Chlamydiae from Down Under: The Curious Cases of Chlamydial Infections in Australia
Abstract
:1. Introduction
1.1. Chlamydia trachomatis: An Australian Public Health Challenge
1.1.1. Ct still a Major Cause of Trachoma in a High-Income Developed Country
1.1.2. Sexually Transmitted Ct Infections–STI Infections with No Intention of Slowing Down
1.1.3. The Curious Molecular Epidemiology of Ocular and Sexually Transmitted Ct Infections
1.2. Chlamydia pneumoniae—An Elusive Human Cosmopolitan Pathogen
Limited Molecular Epidemiology of Australian Cpn Strains Reveals Distinction between Animal and Human Strains
2. Chlamydial Infections at the Human–Animal Interface
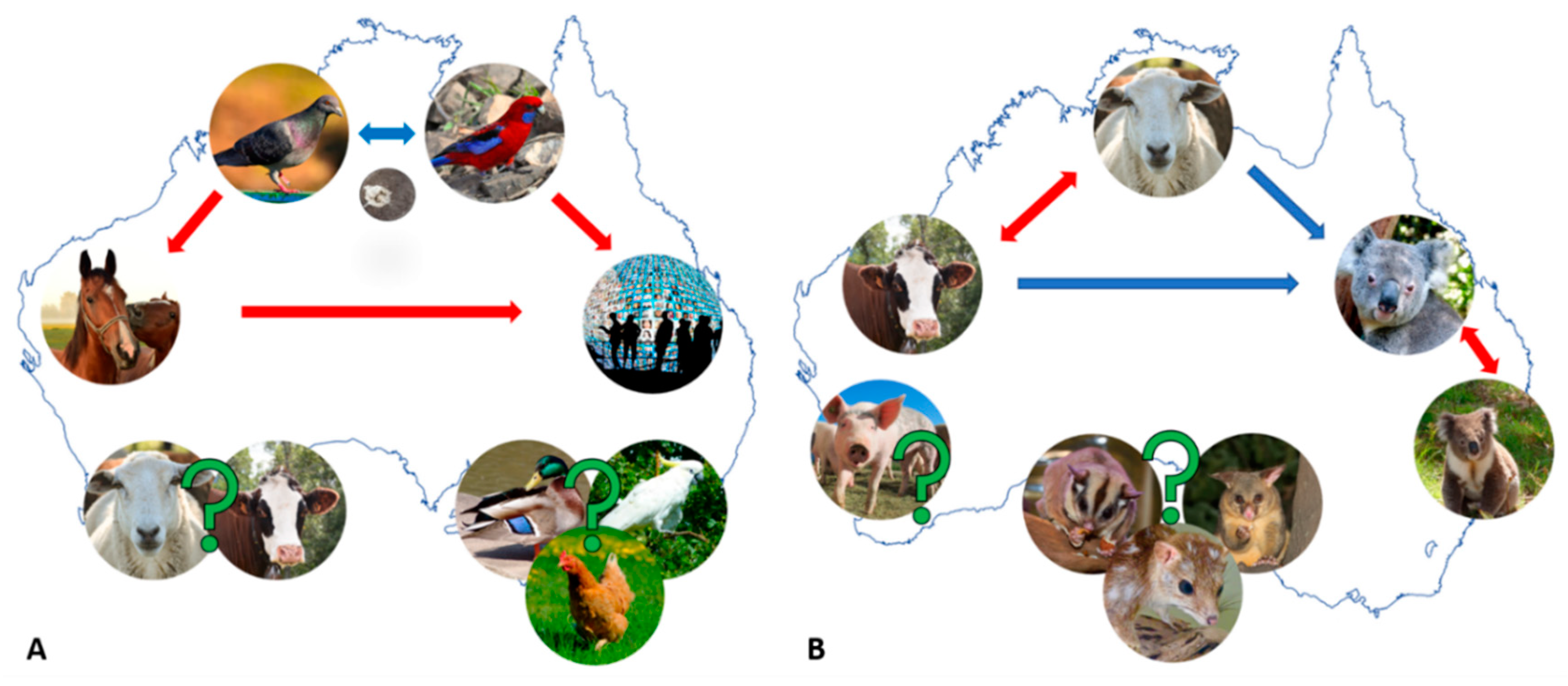
2.1. Chlamydia psittaci—A Parrot Pathogen with Zoonotic Potential
2.1.1. Equine and Avian Psittacosis: On the Heels of Cps Infection “Spill-Over”
2.1.2. Are Australian Birds Really the Culprit in the Cps Infection “Spill-Over”?
3. Chlamydial Infections at the Domesticated–Wildlife Animal Interface
3.1. Chlamydia Pecorum—The Infamous Koala or Economically Significant Livestock Pathogen?
3.1.1. Cpec Infections in Koalas—(Still) a Major Cause of Debilitating Disease
3.1.2. Cpec Infections in Livestock—A Contributor to Economic and Production Losses
3.1.3. The Complex Molecular Epidemiology of Koala and Livestock Infections
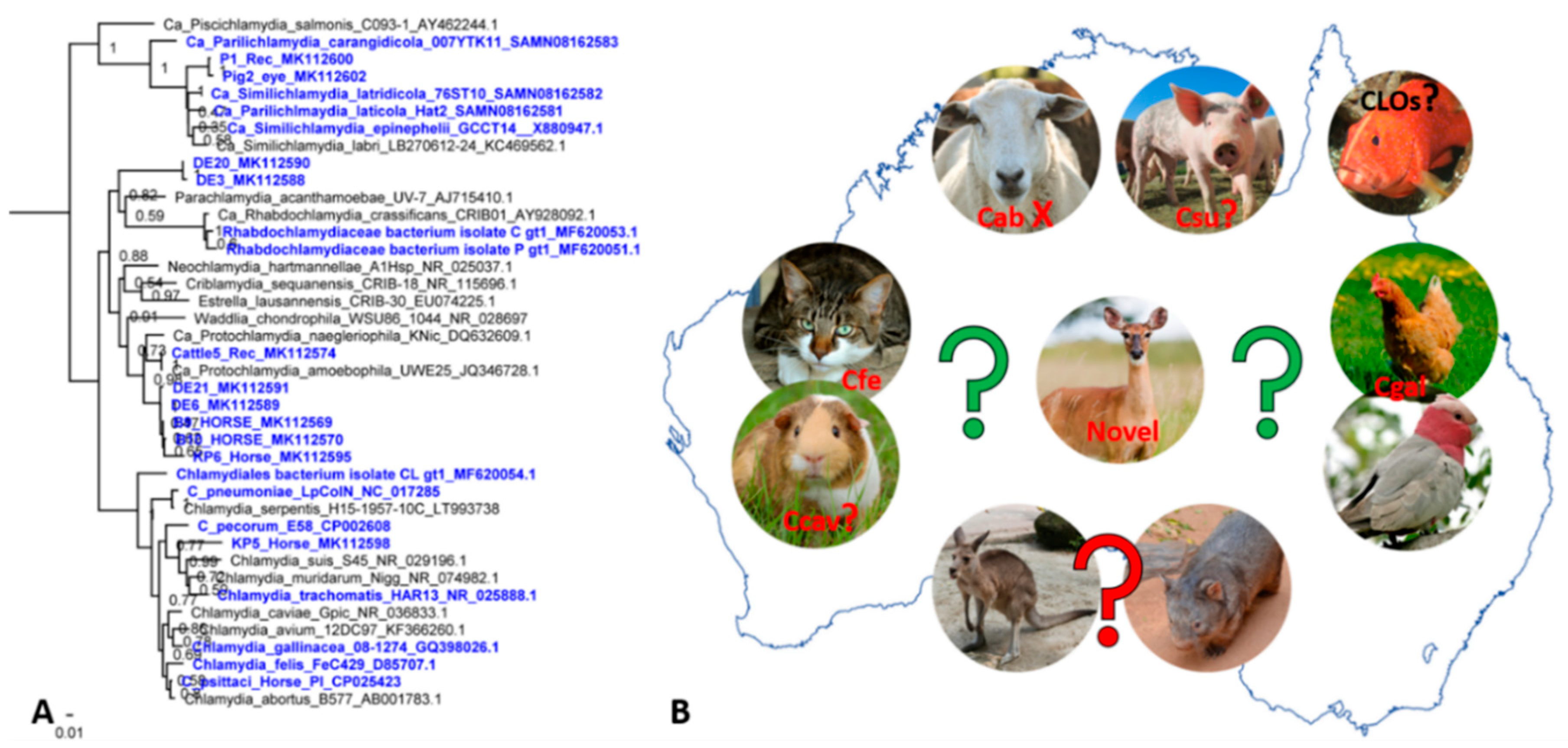
4. Other Chlamydial Infections in Australian Animals: We Can’t Find What We Don’t Seek
5. Present Challenges and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cheong, H.C.; Lee, C.Y.Q.; Cheok, Y.Y.; Tan, G.M.Y.; Looi, C.Y.; Wong, W.F. Chlamydiaceae: Diseases in primary hosts and zoonosis. Microorganisms 2019, 7, 146. [Google Scholar] [CrossRef]
- Bastidas, R.J.; Valdivia, R.H. Emancipating Chlamydia: Advances in the genetic manipulation of a recalcitrant intracellular pathogen. Microbiol. Mol. Biol. Rev. 2016, 80, 411–427. [Google Scholar] [CrossRef]
- Brothwell, J.A.; Muramatsu, M.K.; Zhong, G.; Nelson, D.E. Advances and obstacles in the genetic dissection of chlamydial virulence. Curr. Top. Microbiol. Immunol. 2018, 412, 133–158. [Google Scholar] [CrossRef]
- Thng, C.C.M. A review of sexually transmitted infections in Australia—Considerations in 2018. Acad. Forensic Pathol. 2018, 8, 938–946. [Google Scholar] [CrossRef]
- Chen, M.Y.; Donovan, B. Genital Chlamydia trachomatis infection in Australia: Epidemiology and clinical implications. Sex. Health 2004, 1, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.R.; Anjou, M.D. Trachoma in Australia: An update. Clin. Exp. Ophthalmol. 2013, 41, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.R.; Burton, M.J.; Haddad, D.; West, S.; Wright, H. Trachoma. Lancet 2014, 384, 2142–2152. [Google Scholar] [CrossRef]
- Debattista, J.; Susan, H.; Peter, T. Chlamydial infections and Indigenous health. Microbiol. Aust. 2009, 30, 197–199. [Google Scholar]
- World Health Organisation. Trachoma. Available online: https://www.who.int/news-room/fact-sheets/detail/trachoma (accessed on 30 October 2019).
- Taylor, H.R.; Fox, S.S.; Xie, J.; Dunn, R.A.; Arnold, A.L.; Keeffe, J.E. The prevalence of trachoma in Australia: The National Indigenous Eye Health Survey. Med. J. Aust. 2010, 192, 248–253. [Google Scholar] [CrossRef]
- Warren, J.M.; Birrell, A.L. Trachoma in remote Indigenous Australia: A review and public health perspective. Aust. N. Z. J. Public Health 2016, 40, S48–S52. [Google Scholar] [CrossRef] [PubMed]
- Lange, F.D.; Baunach, E.; McKenzie, R.; Taylor, H.R. Trachoma elimination in remote Indigenous Northern Territory communities: Baseline health-promotion study. Aust. J. Prim. Health 2014, 20, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Cowling, C.S.; Liu, B.C.; Snelling, T.L.; Ward, J.S.; Kaldor, J.M.; Wilson, D.P. National trachoma surveillance annual report, 2012. Commun. Dis. Intell. Q. Rep. 2015, 39, E146–E157. [Google Scholar] [PubMed]
- The Kirby Institute. Australian Trachoma Surveillance Report 2017; University of New South Wales: Sydney, Australia, 2018. [Google Scholar]
- Lange, F.D.; Jones, K.; Ritte, R.; Brown, H.E.; Taylor, H.R. The impact of health promotion on trachoma knowledge, attitudes and practice (KAP) of staff in three work settings in remote Indigenous communities in the Northern Territory. PLoS Negl. Trop. Dis. 2017, 11, e0005503. [Google Scholar] [CrossRef] [PubMed]
- The Kirby Institute. HIV, Viral Hepatitis and Sexually Transmissible Infections in Australia: Annual Surveillance Report 2018; The Kirby Institute, University of New South Wales: Sydney, Australia, 2018. [Google Scholar]
- Garton, L.; Dyda, A.; Guy, R.; Silver, B.; McGregor, S.; Hengel, B.; Rumbold, A.; Taylor-Thomson, D.; Knox, J.; Maher, L.; et al. High chlamydia and gonorrhoea repeat positivity in remote Aboriginal communities 2009-2011: Longitudinal analysis of testing for re-infection at 3 months suggests the need for more frequent screening. Sex. Health 2016, 13, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Guy, R.J.; Ward, J.; Causer, L.M.; Natoli, L.; Badman, S.G.; Tangey, A.; Hengel, B.; Wand, H.; Whiley, D.; Tabrizi, S.N.; et al. Molecular point-of-care testing for chlamydia and gonorrhoea in Indigenous Australians attending remote primary health services (TTANGO): A cluster-randomised, controlled, crossover trial. Lancet Infect. Dis. 2018, 18, 1117–1126. [Google Scholar] [CrossRef]
