Quantification and Multidrug Resistance Profiles of Vancomycin-Resistant Enterococci Isolated from Two Wastewater Treatment Plants in the Same Municipality
Abstract
:1. Introduction
2. Materials and Methods
2.1. Wastewater Treatment Plant Parameters, VRE Surveillance and Antimicrobial Use Information
2.2. Land Use
2.3. Chemical and Environmental Information
2.4. Sample Collection, Enumeration and Isolation of Enterococcus
2.5. Quantitative TaqMan Real-Time PCR
2.6. MIC for Vancomycin Resistant
2.7. Speciation of Vancomycin-Resistant Enterococci
2.8. Antibiotic Susceptibility Testing
2.9. Data Analysis
3. Results
3.1. Microbiological Quantification of Enterococci and Vancomycin-Resistant Enterococci
3.2. Molecular Quantification of Enterococci, Vancomycin Resistance (vanA) and Class I Integrons
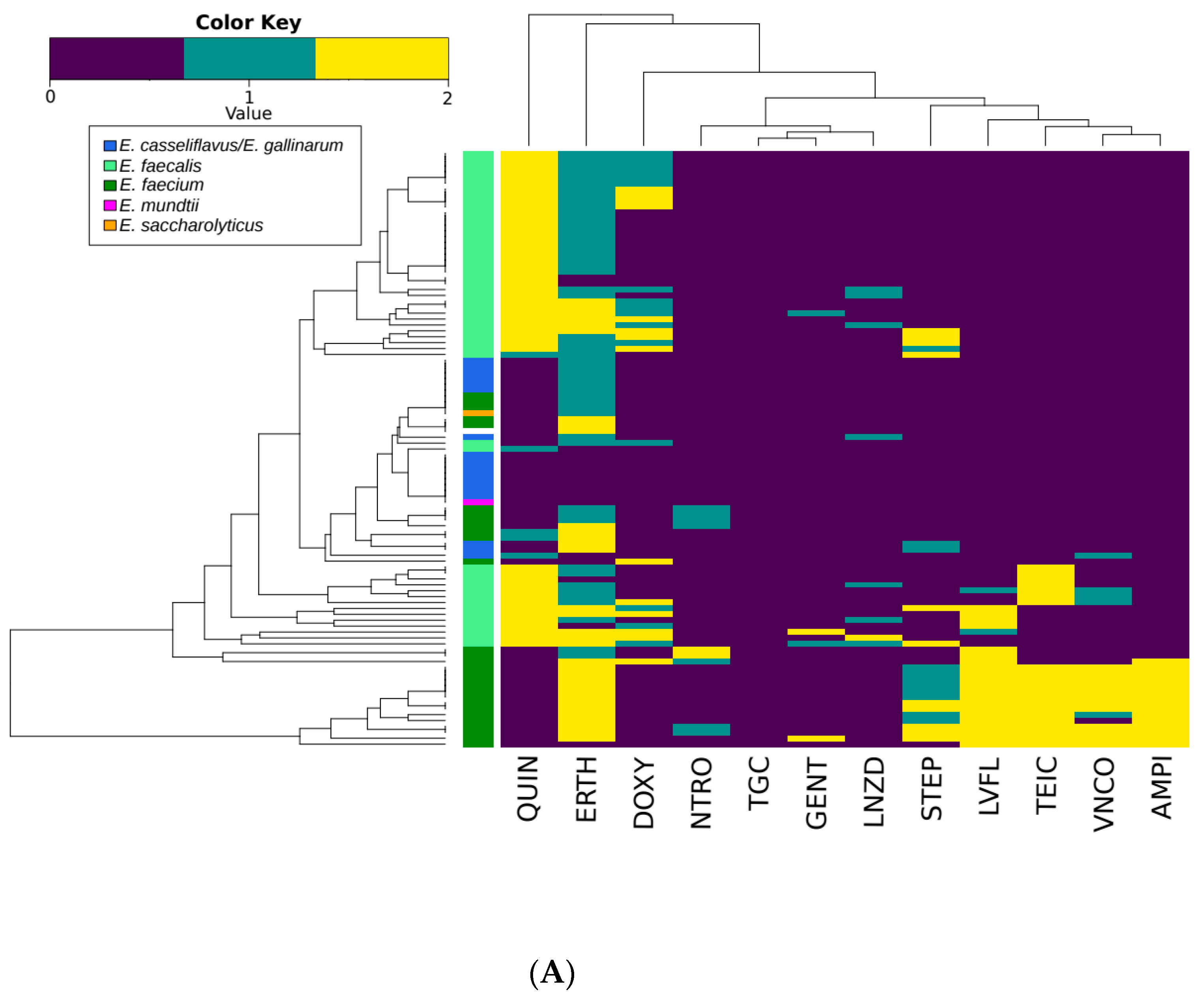
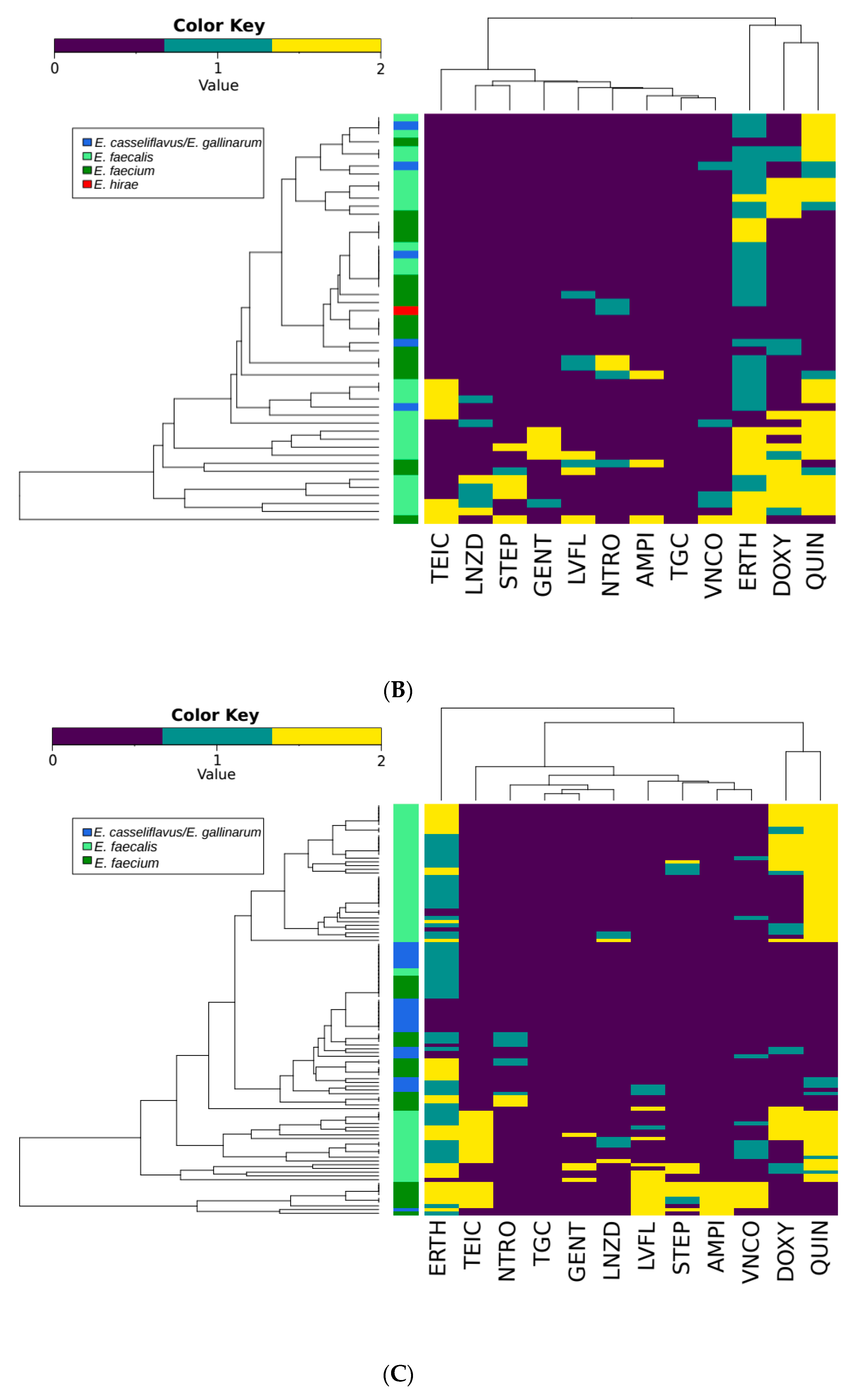
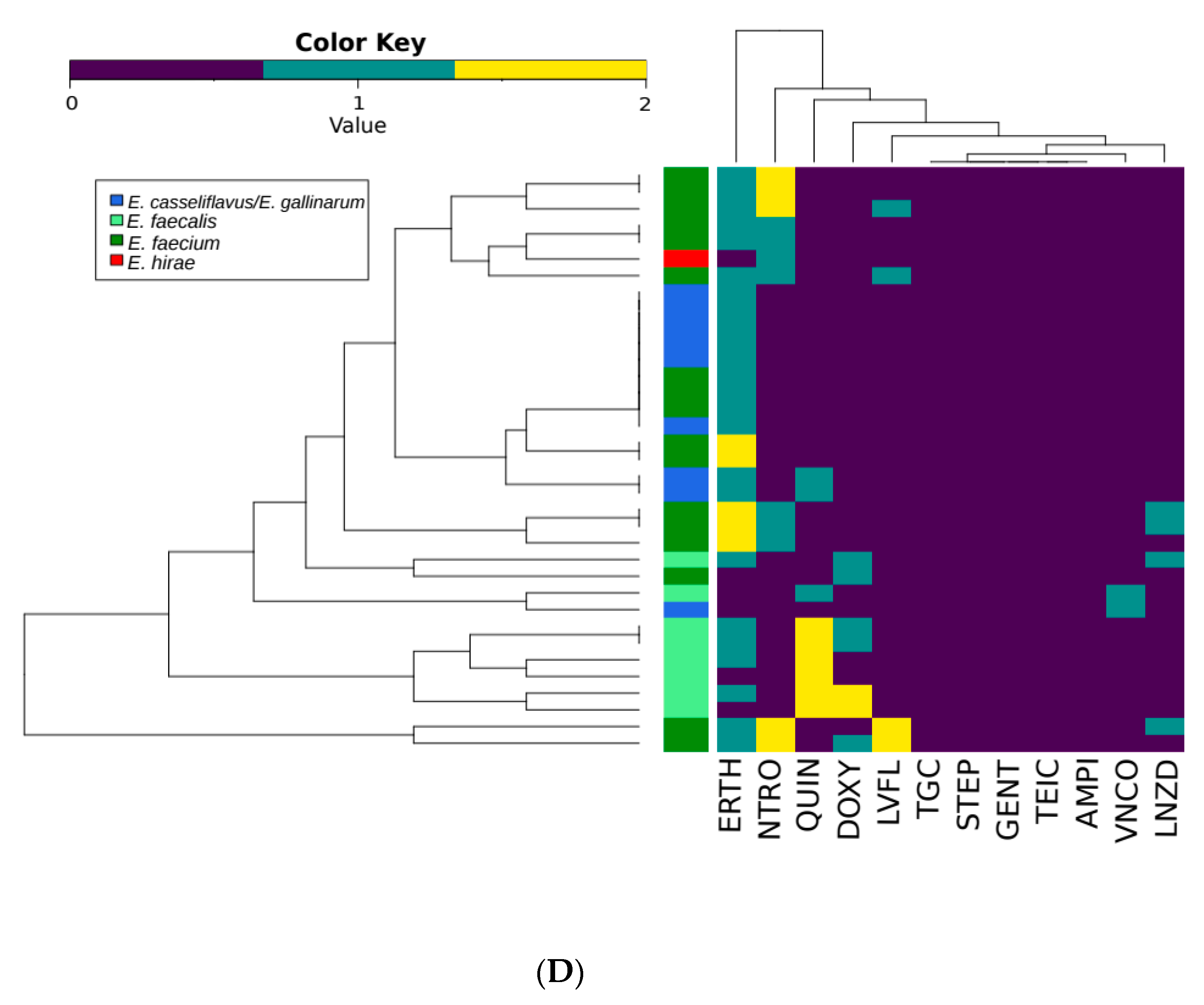
3.3. Antimicrobial Susceptibility Testing
3.4. Principal Component Analysis of Chemical and Environmental Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Canadian Antimicrobial Resistance Surveillance System, 2017. Annual Report. Available online: https://www.canada.ca/en/public-health/services/publications/drugs-health-products/canadian-antimicrobial-resistance-surveillance-system-2017-report-executive-summary.html (accessed on 26 November 2019).
