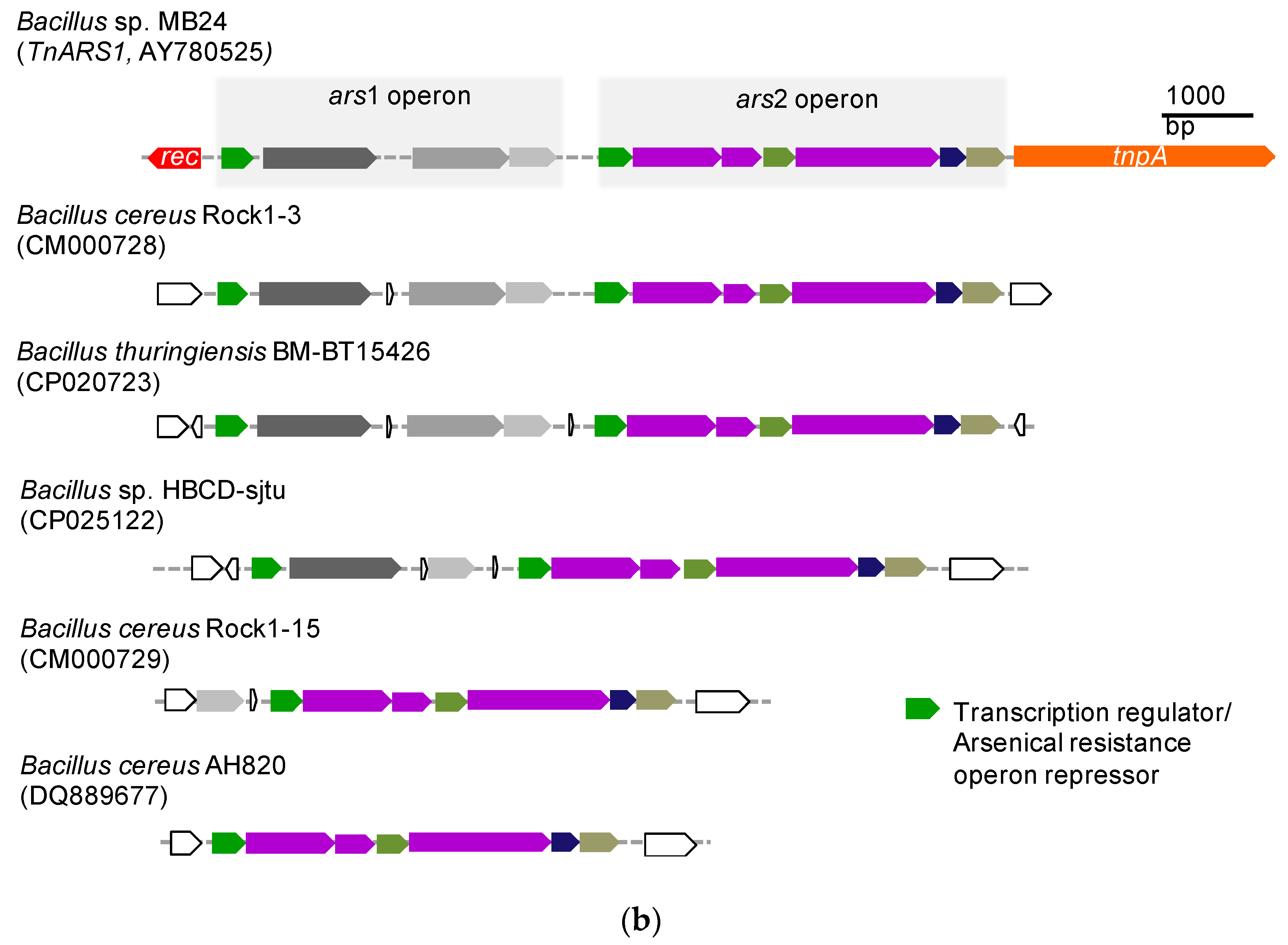
3.3. Genome Mining of Tn5084-like and TnARS1-like Regions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bogdanova, E.; Bass, I.; Minakhin, L.; Petrova, M.; Mindlin, S.; Volodin, A.; Kalyaeva, E.; Tiedje, J.; Hobman, J.; Brown, N. Horizontal spread of mer operons among Gram-positive bacteria in natural environments. Microbiology 1998, 144, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Osborn, A.M.; Bruce, K.D.; Strike, P.; Ritchie, D.A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 1997, 19, 239–262. [Google Scholar] [CrossRef] [PubMed]
- Mindlin, S.; Bass, I.; Bogdanova, E.; Gorlenko, Z.; Kaliaeva, E.; Petrova, M.; Nikiforov, V. Horizontal transfer of mercury resistance genes in natural bacterial populations. Mol. Biol. 2002, 36, 216–227. [Google Scholar] [CrossRef]
- Huang, C.-C.; Narita, M.; Yamagata, T.; Itoh, Y.; Endo, G. Structure analysis of a class II transposon encoding the mercury resistance of the Gram-positive bacterium Bacillus megaterium MB1, a strain isolated from Minamata Bay, Japan. Gene 1999, 234, 361–369. [Google Scholar] [CrossRef]
- Narita, M.; Chiba, K.; Nishizawa, H.; Ishii, H.; Huang, C.-C.; Kawabata, Z.i.; Silver, S.; Endo, G. Diversity of mercury resistance determinants among Bacillus strains isolated from sediment of Minamata Bay. FEMS Microbiol. Lett. 2003, 223, 73–82. [Google Scholar] [CrossRef]
- Huang, C.-C.; Narita, M.; Yamagata, T.; Endo, G. Identification of three merB genes and characterization of a broad-spectrum mercury resistance module encoded by a class II transposon of Bacillus megaterium strain MB1. Gene 1999, 239, 361–366. [Google Scholar] [CrossRef]
- Chien, M.-F.; Ho, Y.-N.; Lin, H.-T.; Lin, K.-H.; Endo, G.; Huang, C.-C. MerB3, an Organomercurial Lyase of Bacillus as an Antidote against Organomercurial Poisoning. J. Environ. Biotechnol. 2019, 19, 73–80. [Google Scholar]
- Rosen, B.P. Families of arsenic transporters. Trends Microbiol. 1999, 7, 207–212. [Google Scholar] [CrossRef]
- Silver, S.; Phung, L.T. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 2005, 71, 599–608. [Google Scholar] [CrossRef]