- Nattabi, B.; Matthews, V.; Bailie, J.; Rumbold, A.; Scrimgeour, D.; Schierhout, G.; Ward, J.; Guy, R.; Kaldor, J.; Thompson, S.C.; et al. Wide variation in sexually transmitted infection testing and counselling at Aboriginal primary health care centres in Australia: Analysis of longitudinal continuous quality improvement data. BMC Infect. Dis. 2017, 17, 148. [Google Scholar] [CrossRef]
- Rowley, J.; Vander Hoorn, S.; Korenromp, E.; Low, N.; Unemo, M.; Abu-Raddad, L.J.; Chico, R.M.; Smolak, A.; Newman, L.; Gottlieb, S.; et al. Chlamydia, gonorrhoea, trichomoniasis and syphilis: Global prevalence and incidence estimates, 2016. Bull. World Health Org. 2019, 97, 548–562. [Google Scholar] [CrossRef]
- Vodstrcil, L.A.; McIver, R.; Huston, W.M.; Tabrizi, S.N.; Timms, P.; Hocking, J.S. The epidemiology of Chlamydia trachomatis organism load during genital infection: A systematic review. J. Infect. Dis. 2014, 211, 1628–1645. [Google Scholar] [CrossRef]
- Phillips, S.; Vodstrcil, L.A.; Huston, W.M.; Lawerence, A.; Timms, P.; Chen, M.Y.; Worthington, K.; McIver, R.; Bradshaw, C.S.; Garland, S.M.; et al. Detection of Chlamydia trachomatis mRNA using digital PCR as a more accurate marker of viable organism. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2117–2122. [Google Scholar] [CrossRef]
- Danielewski, J.A.; Phillips, S.; Kong, F.Y.S.; Smith, K.S.; Hocking, J.S.; Guy, R.; Fairley, C.K.; Garland, S.M.; Tabrizi, S.N. A snapshot of Chlamydia trachomatis genetic diversity using multilocus sequence type analysis in an Australian metropolitan setting. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1297–1303. [Google Scholar] [CrossRef]
- Huffam, S.; Chow, E.P.F.; Leeyaphan, C.; Fairley, C.K.; Hocking, J.S.; Phillips, S.; Tabrizi, S.N.; Bellhouse, C.; Bradshaw, C.S.; Fehler, G.; et al. Chlamydia infection between men and women: A cross-sectional study of heterosexual partnerships. In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2017; Volume 4. [Google Scholar] [CrossRef]
- Goller, J.L.; De Livera, A.M.; Guy, R.J.; Low, N.; Donovan, B.; Law, M.; Kaldor, J.M.; Fairley, C.K.; Hocking, J.S. Rates of pelvic inflammatory disease and ectopic pregnancy in Australia, 2009-2014: Ecological analysis of hospital data. Sex. Transm. Infect. 2018, 94, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Bryan, E.R.; McLachlan, R.I.; Rombauts, L.; Katz, D.J.; Yazdani, A.; Bogoevski, K.; Chang, C.; Giles, M.L.; Carey, A.J.; Armitage, C.W.; et al. Detection of chlamydia infection within human testicular biopsies. Hum. Reprod. 2019. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Kong, F.; Fairley, C.K.; Donovan, B.; Chen, M.; Bradshaw, C.; Boyd, M.; Amin, J.; Timms, P.; Tabrizi, S.; et al. Treatment efficacy of azithromycin 1 g single dose versus doxycycline 100 mg twice daily for 7 days for the treatment of rectal chlamydia among men who have sex with men—A double-blind randomised controlled trial protocol. BMC Infect. Dis. 2017, 17, 35. [Google Scholar] [CrossRef] [PubMed]
- Stark, D.; van Hal, S.; Hillman, R.; Harkness, J.; Marriott, D. Lymphogranuloma venereum in Australia: Anorectal Chlamydia trachomatis serovar L2b in men who have sex with men. J. Clin. Microbiol. 2007, 45, 1029–1031. [Google Scholar] [CrossRef]
- Templeton, D.J.; Ressler, K.A.; Hope, K.; Poynten, I.M. Enhanced surveillance of a lymphogranuloma venereum outbreak in Sydney 2010-2012. Aust. N. Z. J. Public Health 2016, 40, 368–370. [Google Scholar] [CrossRef]
- Davies, S.C.; Shapiro, J.; Comninos, N.B.; Templeton, D.J. Lymphogranuloma venereum presenting as penile ulcer in two HIV-negative gay men. Int. J. STD AIDS 2019, 30, 515–518. [Google Scholar] [CrossRef]
- Lau, A.; Kong, F.Y.S.; Huston, W.; Chow, E.P.F.; Fairley, C.K.; Hocking, J.S. Factors associated with anorectal Chlamydia trachomatis or Neisseria gonorrhoeae test positivity in women: A systematic review and meta-analysis. Sex. Transm. Infect. 2019. [Google Scholar] [CrossRef]
- Callander, D.; McManus, H.; Guy, R.; Hellard, M.; O′Connor, C.C.; Fairley, C.K.; Chow, E.P.F.; McNulty, A.; Lewis, D.A.; Carmody, C.; et al. Rising Chlamydia and Gonorrhoea incidence and associated risk factors among female sex workers in Australia: A retrospective cohort study. Sex. Transm. Dis. 2018, 45, 199–206. [Google Scholar] [CrossRef]
- Phillips, T.R.; Fairley, C.K.; Maddaford, K.; Danielewski, J.; Hocking, J.S.; Lee, D.; Williamson, D.A.; Murray, G.; Kong, F.; De Petra, V.; et al. Bacterial load of Chlamydia trachomatis in the posterior oropharynx, tonsillar fossae and saliva among gay and bisexual men with untreated oropharyngeal chlamydia. J. Clin. Microbiol. 2019. [Google Scholar] [CrossRef]
- Kong, F.Y.S.; Hocking, J.S. Treatment challenges for urogenital and anorectal Chlamydia trachomatis. BMC Infect. Dis. 2015, 15, 293. [Google Scholar] [CrossRef]