- Kim, S.; Aga, D.S. Potential ecological and human health impacts of antibiotics and antibiotic-resistant bacteria from wastewater treatment plants. J. Toxic. Environ. Health Part B 2007, 10, 559–573. [Google Scholar] [CrossRef] [PubMed]
- Kümmerer, K. Antibiotics in the aquatic environment—A review–part I and part II. Chemosphere 2009, 75, 417–434. [Google Scholar] [CrossRef] [PubMed]
- Marcinek, H.; Wirth, R.; Muscholl-Silberhorn, A.; Gauer, M. Enterococcus faecalis gene transfer under natural conditions in municipal sewage water treatment plants. Appl. Environ. Microbiol. 1998, 64, 626–632. [Google Scholar] [PubMed]
- Hayes, J.R.; English, L.L.; Carter, P.J.; Proescholdt, T.; Lee, K.Y.; Wagner, D.D.; White, D.G. Prevalence and antimicrobial resistance of Enterococcus species isolated from retail meats. Appl. Environ. Microbiol. 2003, 69, 7153–7160. [Google Scholar] [CrossRef] [PubMed]
- Witte, W. Ecological impact of antibiotic use in animals on different complex microflora: Environment. Int. J. Agents 2000, 14, 321–325. [Google Scholar] [CrossRef]
- Da Silva, M.F.; Tiago, I.; Veríssimo, A.; Boaventura, R.A.; Nunes, O.C.; Manaia, C.M. Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2005, 55, 322–329. [Google Scholar] [CrossRef]
- Hoyle, D.V.; Knight, H.I.; Shaw, D.J.; Hillman, K.; Pearce, M.C.; Low, J.C.; Gunn, G.J.; Woolhouse, M.E. Acquisition and epidemiology of antibiotic-resistant Escherichia coli in a cohort of newborn calves. J. Antimicrob. Chemother. 2004, 53, 867–871. [Google Scholar] [CrossRef]
- Castiglioni, S.; Thomas, K.V.; Kasprzyk-Hordern, B.; Vandam, L.; Griffiths, P. Testing wastewater to detect illicit drugs: State of the art, potential and research needs. Sci. Total Environ. 2014, 487, 613–620. [Google Scholar] [CrossRef]
- Iversen, A.; Kühn, I.; Franklin, A.; Möllby, R. High prevalence of vancomycin-resistant enterococci in Swedish sewage. Appl. Environ. Microbiol. 2002, 68, 2838–2842. [Google Scholar] [CrossRef]
- Klare, I.; Konstabel, C.; Badstübner, D.; Werner, G.; Witte, W. Occurrence and spread of antibiotic resistances in Enterococcus faecium. Int. J. Food Microbiol. 2003, 88, 269–290. [Google Scholar] [CrossRef]
- Boneca, I.G.; Chiosis, G. Vancomycin resistance: Occurrence, mechanisms and strategies to combat it. Expert Opin. Ther. Targets 2003, 7, 311–328. [Google Scholar] [CrossRef] [PubMed]
- Eliopoulos, G.M. Vancomycin-resistant enterococci: Mechanism and clinical relevance. Infect. Dis. Clin. N. Am. 1997, 11, 851–865. [Google Scholar] [CrossRef]
- Klein, G. Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. Int. J. Food Microbiol. 2003, 88, 123–131. [Google Scholar] [CrossRef]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Kingston General Hospital. Patient Safety Indicator Reports. 2017. Available online: http://www.kgh.on.ca/about-kgh/achieving-our-aim/quality-and-patient-safety/patient-safety-indicator-reports (accessed on 26 November 2019).
- Canadian Antimicrobial Resistance Surveillance System, 2016. Annual Report. Available online: https://www.canada.ca/en/public-health/services/publications/drugs-health-products/canadian-antimicrobial-resistance-surveillance-system-report-2016.html (accessed on 26 November 2019).
- Burm, R.J.; Vaughan, R.D. Bacteriological comparison between combined and separate sewer discharges in southeastern Michigan. J. Water Pollut. Control Fed. 1966, 38, 400–409. [Google Scholar] [PubMed]
- Environmental Protection Agency (EPA). Method 1611: Enterococci in Water by TaqMan® Quantitative Polymerase Chain Reaction (qPCR) Assay. 2012. Available online: https://www.epa.gov/sites/production/files/2015-08/.../method_1611_2012.pdf (accessed on 26 November 2019).