- Chen, C.-M.; Misra, T.; Silver, S.; Rosen, B. Nucleotide sequence of the structural genes for an anion pump. The plasmid-encoded arsenical resistance operon. J. Biol. Chem. 1986, 261, 15030–15038. [Google Scholar]
- Bruhn, D.F.; Li, J.; Silver, S.; Roberta, F.; Rosen, B.P. The arsenical resistance operon of IncN plasmid R46. FEMS Microbiol. Lett. 1996, 139, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.; Silver, S. Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc. Natl. Acad. Sci. USA 1992, 89, 9474–9478. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, R.; Rosen, B.P.; Phung, L.T.; Silver, S. Microbial arsenic: From geocycles to genes and enzymes. FEMS Microbiol. Rev. 2002, 26, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Gladysheva, T.B.; Oden, K.L.; Rosen, B.P. Properties of the arsenate reductase of plasmid R773. Biochemistry 1994, 33, 7288–7293. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, R.; Peschel, A.; Wieland, B.; Götz, F. Expression and regulation of the antimonite, arsenite, and arsenate resistance operon of Staphylococcus xylosus plasmid pSX267. J. Bacteriol. 1992, 174, 3676–3683. [Google Scholar] [CrossRef]
- Cai, J.; DuBow, M.S. Expression of the Escherichia coli chromosomal ars operon. Can. J. Microbiol. 1996, 42, 662–671. [Google Scholar] [CrossRef]
- Silver, S.; Ji, G.; Bröer, S.; Dey, S.; Dou, D.; Rosen, B.P. Orphan enzyme or patriarch of a new tribe: The arsenic resistance ATPase of bacterial plasmids. Mol. Microbiol. 1993, 8, 637–642. [Google Scholar] [CrossRef]
- Xu, C.; Rosen, B.P. Dimerization is essential for DNA binding and repression by the ArsR metalloregulatory protein of Escherichia coli. J. Biol. Chem. 1997, 272, 15734–15738. [Google Scholar] [CrossRef]
- Achour-Rokbani, A.; Cordi, A.; Poupin, P.; Bauda, P.; Billard, P. Characterization of the ars gene cluster from extremely arsenic-resistant Microbacterium sp. strain A33. Appl. Environ. Microbiol. 2010, 76, 948–955. [Google Scholar] [CrossRef]
- Branco, R.; Chung, A.-P.; Morais, P.V. Sequencing and expression of two arsenic resistance operons with different functions in the highly arsenic-resistant strain Ochrobactrum tritici SCII24 T. BMC Microbiol. 2008, 8, 95. [Google Scholar] [CrossRef]