- Vodstrcil, L.A.; Rupasinghe, T.W.T.; Kong, F.Y.S.; Tull, D.; Worthington, K.; Chen, M.Y.; Huston, W.M.; Timms, P.; McConville, M.J.; Fairley, C.K.; et al. Measurement of tissue azithromycin levels in self-collected vaginal swabs post treatment using liquid chromatography and tandem mass spectrometry (LC-MS/MS). PLoS ONE 2017, 12, e0177615. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.T.; Callander, D.; Hocking, J.S.; McGregor, S.; McManus, H.; Dyda, A.; Moreira, C.; Braat, S.; Hengel, B.; Ward, J.; et al. Population-level diagnosis and care cascade for chlamydia in Australia. Sex. Transm. Infect. 2019. [Google Scholar] [CrossRef] [PubMed]
- Guy, R.J.; Micallef, J.M.; Mooney-Somers, J.; Jamil, M.S.; Harvey, C.; Bateson, D.; van Gemert, C.; Wand, H.; Kaldor, J. Evaluation of Chlamydia partner notification practices and use of the “let them know” website by family planning clinicians in Australia: Cross-sectional study. J. Med. Internet Res. 2016, 18, e173. [Google Scholar] [CrossRef] [PubMed]
- Lorch, R.; Hocking, J.; Temple-Smith, M.; Law, M.; Yeung, A.; Wood, A.; Vaisey, A.; Donovan, B.; Fairley, C.K.; Kaldor, J.; et al. The chlamydia knowledge, awareness and testing practices of Australian general practitioners and practice nurses: Survey findings from the Australian chlamydia control effectiveness pilot (ACCEPt). BMC Fam. Pract 2013, 14, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, P.; Harris, S.R.; Smith, H.M.B.S.; Hadfield, J.; O′Neill, C.; Cutcliffe, L.T.; Douglas, F.P.; Asche, L.V.; Mathews, J.D.; Hutton, S.I.; et al. Chlamydia trachomatis from Australian Aboriginal people with trachoma are polyphyletic composed of multiple distinctive lineages. Nat. Commun. 2016, 7, 10688. [Google Scholar] [CrossRef]
- Giffard, P.M.; Brenner, N.C.; Tabrizi, S.N.; Garland, S.M.; Holt, D.C.; Andersson, P.; Lilliebridge, R.A.; Tong, S.Y.; Karimi, M.; Boylan, P.; et al. Chlamydia trachomatis genotypes in a cross-sectional study of urogenital samples from remote Northern and Central Australia. BMJ Open 2016, 6, e009624. [Google Scholar] [CrossRef] [Green Version]
- Giffard, P.M.; Andersson, P.; Wilson, J.; Buckley, C.; Lilliebridge, R.; Harris, T.M.; Kleinecke, M.; O′Grady, K.F.; Huston, W.M.; Lambert, S.B.; et al. CtGEM typing: Discrimination of Chlamydia trachomatis ocular and urogenital strains and major evolutionary lineages by high resolution melting analysis of two amplified DNA fragments. PLoS ONE 2018, 13, e0195454. [Google Scholar] [CrossRef] [Green Version]
- Hadfield, J.; Harris, S.R.; Seth-Smith, H.M.B.; Parmar, S.; Andersson, P.; Giffard, P.M.; Schachter, J.; Moncada, J.; Ellison, L.; Vaulet, M.L.G.; et al. Comprehensive global genome dynamics of Chlamydia trachomatis show ancient diversification followed by contemporary mixing and recent lineage expansion. Genome Res. 2017, 27, 1220–1229. [Google Scholar] [CrossRef] [Green Version]
- Versteeg, B.; Bruisten, S.M.; Pannekoek, Y.; Jolley, K.A.; Maiden, M.C.J.; van der Ende, A.; Harrison, O.B. Genomic analyses of the Chlamydia trachomatis core genome show an association between chromosomal genome, plasmid type and disease. BMC Genom. 2018, 19, 130. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, B.; Isaksson, J.; Ryberg, M.; Tangrot, J.; Saleh, I.; Versteeg, B.; Gravningen, K.; Bruisten, S. Global multilocus sequence type analysis of Chlamydia trachomatis strains from 16 countries. J. Clin. Microbiol. 2015, 53, 2172–2179. [Google Scholar] [CrossRef] [Green Version]
- de Vries, H.J.; Schim van der Loeff, M.F.; Bruisten, S.M. High-resolution typing of Chlamydia trachomatis: Epidemiological and clinical uses. Curr. Opin. Infect. Dis. 2015, 28, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Molano, M.; Tabrizi, S.N.; Phillips, S.; Danielewski, J.; Cornall, A.; Morre, S.A.; Garland, S.M. Development of a rapid colorimetric multiplex PCR–reverse line blot for the detection and typing of 14 Chlamydia trachomatis genovars. J. Med. Microbiol. 2018, 67, 1560–1570. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, M.; Filardo, S.; Romano, S.; Sessa, R. Chlamydia trachomatis and Chlamydia pneumoniae interaction with the host: Latest advances and future prospective. Microorganisms 2019, 7, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roulis, E.; Polkinghorne, A.; Timms, P. Chlamydia pneumoniae: Modern insights into an ancient pathogen. Trends Microbiol. 2013, 21, 120–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charles, P.G.P.; Whitby, M.; Fuller, A.J.; Stirling, R.; Wright, A.A.; Korman, T.M.; Holmes, P.W.; Christiansen, K.J.; Waterer, G.W.; Pierce, R.J.P.; et al. The etiology of community-acquired pneumonia in Australia: Why penicillin plus doxycycline or a macrolide is the most appropriate therapy. Clin. Infect. Dis. 2008, 46, 1513–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmu, A.A.; Ware, R.S.; Lambert, S.B.; Sarna, M.; Bialasiewicz, S.; Seib, K.L.; Atack, J.M.; Nissen, M.D.; Grimwood, K. Nasal swab bacteriology by PCR during the first 24-months of life: A prospective birth cohort study. Pediatr. Pulmonol. 2019, 54, 289–296. [Google Scholar] [CrossRef]
- Rane, V.; Khailin, K.; Williams, J.; Francis, M.; Kotsanas, D.; Korman, T.M.; Graham, M. Underdiagnosis of Chlamydia trachomatis and Chlamydia psittaci revealed by introduction of respiratory multiplex PCR assay with Chlamydiaceae family primers. Diagn. Microbiol. Infect. Dis. 2018, 90, 163–166. [Google Scholar] [CrossRef]