- Harwood, V.J.; Brownell, M.; Perusek, W.; Whitlock, J.E. Vancomycin-resistant Enterococcus spp. isolated from wastewater and chicken feces in the United States. Appl. Environ. Microbiol. 2001, 67, 4930–4933. [Google Scholar] [CrossRef] [PubMed]
- Gaze, W.H.; Zhang, L.; Abdouslam, N.A.; Hawkey, P.M.; Calvo-Bado, L.; Royle, J.; Brown, H.; Davis, S.; Kay, P.; Boxall, A.B.A.; et al. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J. 2011, 5, 1253. [Google Scholar] [CrossRef]
- Zaheer, R.; Yanke, L.J.; Church, D.; Topp, E.; Read, R.R.; McAllister, T.A. High-throughput species identification of enterococci using pyrosequencing. J. Microbiol. Methods 2012, 89, 174–178. [Google Scholar] [CrossRef]
- Sanderson, H.; Ortega-Polo, R.; McDermott, K.; Zaheer, R.; Brown, R.S.; Majury, A.; TAMcAllister; Liss, S.N. Comparison of biochemical and genotypic speciation methods for vancomycin-resistant enterococci isolated from urban wastewater treatment plants. J. Microbiol. Methods 2019, 161, 102–110. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). M100 Performance Standards for Antimicrobial Susceptibility Testing, 28th Edition. 2018. Available online: https://clsi.org/standards/products/microbiology/documents/m100/ (accessed on 26 November 2019).
- European Union Committee on Antimicrobial Susceptibility Testing. The European Committee on Antimicrobial Susceptibility Testing—EUCAST. 2017. Available online: http://www.eucast.org/ (accessed on 26 November 2019).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Wien, Austria, 2017. [Google Scholar]
- Hammer, Ř.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Da Costa, P.M.; Vaz-Pires, P.; Bernardo, F. Antimicrobial resistance in Enterococcus spp. isolated in inflow, effluent and sludge from municipal sewage water treatment plants. Water Res. 2006, 40, 1735–1740. [Google Scholar] [CrossRef] [PubMed]
- Martins da Costa, P.M.; Vaz-Pires, P.M.; Bernardo, F.M. Antibiotic resistance of Enterococcus spp. isolated from wastewater and sludge of poultry slaughterhouses. J. Environ. Sci. Health Part B 2006, 41, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Hamming, R.W. Error detecting and error correcting codes. Bell Syst. Tech. J. 1950, 29, 147–160. [Google Scholar] [CrossRef]
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. E1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071); TU Wien: Wien, Austria, 2017. [Google Scholar]
- Galili, T.; O’Callaghan, A.; Sidi, J.; Sievert, C. Heatmaply: An R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 2017, 1, 3. [Google Scholar] [CrossRef]
- Blanch, A.R.; Caplin, J.L.; Iversen, A.; Kühn, I.; Manero, A.; Taylor, H.D.; Vilanova, X. Comparison of enterococcal populations related to urban and hospital wastewater in various climatic and geographic European regions. J. Appl. Microbiol. 2003, 94, 994–1002. [Google Scholar] [CrossRef]
- Sinton, L.W.; Donnison, A.M. Characterisation of faecal streptococci from some New Zealand effluents and receiving waters. N. Z. J. Mar. Freshw. Res. 1994, 28, 145–158. [Google Scholar] [CrossRef]
- Łuczkiewicz, A.; Jankowska, K.; Fudala-Książek, S.; Olańczuk-Neyman, K. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 2010, 44, 5089–5097. [Google Scholar] [CrossRef]
- Varela, A.R.; Ferro, G.; Vredenburg, J.; Yanık, M.; Vieira, L.; Rizzo, L.; Manaia, C.M. Vancomycin resistant enterococci: From the hospital effluent to the urban wastewater treatment plant. Sci. Total Environ. 2013, 450, 155–161. [Google Scholar] [CrossRef]
- Manafi, M. New developments in chromogenic and fluorogenic culture media. Int. J. Food Microbiol. 2000, 60, 205–218. [Google Scholar] [CrossRef]