- Li, X.; Krumholz, L.R. Regulation of arsenate resistance in Desulfovibrio desulfuricans G20 by an arsRBCC operon and an arsC gene. J. Bacteriol. 2007, 189, 3705–3711. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.N.; Saltikov, C.W. The ArsR repressor mediates arsenite-dependent regulation of arsenate respiration and detoxification operons of Shewanella sp. strain ANA-3. J. Bacteriol. 2009, 191, 6722–6731. [Google Scholar] [CrossRef] [PubMed]
- Ordóñez, E.; Letek, M.; Valbuena, N.; Gil, J.A.; Mateos, L.M. Analysis of genes involved in arsenic resistance in Corynebacterium glutamicum ATCC 13032. Appl. Environ. Microbiol. 2005, 71, 6206–6215. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; DeMel, S.; Shi, J.; Gladysheva, T.; Gatti, D.L.; Rosen, B.P.; Edwards, B.F. Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure 2001, 9, 1071–1081. [Google Scholar] [CrossRef]
- Gihring, T.M.; Bond, P.L.; Peters, S.C.; Banfield, J.F. Arsenic resistance in the archaeon” Ferroplasma acidarmanus”: New insights into the structure and evolution of the ars genes. Extremophiles 2003, 7, 123–130. [Google Scholar] [CrossRef]
- Dedonder, R.; Lepesant, J.; Lepesant-Kejzlarova, J.; Billault, A.; Steinmetz, M.; Kunst, F. Construction of a kit of reference strains for rapid genetic mapping in Bacillus subtilis 168. Appl. Environ. Microbiol. 1977, 33, 989. [Google Scholar]
- Matsui, K.; Yoshinami, S.; Narita, M.; Chien, M.-F.; Phung, L.T.; Silver, S.; Endo, G. Mercury resistance transposons in Bacilli strains from different geographical regions. FEMS Microbiol. Lett. 2016, 363, fnw013. [Google Scholar] [CrossRef]
- Medema, M.H.; Takano, E.; Breitling, R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 2013, 30, 1218–1223. [Google Scholar] [CrossRef]
- Huang, C.-C.; Chen, M.-W.; Hsieh, J.-L.; Lin, W.-H.; Chen, P.-C.; Chien, L.-F. Expression of mercuric reductase from Bacillus megaterium MB1 in eukaryotic microalga Chlorella sp. DT: An approach for mercury phytoremediation. Appl. Microbiol. Biotechnol. 2006, 72, 197–205. [Google Scholar] [CrossRef]
- Chien, M.-F.; Huang, C.-C.; Kusano, T.; Endo, G. Facilities for transcription and mobilization of an exon-less bacterial group II intron nested in transposon TnMERI1. Gene 2008, 408, 164–171. [Google Scholar] [CrossRef]