- Teoh, L.; Mackay, I.M.; Van Asperen, P.P.; Acworth, J.P.; Hurwitz, M.; Upham, J.W.; Siew, W.H.; Wang, C.Y.T.; Sloots, T.P.; Neeman, T.; et al. Presence of atopy increases the risk of asthma relapse. Arch. Dis. Child. 2018, 103, 346–351. [Google Scholar] [CrossRef]
- Wark, P.A.B.; Johnston, S.L.; Simpson, J.L.; Hensley, M.J.; Gibson, P.G. Chlamydia pneumoniae immunoglobulin A reactivation and airway inflammation in acute asthma. Eur. Respir. J. 2002, 20, 834. [Google Scholar] [CrossRef] [Green Version]
- Glanville, A.R.; Gencay, M.; Tamm, M.; Chhajed, P.; Plit, M.; Hopkins, P.; Aboyoun, C.; Roth, M.; Malouf, M. Chlamydia pneumoniae infection after lung transplantation. J. Heart Lung Transplant. 2005, 24, 131–136. [Google Scholar] [CrossRef]
- Campbell, L.A.; Kuo, C.C. Chlamydia pneumonia—An infectious risk factor for atherosclerosis? Nat. Rev. Microbiol. 2004, 2, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, M.; Walker, P.; Gibbs, H.; Timms, P. Multiple genotypes of Chlamydia pneumoniae identified in human carotid plaque. Microbiology 2005, 151, 2285–2290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cochrane, M.; Kalle, W.H.J.; Roffey, P.; Moriarty, H.T. The detection of Chlamydia pneumoniae in atherosclerotic plaques of Australian subjects. Pathology 2002, 34, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.K.C.; Sintchenko, V.; Meldrum-Hanna, W.; Skinner, M.P.; Ross, D.L.; Gilbert, G.L. Failure to detect Chlamydia pneumoniae in coronary atheromas of Australian patients undergoing coronary artery bypass grafting. Heart Lung Circ. 2000, 9, A124. [Google Scholar] [CrossRef]
- Coles, K.A.; Plant, A.J.; Riley, T.V.; Smith, D.W.; McQuillan, B.M.; Thompson, P.L. Lack of association between seropositivity to Chlamydia pneumoniae and carotid atherosclerosis. Am. J. Cardiol. 1999, 84, 825–828. [Google Scholar] [CrossRef]
- Thibault, P.; Attia, J.; Oldmeadow, C. A prolonged antibiotic protocol to treat persistent Chlamydophila pneumoniae infection improves the extracranial venous circulation in multiple sclerosis. Phlebology 2018, 33, 397–406. [Google Scholar] [CrossRef]
- Woolley, I.J.; Larsen, M.; Jones, S.; Gahan, M.E.; Jasenko, I.; Johnsen, S.P.; Wesselingh, S.; Fuller, A.; Ostergaard, L. Chlamydia pneumoniae in HIV-infected patients and controls assessed by a novel whole blood interferon-gamma assay, serology and PCR. Clin. Microbiol. Infect. 2004, 10, 820–825. [Google Scholar] [CrossRef] [Green Version]
- Robman, L.; Mahdi, O.; McCarty, C.; Dimitrov, P.; Tikellis, G.; McNeil, J.; Byrne, G.; Taylor, H.; Guymer, R. Exposure to Chlamydia pneumoniae infection and progression of age-related macular degeneration. Am. J. Epidemiol. 2005, 161, 1013–1019. [Google Scholar] [CrossRef]
- Guymer, R.; Robman, L. Chlamydia pneumoniae and age-related macular degeneration: A role in pathogenesis or merely a chance association? Clin. Exp. Ophthalmol. 2007, 35, 89–93. [Google Scholar] [CrossRef]
- Roulis, E.; Bachmann, N.; Humphrys, M.; Myers, G.; Huston, W.; Polkinghorne, A.; Timms, P. Phylogenetic analysis of human Chlamydia pneumoniae strains reveals a distinct Australian indigenous clade that predates European exploration of the continent. BMC Genom. 2015, 16, 1094. [Google Scholar] [CrossRef]
- Desmond, L.A.; Lloyd, M.A.; Ryan, S.A.; Janus, E.D.; Karunajeewa, H.A. Respiratory viruses in adults hospitalised with Community-Acquired Pneumonia during the non-winter months in Melbourne: Routine diagnostic practice may miss large numbers of influenza and respiratory syncytial virus infections. Commun. Dis. Intell. 2019, 43. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.M.; Hutton, S.; Myers, G.S.A.; Brunham, R.; Timms, P. Chlamydia pneumoniae is genetically diverse in animals and appears to have crossed the host barrier to humans on (at least) two occasions. PLoS Pathog. 2010, 6, e1000903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, C.; Hovis, K.; Bavoil, P.; Myers, G.; Carrasco, J.; Timms, P. Comparison of koala LPCoLN and human strains of Chlamydia pneumoniae highlights extended genetic diversity in the species. BMC Genom. 2010, 11, 442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myers, G.S.A.; Mathews, S.A.; Eppinger, M.; Mitchell, C.; O′Brien, K.K.; White, O.R.; Benahmed, F.; Brunham, R.C.; Read, T.D.; Ravel, J.; et al. Evidence that Human Chlamydia pneumoniae was zoonotically acquired. J. Bacteriol. 2009, 191, 7225–7233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenner, F. Frank Macfarlane Burnet 1899–1985. Hist. Rec. Aust. Sci. 1987, 7, 39–77. [Google Scholar] [CrossRef]
- Burnet, F.M. Psittacosis in Australian parrots. Med. J. Aust. 1934, 2, 743–746. [Google Scholar] [CrossRef]
- Burnet, F.M. Enzootic Psittacosis amongst Wild Australian Parrots. J. Hyg. 1935, 35, 412–420. [Google Scholar] [CrossRef] [Green Version]
- Burnet, F.M. A note of the occurrence of fatal psittacosis in parrots living in the wild state. Med. J. Aust. 1939, 1, 545–546. [Google Scholar] [CrossRef]
- Taylor, K.; Durrheim, D.; Massey, P.; Hughes, K.; Heller, J.; Jones, B. An atypical case of typical pneumonia. Aust. J. Gen. Pract. 2018, 47, 119–121. [Google Scholar] [CrossRef]