- Perry, J.D.; Morris, K.A.; James, A.L.; Oliver, M.; Gould, F.K. Evaluation of novel chromogenic substrates for the detection of bacterial β-glucosidase. J. Appl. Microbiol. 2007, 102, 410–415. [Google Scholar] [CrossRef]
- Sadar, H.S.; Biedenbach, D.; Jones, R.N. Evalution of Vitek and API 20S for species identification of enterococci. Diagn. Microbiol. Infect. Dis. 1995, 22, 315–319. [Google Scholar] [CrossRef]
- Winston, L.G.; Pang, S.; Haller, B.L.; Wong, M.; Chambers, H.F.; Perdreau-Remington, F. API 20 strep identification system may incorrectly speciate enterococci with low level resistance to vancomycin. Diagn. Microbiol. Infect. Dis. 2004, 48, 287–288. [Google Scholar] [CrossRef] [PubMed]
- Naser, S.M.; Thompson, F.L.; Hoste, B.; Gevers, D.; Dawyndt, P.; Vancanneyt, M.; Swings, J. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 2005, 151, 2141–2150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naser, S.M.; Dawyndt, P.; Hoste, B.; Gevers, D.; Vandemeulebroecke, K.; Cleenwerck, I.; Vancanneyt, M.; Swings, J. Identification of lactobacilli by pheS and rpoA gene sequence analyses. Int. J. Syst. Evol. Microbiol. 2007, 57, 2777–2789. [Google Scholar] [CrossRef] [Green Version]
- Glaeser, S.P.; Kämpfer, P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015, 38, 237–245. [Google Scholar] [CrossRef]
- Manero, A.; Vilanova, X.; Cerdà-Cuéllar, M.; Blanch, A.R. Characterization of sewage waters by biochemical fingerprinting of Enterococci. Water Res. 2002, 36, 2831–2835. [Google Scholar] [CrossRef]
- Leclercq, R.; Oberlé, K.; Galopin, S.; Cattoir, V.; Budzinski, H.; Petit, F. Changes in enterococcal populations and related antibiotic resistance along a medical center-wastewater treatment plant-river continuum. Appl. Environ. Microbiol. 2013, 79, 2428–2434. [Google Scholar] [CrossRef] [Green Version]
- Araújo, C.; Torres, C.; Silva, N.; Carneiro, C.; Gonçalves, A.; Radhouani, H.; Ruiz-Larrea, F. Vancomycin-resistant enterococci from Portuguese wastewater treatment plants. J. Basic Microbiol. 2010, 50, 605–609. [Google Scholar] [CrossRef]
- Goldstein, R.R.E.; Micallef, S.A.; Gibbs, S.G.; George, A.; Claye, E.; Sapkota, A.; Sapkota, A.R. Detection of vancomycin-resistant enterococci (VRE) at four US wastewater treatment plants that provide effluent for reuse. Sci. Total Environ. 2014, 466, 404–411. [Google Scholar] [CrossRef]
- Nagulapally, S.R.; Ahmad, A.; Henry, A.; Marchin, G.L.; Zurek, L.; Bhandari, A. Occurrence of ciprofloxacin-, trimethoprim-sulfamethoxazole-, and vancomycin-resistant bacteria in a municipal wastewater treatment plant. Water Environ. Res. 2009, 81, 82–90. [Google Scholar] [CrossRef]
- Schwartz, T.; Kohnen, W.; Jansen, B.; Obst, U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 2003, 43, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Lanthier, M.; Scott, A.; Lapen, D.R.; Zhang, Y.; Topp, E. Frequency of virulence genes and antibiotic resistances in Enterococcus spp. isolates from wastewater and feces of domesticated mammals and birds, and wildlife. Can. J. Microbiol. 2010, 56, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Lanthier, M.; Scott, A.; Zhang, Y.; Cloutier, M.; Durie, D.; Henderson, V.C.; Topp, E. Distribution of selected virulence genes and antibiotic resistance in Enterococcus species isolated from the South Nation River drainage basin, Ontario, Canada. J. Appl. Microbiol. 2011, 110, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Shanthivunguturi, P.S.R.; Kumar, Y.A. Estimation of Carbonate-Bicarbonate Alkalinity in Water by Volumetric and Electro-Analytical Methods: A Comparative Study. Int. J. Curr. Res. Chem. Pharma. Sci. 2014, 1, 151–157. [Google Scholar]