- McClintock, B. The Significance of Responses of the Genome to Challenge; World Scientific: Singapore, 1983. [Google Scholar]
- Shah, S.B.; Ali, F.; Huang, L.; Wang, W.; Xu, P.; Tang, H. Complete genome sequence of Bacillus sp. HBCD-sjtu, an efficient HBCD-degrading bacterium. 3 Biotech 2018, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, L.; Peters, B.M.; Li, B.; Chen, D.; Xu, Z.; Shirtliff, M.E. Complete genome sequence and bioinformatics analyses of Bacillus thuringiensis strain BM-BT15426. Microb. Pathog. 2017, 108, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Rasko, D.A.; Rosovitz, M.; Økstad, O.A.; Fouts, D.E.; Jiang, L.; Cer, R.Z.; Kolstø, A.-B.; Gill, S.R.; Ravel, J. Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1. J. Bacteriol. 2007, 189, 52–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]

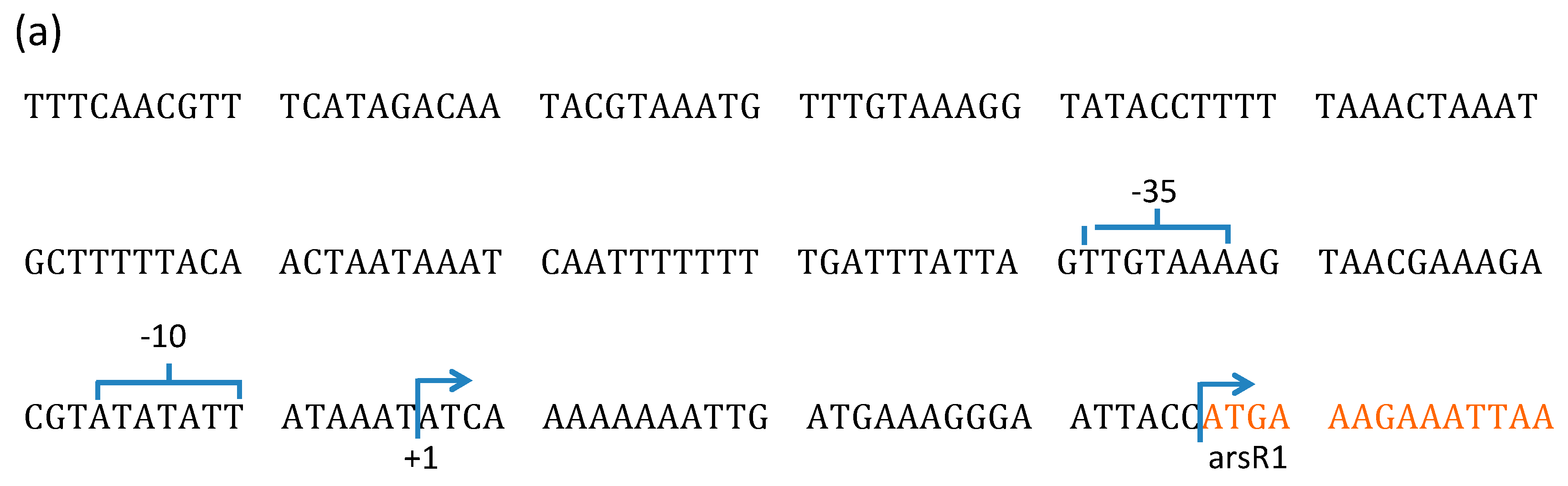


| Arsenic Resistance (mM) | ||||
|---|---|---|---|---|
| As(III) | As(V) | |||
| MTC | MIC | MTC | MIC | |
| Bacillus subtilis 168 | 4 | 6 | 38 | 40 |
| Bacillus subtilis 168ΔarsB | 2 | 4 | 2 | 4 |
| Bacillus subtilis 168ΔarsB/pHYARS1 | 2 | 4 | 6 | 8 |
| Bacillus subtilis 168ΔarsB/pHYARS2 | 4 | 6 | 20 | 30 |
| Bacillus subtilis 168ΔarsB/pHYARS3 | 14 | 16 | 10 | 20 |
| Bacillus sp. MB24 | 12 | 14 | 28 | 30 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chien, M.-F.; Ho, Y.-N.; Yang, H.-E.; Narita, M.; Miyauchi, K.; Endo, G.; Huang, C.-C. Identification of A Novel Arsenic Resistance Transposon Nested in A Mercury Resistance Transposon of Bacillus sp. MB24. Microorganisms 2019, 7, 566. https://doi.org/10.3390/microorganisms7110566
Chien M-F, Ho Y-N, Yang H-E, Narita M, Miyauchi K, Endo G, Huang C-C. Identification of A Novel Arsenic Resistance Transposon Nested in A Mercury Resistance Transposon of Bacillus sp. MB24. Microorganisms. 2019; 7(11):566. https://doi.org/10.3390/microorganisms7110566
Chicago/Turabian StyleChien, Mei-Fang, Ying-Ning Ho, Hui-Erh Yang, Masaru Narita, Keisuke Miyauchi, Ginro Endo, and Chieh-Chen Huang. 2019. "Identification of A Novel Arsenic Resistance Transposon Nested in A Mercury Resistance Transposon of Bacillus sp. MB24" Microorganisms 7, no. 11: 566. https://doi.org/10.3390/microorganisms7110566