- Branley, J.M.; Roy, B.; Dwyer, D.E.; Sorrell, T.C. Real-time PCR detection and quantitation of Chlamydophila psittaci in human and avian specimens from a veterinary clinic cluster. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 27, 269–273. [Google Scholar] [CrossRef]
- Branley, J.M.; Weston, K.M.; England, J.; Dwyer, D.E.; Sorrell, T.C. Clinical features of endemic community-acquired psittacosis. New Microbes New Infect. 2014, 2, 7–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Telfer, B.L.; Moberley, S.A.; Hort, K.P.; Branley, J.M.; Dwyer, D.E.; Muscatello, D.J.; Correll, P.K.; England, J.; McAnulty, J.M. Probable psittacosis outbreak linked to wild birds. Emerg. Infect. Dis. 2005, 11, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Yung, A.P.; Grayson, M.L. Psittacosis—A review of 135 cases. Med. J. Aust. 1988, 148, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Knittler, M.R.; Sachse, K. Chlamydia psittaci: Update on an underestimated zoonotic agent. Pathog. Dis. 2015, 73, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogerwerf, L.; De Gier, B.; Baan, B.; Van Der Hoek, W. Chlamydia psittaci (psittacosis) as a cause of community-acquired pneumonia: A systematic review and meta-analysis. Epidemiol. Infect. 2017, 145, 3096–3105. [Google Scholar] [CrossRef] [Green Version]
- De Boeck, C.; Dehollogne, C.; Dumont, A.; Spierenburg, M.; Heijne, M.; Gyssens, I.; Van der Hilst, J.; Vanrompay, D. Managing a cluster outbreak of psittacosis in Belgium linked to a pet shop visit in The Netherlands. Epidemiol. Infect. 2016, 144, 1710–1716. [Google Scholar] [CrossRef]
- Taylor, K.A.; Durrheim, D.; Heller, J.; O′Rourke, B.; Hope, K.; Merritt, T.; Freeman, P.; Chicken, C.; Carrick, J.; Branley, J.; et al. Equine chlamydiosis-An emerging infectious disease requiring a one health surveillance approach. Zoonoses Public Health 2017, 65, 218–221. [Google Scholar] [CrossRef]
- Chan, J.; Doyle, B.; Branley, J.; Sheppeard, V.; Gabor, M.; Viney, K.; Quinn, H.; Janover, O.; McCready, M.; Heller, J. An outbreak of psittacosis at a veterinary school demonstrating a novel source of infection. One Health 2017, 3, 29–33. [Google Scholar] [CrossRef]
- Jenkins, C.; Jelocnik, M.; Micallef, M.L.; Galea, F.; Taylor-Brown, A.; Bogema, D.R.; Liu, M.; O′Rourke, B.; Chicken, C.; Carrick, J.; et al. An epizootic of Chlamydia psittaci equine reproductive loss associated with suspected spillover from native Australian parrots. Emerg. Microbes Infect. 2018, 7, 88. [Google Scholar] [CrossRef] [Green Version]
- Jelocnik, M.; Branley, J.; Heller, J.; Raidal, S.; Alderson, S.; Galea, F.; Gabor, M.; Polkinghorne, A. Multilocus sequence typing identifies an avian-like Chlamydia psittaci strain involved in equine placentitis and associated with subsequent human psittacosis. Emerg. Microbes Infect. 2017, 6, e7. [Google Scholar] [CrossRef] [Green Version]
- Gough, S.L.; Carrick, J.; Raidal, S.L.; Keane, S.; Collins, N.; Cudmore, L.; Russell, C.M.; Raidal, S.; Hughes, K.J. Chlamydia psittaci infection as a cause of respiratory disease in neonatal foals. Equine Vet. J. 2019, 24. [Google Scholar] [CrossRef] [PubMed]
- Jelocnik, M.; Jenkins, C.; O′Rourke, B.; Barnwell, J.; Polkinghorne, A. Molecular evidence to suggest pigeon-type Chlamydia psittaci in association with an equine foal loss. Transbound. Emerg. Dis. 2018, 65, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, P.; Sachse, K.; Kaltenboeck, B. Chlamydiaceae in cattle: Commensals, trigger organisms, or pathogens? Vet. J. 2011, 189, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Jelocnik, M.; Taylor-Brown, A.; O′Dea, C.; Anstey, S.; Bommana, S.; Masters, N.; Katouli, M.; Jenkins, C.; Polkinghorne, A. Detection of a range of genetically diverse chlamydiae in Australian domesticated and wild ungulates. Transbound. Emerg. Dis. 2019, 66, 1132–1137. [Google Scholar] [CrossRef] [PubMed]
- Jelocnik, M.; Laurence, M.; Murdoch, F.; Polkinghorne, A. Detection of Chlamydiaceae in ocular swabs from Australian pre-export feedlot sheep. Aust. Vet. J. 2019, 97, 401–403. [Google Scholar] [CrossRef]
- Branley, J.; Bachmann, N.L.; Jelocnik, M.; Myers, G.S.; Polkinghorne, A. Australian human and parrot Chlamydia psittaci strains cluster within the highly virulent 6BC clade of this important zoonotic pathogen. Sci. Rep. 2016, 6, 30019. [Google Scholar] [CrossRef] [Green Version]
- Amery-Gale, J.; Legione, A.R.; Marenda, M.S.; Owens, J.; Eden, P.A.; Konsak-Ilievski, B.M.; Whiteley, P.L.; Dobson, E.C.; Browne, E.A.; Slocombe, R.F.; et al. Surveillance for Chlamydia spp. with multilocus sequence typing analysis in wild and captive birds in Victoria, Australia. J. Wildl. Dis. 2019. [Google Scholar]
- Sutherland, M.; Sarker, S.; Vaz, P.K.; Legione, A.R.; Devlin, J.M.; Macwhirter, P.L.; Whiteley, P.L.; Raidal, S.R. Disease surveillance in wild Victorian cacatuids reveals co-infection with multiple agents and detection of novel avian viruses. Vet. Microbiol. 2019, 235, 257–264. [Google Scholar] [CrossRef]