- Lechevallier, M.W.; Cawthon, C.D.; Lee, R.G. Inactivation of biofilm bacteria. Appl. Environ. Microbiol. 1988, 54, 2492–2499. [Google Scholar]
- Kim, S.; Jensen, J.N.; Aga, D.S.; Weber, A.S. Fate of tetracycline resistant bacteria as a function of activated sludge process organic loading and growth rate. Water Sci. Technol. 2007, 55, 291–297. [Google Scholar] [CrossRef]
- Conley, D.J.; Paerl, H.W.; Howarth, R.W.; Boesch, D.F.; Seitzinger, S.P.; Havens, K.E.; Lancelot, C.; Likens, G.E. Controlling eutrophication: Nitrogen and phosphorus. Science 2009, 323, 1014–1015. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhuang, Y.; Geng, J.; Ren, H.; Zhang, Y.; Ding, L.; Xu, K. Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection. Sci. Total Environ. 2015, 512, 125–132. [Google Scholar] [CrossRef]
- Okabe, S.; Aoi, Y.; Satoh, H.; Suwa, Y. Nitrification in wastewater treatment. In Nitrification; American Society of Microbiology: Washington, DC, USA, 2011; pp. 405–433. [Google Scholar]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Al-Saydeh, S.A.; El-Naas, M.H.; Zaidi, S.J. Copper removal from industrial wastewater: A comprehensive review. J. Ind. Eng. Chem. 2017, 56, 35–44. [Google Scholar] [CrossRef]
- Hasman, H.; Aarestrup, F.M. TcrB, a gene conferring transferable copper resistance in Enterococcus faecium: Occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Antimicrob. Agents Chemother. 2002, 46, 1410–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renner, P.; Peters, J. Resistance of enterococci to heat and chemical agents. Zentralblatt Hyg. Umweltmed. 1999, 202, 41–50. [Google Scholar] [CrossRef]
- Mohamed, J.A.; Huang, D.B. Biofilm formation by enterococci. J. Med. Microbiol. 2007, 56, 1581–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrovich, M.; Chu, B.; Wright, D.; Griffin, J.; Elfeki, M.; Murphy, B.T.; Wells, G. Antibiotic resistance genes show enhanced mobilization through suspended growth and biofilm-based wastewater treatment processes. FEMS Microbiol. Ecol. 2018, 94, fiy041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalhoff, A. Global fluoroquinolone resistance epidemiology and implictions for clinical use. Interdiscip. Perspect. Infect. Dis. 2012. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, H.U.K.; Hammerum, A.M.; Ekelund, K.; Bang, D.; Pallesen, L.V.; Frimodt-Møller, N. Tetracycline and macrolide co-resistance in Streptococcus pyogenes: Co-selection as a reason for increase in macrolide-resistant S. pyogenes? Microb. Drug Resist. 2004, 10, 231–238. [Google Scholar] [CrossRef]
- Livermore, D.M.; Winstanley, T.G.; Shannon, K.P. Interpretative reading: Recognizing the unusual and inferring resistance mechanisms from resistance phenotypes. J. Antimicrob. Chemother. 2001, 48, 87–102. [Google Scholar] [CrossRef] [Green Version]
- Hammerum, A.M. Enterococci of animal origin and their significance for public health. Clin. Microbiol. Infect. 2012, 18, 619–625. [Google Scholar] [CrossRef]
- Li, D.; Yang, M.; Hu, J.; Zhang, Y.; Chang, H.; Jin, F. Determination of penicillin G and its degradation products in a penicillin production wastewater treatment plant and the receiving river. Water Res. 2008, 42, 307–317. [Google Scholar] [CrossRef]
- Tran, N.H.; Chen, H.; Reinhard, M.; Mao, F.; Gin, K.Y.H. Occurrence and removal of multiple classes of antibiotics and antimicrobial agents in biological wastewater treatment processes. Water Res. 2016, 104, 461–472. [Google Scholar] [CrossRef]