- Read, T.D.; Joseph, S.J.; Didelot, X.; Liang, B.; Patel, L.; Dean, D. Comparative analysis of Chlamydia psittaci genomes reveals the recent emergence of a pathogenic lineage with a broad host range. MBio 2013, 4, e00604-12. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Astudillo, V.; Allavena, R.; McKinnon, A.; Larkin, R.; Henning, J. Decline causes of Koalas in South East Queensland, Australia: A 17-year retrospective study of mortality and morbidity. Sci. Rep. 2017, 7, 42587. [Google Scholar] [CrossRef] [Green Version]
- Narayan, E. Physiological stress levels in wild koala sub-populations facing anthropogenic induced environmental trauma and disease. Sci. Rep. 2019, 9, 6031. [Google Scholar] [CrossRef] [PubMed]
- Polkinghorne, A.; Hanger, J.; Timms, P. Recent advances in understanding the biology, epidemiology and control of chlamydial infections in koalas. Vet. Microbiol. 2013, 165, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Walker, E.; Moore, C.; Shearer, P.; Jelocnik, M.; Bommana, S.; Timms, P.; Polkinghorne, A. Clinical, diagnostic and pathologic features of presumptive cases of Chlamydia pecorum-associated arthritis in Australian sheep flocks. BMC Vet. Res. 2016, 12, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legione, A.R.; Patterson, J.L.; Whiteley, P.L.; Amery-Gale, J.; Lynch, M.; Haynes, L.; Gilkerson, J.R.; Polkinghorne, A.; Devlin, J.M.; Sansom, F.M. Identification of unusual Chlamydia pecorum genotypes in Victorian koalas (Phascolarctos cinereus) and clinical variables associated with infection. J. Med. Microbiol. 2016, 65, 420–428. [Google Scholar] [CrossRef]
- Nyari, S.; Waugh, C.A.; Dong, J.; Quigley, B.L.; Hanger, J.; Loader, J.; Polkinghorne, A.; Timms, P. Epidemiology of chlamydial infection and disease in a free-ranging koala (Phascolarctos cinereus) population. PLoS ONE 2017, 12, e0190114. [Google Scholar] [CrossRef]
- Robbins, A.; Hanger, J.; Jelocnik, M.; Quigley, B.L.; Timms, P. Longitudinal study of wild koalas (Phascolarctos cinereus) reveals chlamydial disease progression in two thirds of infected animals. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Speight, K.N.; Polkinghorne, A.; Penn, R.; Boardman, W.; Timms, P.; Fraser, T.; Johnson, K.; Faull, R.; Bate, S.; Woolford, L. Prevalence and pathologic features of Chlamydia pecorum infections in south australian koalas (Phascolarctos cinereus). J. Wildl. Dis. 2016, 52, 301–306. [Google Scholar] [CrossRef]
- Patterson, J.L.; Lynch, M.; Anderson, G.A.; Noormohammadi, A.H.; Legione, A.; Gilkerson, J.R.; Devlin, J.M. The prevalence and clinical significance of Chlamydia infection in island and mainland populations of Victorian koalas (Phascolarctos cinereus). J. Wildl. Dis. 2015, 51, 309–317. [Google Scholar] [CrossRef]
- Fabijan, J.; Caraguel, C.; Jelocnik, M.; Polkinghorne, A.; Boardman, W.S.J.; Nishimoto, E.; Johnsson, G.; Molsher, R.; Woolford, L.; Timms, P.; et al. Chlamydia pecorum prevalence in South Australian koala (Phascolarctos cinereus) populations: Identification and modelling of a population free from infection. Sci. Rep. 2019, 9, 6261. [Google Scholar] [CrossRef] [Green Version]
- Hulse, L.S.; Beagley, K.; Ellis, W.; Fitzgibbon, S.; Gillett, A.; Barth, B.; Robbins, A.; Pyne, M.; Larkin, R.; Johnston, S.D. Epidemiology of chlamydia-induced reproductive disease in male koalas (Phascolarctos cinereus) from southeast Queensland, Australia as assessed from penile urethral swabs and semen. J. Wildl. Dis. 2019. [Google Scholar]
- Phillips, S.; Robbins, A.; Loader, J.; Hanger, J.; Booth, R.; Jelocnik, M.; Polkinghorne, A.; Timms, P. Chlamydia pecorum gastrointestinal tract infection associations with urogenital tract infections in the koala (Phascolarctos cinereus). PLoS ONE 2018, 13, e0206471. [Google Scholar] [CrossRef]
- Russell, I.; Timms, P.; Hanger, J.; Loader, J.; Gillett, A.; Waugh, C. Prevalence of Chlamydia pecorum in juvenile koalas (Phascolarctos cinereus) and evidence for protection from infection via maternal immunization. J. Wildl. Dis. 2018, 54, 863–865. [Google Scholar] [CrossRef]
- Jelocnik, M.; Gillett, A.; Hanger, J.; Polkinghorne, A. Chlamydiosis in koalas. In Current Therapy in Medicine of Australian Mammals; Vogelnest, L., Portas, T., Eds.; CSIRO Publishing: Clayton, Australia, 2019. [Google Scholar]
- Robbins, A.; Loader, J.; Timms, P.; Hanger, J. Optimising the short and long-term clinical outcomes for koalas (Phascolarctos cinereus) during treatment for chlamydial infection and disease. PLoS ONE 2018, 13, e0209679. [Google Scholar] [CrossRef]
- Mackie, J.T.; Gillett, A.K.; Palmieri, C.; Feng, T.; Higgins, D.P. Pneumonia due to Chlamydia pecorum in a Koala (Phascolarctos cinereus). J. Comp. Pathol. 2016, 155, 356–360. [Google Scholar] [CrossRef]
- Burnard, D.; Gillett, A.; Polkinghorne, A. Chlamydia pecorum in joint tissue and synovial fluid of a Koala (Phascolarctos cinereus) with arthritis. J. Wildl. Dis. 2018, 54, 646–649. [Google Scholar] [CrossRef]
- Waugh, C.A.; Hanger, J.; Loader, J.; King, A.; Hobbs, M.; Johnson, R.; Timms, P. Infection with koala retrovirus subgroup B (KoRV-B), but not KoRV-A, is associated with chlamydial disease in free-ranging koalas (Phascolarctos cinereus). Sci. Rep. 2017, 7, 134. [Google Scholar] [CrossRef] [Green Version]
- Quigley, B.L.; Ong, V.A.; Hanger, J.; Timms, P. Molecular dynamics and mode of transmission of koala retrovirus as it invades and spreads through a wild Queensland koala population. J. Virol. 2018, 92, e01871-17. [Google Scholar] [CrossRef] [Green Version]
- Fabijan, J.; Miller, D.; Olagoke, O.; Woolford, L.; Boardman, W.; Timms, P.; Polkinghorne, A.; Simmons, G.; Hemmatzadeh, F.; Trott, D.J.; et al. Prevalence and clinical significance of koala retrovirus in two South Australian koala (Phascolarctos cinereus) populations. J. Med. Microbiol. 2019, 68, 1072–1080. [Google Scholar] [CrossRef]
- Lawrence, A.; Fraser, T.; Gillett, A.; Tyndall, J.D.; Timms, P.; Polkinghorne, A.; Huston, W.M. Chlamydia serine protease inhibitor, targeting HtrA, as a new treatment for koala Chlamydia infection. Sci. Rep. 2016, 6, 31466. [Google Scholar] [CrossRef] [Green Version]
- Phillips, S.; Quigley, B.L.; Timms, P. Seventy years of Chlamydia vaccine research—Limitations of the past and directions for the future. Front. Microbiol. 2019, 10, 70. [Google Scholar] [CrossRef] [Green Version]
- Nyari, S.; Khan, S.A.; Rawlinson, G.; Waugh, C.A.; Potter, A.; Gerdts, V.; Timms, P. Vaccination of koalas (Phascolarctos cinereus) against Chlamydia pecorum using synthetic peptides derived from the major outer membrane protein. PLoS ONE 2018, 13, e0200112. [Google Scholar] [CrossRef] [PubMed]
- Desclozeaux, M.; Robbins, A.; Jelocnik, M.; Khan, S.A.; Hanger, J.; Gerdts, V.; Potter, A.; Polkinghorne, A.; Timms, P. Immunization of a wild koala population with a recombinant Chlamydia pecorum Major Outer Membrane Protein (MOMP) or Polymorphic Membrane Protein (PMP) based vaccine: New insights into immune response, protection and clearance. PLoS ONE 2017, 12, e0178786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.A.; Polkinghorne, A.; Waugh, C.; Hanger, J.; Loader, J.; Beagley, K.; Timms, P. Humoral immune responses in koalas (Phascolarctos cinereus) either naturally infected with Chlamydia pecorum or following administration of a recombinant chlamydial major outer membrane protein vaccine. Vaccine 2016, 34, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Kollipara, A.; Wan, C.; Rawlinson, G.; Brumm, J.; Nilsson, K.; Polkinghorne, A.; Beagley, K.; Timms, P. Antigenic specificity of a monovalent versus polyvalent MOMP based Chlamydia pecorum vaccine in koalas (Phascolarctos cinereus). Vaccine 2013, 31, 1217–1223. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.A.; Desclozeaux, M.; Waugh, C.; Hanger, J.; Loader, J.; Gerdts, V.; Potter, A.; Polkinghorne, A.; Beagley, K.; Timms, P. Antibody and cytokine responses of koalas (Phascolarctos cinereus) vaccinated with recombinant Chlamydial Major Outer Membrane Protein (MOMP) with two different adjuvants. PLoS ONE 2016, 11, e0156094. [Google Scholar] [CrossRef]
- Nyari, S.; Booth, R.; Quigley, B.L.; Waugh, C.A.; Timms, P. Therapeutic effect of a Chlamydia pecorum recombinant major outer membrane protein vaccine on ocular disease in koalas (Phascolarctos cinereus). PLoS ONE 2019, 14, e0210245. [Google Scholar] [CrossRef] [Green Version]
- Waugh, C.; Khan, S.A.; Carver, S.; Hanger, J.; Loader, J.; Polkinghorne, A.; Beagley, K.; Timms, P. A Prototype recombinant-protein based Chlamydia pecorum vaccine results in reduced chlamydial burden and less clinical disease in free-ranging koalas (Phascolarctos cinereus). PLoS ONE 2016, 11, e0146934. [Google Scholar] [CrossRef]
- Burnard, D.; Huston, W.M.; Webb, J.K.; Jelocnik, M.; Reiss, A.; Gillett, A.; Fitzgibbon, S.; Carver, S.; Carrucan, J.; Flanagan, C.; et al. Molecular evidence of Chlamydia pecorum and arthropod-associated Chlamydiae in an expanded range of marsupials. Sci. Rep. 2017, 7, 12844. [Google Scholar] [CrossRef] [Green Version]
- Borel, N.; Polkinghorne, A.; Pospischil, A. A review on chlamydial diseases in animals: Still a challenge for pathologists? Vet. Pathol. 2018, 55, 374–390. [Google Scholar] [CrossRef] [Green Version]
- Bommana, S.; Walker, E.; Desclozeaux, M.; Jelocnik, M.; Timms, P.; Polkinghorne, A.; Carver, S. Molecular and serological dynamics of Chlamydia pecorum infection in a longitudinal study of prime lamb production. PeerJ 2018, 6, e4296. [Google Scholar] [CrossRef] [Green Version]
- Walker, E.; Jelocnik, M.; Bommana, S.; Timms, P.; Carver, S.; Polkinghorne, A. Understanding the health and production impacts of endemic Chlamydia pecorum infections in lambs. Vet. Microbiol. 2018, 217, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Jelocnik, M.; Forshaw, D.; Cotter, J.; Roberts, D.; Timms, P.; Polkinghorne, A. Molecular and pathological insights into Chlamydia pecorum-associated sporadic bovine encephalomyelitis (SBE) in Western Australia. BMC Vet. Res. 2014, 10, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, R.; Jacobson, C.; Gardner, G.; Carmichael, I.; Campbell, A.J.; Ryan, U. Longitudinal prevalence and faecal shedding of Chlamydia pecorum in sheep. Vet. J. 2014, 201, 322–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anstey, S.I.; Quigley, B.L.; Polkinghorne, A.; Jelocnik, M. Chlamydial infection and on-farm risk factors in dairy cattle herds in South East Queensland. Aust. Vet. J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Desclozeaux, M.; Jelocnik, M.; Whitting, K.; Saifzadeh, S.; Bommana, S.; Potter, A.; Gerdts, V.; Timms, P.; Polkinghorne, A. Safety and immunogenicity of a prototype anti-Chlamydia pecorum recombinant protein vaccine in lambs and pregnant ewes. Vaccine 2017, 35, 3461–3465. [Google Scholar] [CrossRef]
- Bommana, S.; Jelocnik, M.; Borel, N.; Marsh, I.; Carver, S.; Polkinghorne, A. The limitations of commercial serological assays for detection of chlamydial infections in Australian livestock. J. Med. Microbiol. 2019, 68, 627–632. [Google Scholar] [CrossRef]
- McCauley, L.M.E.; Lancaster, M.J.; Butler, K.L.; Ainsworth, C.G.V. Serological analysis of Chlamydophila abortus in Australian sheep and implications for the rejection of breeder sheep for export. Aust. Vet. J. 2010, 88, 32–38. [Google Scholar] [CrossRef]
- Bachmann, N.; Fraser, T.; Bertelli, C.; Jelocnik, M.; Gillett, A.; Funnell, O.; Flanagan, C.; Myers, G.S.; Timms, P.; Polkinghorne, A. Comparative genomics of koala, cattle and sheep strains of Chlamydia pecorum. BMC Genom. 2014, 15, 667. [Google Scholar] [CrossRef] [Green Version]
- Bachmann, N.L.; Sullivan, M.J.; Jelocnik, M.; Myers, G.S.A.; Timms, P.; Polkinghorne, A. Culture-independent genome sequencing of clinical samples reveals an unexpected heterogeneity of infections by Chlamydia pecorum. J. Clin. Microbiol. 2015, 53, 1573–1581. [Google Scholar] [CrossRef] [Green Version]
- Jelocnik, M.; Walker, E.; Pannekoek, Y.; Ellem, J.; Timms, P.; Polkinghorne, A. Evaluation of the relationship between Chlamydia pecorum sequence types and disease using a species-specific multi-locus sequence typing scheme (MLST). Vet. Microbiol. 2014, 174, 214–222. [Google Scholar] [CrossRef]
- Jelocnik, M.; Frentiu, F.D.; Timms, P.; Polkinghorne, A. Multi-locus sequence analysis provides insights into the molecular epidemiology of Chlamydia pecorum infections in Australian sheep, cattle and koalas. J. Clin. Microbiol. 2013, 51, 2625–2632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legione, A.R.; Amery-Gale, J.; Lynch, M.; Haynes, L.; Gilkerson, J.R.; Sansom, F.M.; Devlin, J.M. Chlamydia pecorum infection in free-ranging koalas (Phascolarctos cinereus ) on French Island, Victoria, Australia. J. Wildl. Dis. 2016, 52, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.M.; Schmertmann, L.J.; Higgins, D.P.; Casteriano, A.; Irinyi, L.; Mella, V.S.A.; Crowther, M.S.; Meyer, W.; Krockenberger, M.B. Genetic differences in Chlamydia pecorum between neighbouring sub-populations of koalas (Phascolarctos cinereus). Vet. Microbiol. 2019, 231, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Jelocnik, M.; Bachmann, N.L.; Kaltenboeck, B.; Waugh, C.; Woolford, L.; Speight, K.N.; Gillett, A.; Higgins, D.P.; Flanagan, C.; Myers, G.S.; et al. Genetic diversity in the plasticity zone and the presence of the chlamydial plasmid differentiates Chlamydia pecorum strains from pigs, sheep, cattle, and koalas. BMC Genom. 2015, 16, 893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Australian Government Department of Agriculture. Biosecurity in Australia. Available online: https://www.agriculture.gov.au/biosecurity/australia (accessed on 11 November 2019).
- Stokes, H.S.; Martens, J.M.; Chamings, A.; Walder, K.; Berg, M.L.; Segal, Y.; Bennett, A. Identification of Chlamydia gallinacea in a parrot and in free-range chickens in Australia. Aust. Vet. J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Stride, M.C.; Polkinghorne, A.; Miller, T.L.; Nowak, B.F. Molecular characterization of “Candidatus Similichlamydia latridicola” gen. nov., sp. nov. (Chlamydiales: “Candidatus Parilichlamydiaceae”), a novel Chlamydia-like epitheliocystis agent in the striped trumpeter, Latris lineata (Forster). Appl. Environ. Microbiol. 2013, 79, 4914–4920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor-Brown, A.; Pillonel, T.; Bridle, A.; Qi, W.; Bachmann, N.L.; Miller, T.L.; Greub, G.; Nowak, B.; Seth-Smith, H.M.B.; Vaughan, L.; et al. Culture-independent genomics of a novel chlamydial pathogen of fish provides new insight into host-specific adaptations utilized by these intracellular bacteria. Environ. Microbiol. 2017, 19, 1899–1913. [Google Scholar] [CrossRef]
- Robertson, T.; Bibby, S.; O′Rourke, D.; Belfiore, T.; Agnew-Crumpton, R.; Noormohammadi, A.H. Identification of Chlamydial species in crocodiles and chickens by PCR-HRM curve analysis. Vet. Microbiol. 2010, 145, 373–379. [Google Scholar] [CrossRef]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jelocnik, M. Chlamydiae from Down Under: The Curious Cases of Chlamydial Infections in Australia. Microorganisms 2019, 7, 602. https://doi.org/10.3390/microorganisms7120602
Jelocnik M. Chlamydiae from Down Under: The Curious Cases of Chlamydial Infections in Australia. Microorganisms. 2019; 7(12):602. https://doi.org/10.3390/microorganisms7120602
Chicago/Turabian StyleJelocnik, Martina. 2019. "Chlamydiae from Down Under: The Curious Cases of Chlamydial Infections in Australia" Microorganisms 7, no. 12: 602. https://doi.org/10.3390/microorganisms7120602
APA StyleJelocnik, M. (2019). Chlamydiae from Down Under: The Curious Cases of Chlamydial Infections in Australia. Microorganisms, 7(12), 602. https://doi.org/10.3390/microorganisms7120602