- McClellan, K.; Halden, R.U. Pharmaceuticals and personal care products in archived US biosolids from the 2001 EPA national sewage sludge survey. Water Res. 2010, 44, 658–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meena, S.; Mohapatra, S.; Sood, S.; Dhawan, B.; Bas, B.K.; Kapil, A. Revisiting Nitrofurantoin for Vancomycin Resistant Enterococci. J. Clin. Diagn. Res. 2017, 11, DC19. [Google Scholar] [CrossRef] [PubMed]
- Abia, A.L.K.; Ubomba-Jaswa, E.; Momba, M.N.B. High prevalence of multiple-antibiotic-resistant (MAR) Escherichia coli in river bed sediments of the Apies River, South Africa. Environ. Monit. Assess. 2015, 187, 652. [Google Scholar] [CrossRef] [PubMed]
- Osei Sekyere, J. Genomic insights into nitrofurantoin resistance mechanisms and epidemiology in clinical Enterobacteriaceae. Future Sci. OA 2018, 4, FSO293. [Google Scholar] [CrossRef] [Green Version]

| Biological Aerated Filter | Conventional Activated Sludge | SEM | p Value * | ||||
|---|---|---|---|---|---|---|---|
| Primary Effluent | Final Effluent | Primary Effluent | Final Effluent | WWTP | Effluent | ||
| Absolute Quantities (CFU/100mL) | |||||||
| Total Enterococcus ×103 | 72.054 | 109.764 | 0.0718 | 1.381 | 8.638 | 0.233 | <0.001 |
| Low Level VRE ×103 | 17.830 | 28.428 | 0.0169 | 0.356 | 1.820 | 0.113 | <0.001 |
| High Level VRE ×102 | 26.482 | 57.685 | 0.0457 | 0.226 | 5.264 | 0.097 | <0.001 |
| Relative Quantities (%) | |||||||
| Low Level VRE | 35.779 | 31.856 | 31.697 | 28.117 | 1.622 | 0.255 | 0.235 |
| High Level VRE | 9.869 | 11.637 | 8.535 | 4.113 | 1.266 | 0.673 | 0.124 |
| Removal Efficiency (%) | |||||||
| Total Enterococcus | 99.837 | 98.210 | 0.218 | <0.001 | |||
| Low Level VRE | 99.850 | 97.777 | 0.387 | <0.001 | |||
| High Level VRE | 99.588 | 96.817 | 0.736 | 0.059 | |||
| Primary Effluent | Final Effluent | SEM | p Value * | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BAF | CAS | BAF | CAS | WWTP | Effluent | Primary Effluent | Final Effluent | ||
| Absolute Quantities (copies/100mL) | |||||||||
| Total Bacteria (16S rRNA) ×104 | 3.628 | 24.727 | 1.189 | 2.250 | 5.451 | 0.315 | 0.258 | 0.524 | 1.000 |
| Total Enterococcus (23S rRNA) ×103 | 0.0163 | 13.836 | 0.032 | 0.066 | 3.348 | 0.312 | 0.305 | 0.478 | 1.000 |
| Vancomycin resistance gene (vanA) | 32.673 | 16.195 | 8.243 | 3.002 | 3.280 | 0.099 | 0.003 | 0.203 | 0.919 |
| Class I integrons (IntI-1) ×104 | 6.019 | 132.635 | 2.194 | 3.559 | 22.174 | 0.153 | 0.134 | 0.168 | 1.000 |
| Relative Quantities (%) | |||||||||
| Total Enterococcus (23S rRNA) ×10−2 | 0.430 | 2.297 | 0.966 | 1.297 | 0.253 | 0.038 | 0.570 | 0.043 | 0.988 |
| Vancomycin Resistance Gene (vanA) ×10−4 | 0.109 | 0.041 | 0.189 | 0.036 | 0.030 | 0.070 | 0.545 | 0.853 | 0.290 |
| Class I integrons (IntI-1) ×102 | 2.002 | 20.547 | 21.553 | 5.432 | 5.716 | 0.917 | 0.849 | 0.667 | 0.756 |
| Sample Types | Species | % Isolates | Samples Types | Species | % Isolates |
|---|---|---|---|---|---|
| BAF Primary Effluent (n = 101) | E. faecium (n = 30) | 29.7 | CAS Primary Effluent (n = 110) | E. faecium (n = 28) | 25.5 |
| E. faecalis (n = 51) | 50.4 | E. faecalis (n = 58) | 52.7 | ||
| E. casseliflavus/E. gallinarum (n = 18) | 17.8 | E. casseliflavus/E. gallinarum (n = 24) | 21.8 | ||
| Other (n = 2) | 2 | Other (n = 0) | 0 | ||
| BAF Final Effluent (n = 51) | E. faecium (n = 19) | 37.3 | CAS Final Effluent (n = 35) | E. faecium (n = 17) | 48.6 |
| E. faecalis (n = 26) | 51 | E. faecalis (n = 8) | 22.9 | ||
| E. casseliflavus/E. gallinarum (n = 5) | 9.8 | E. casseliflavus/E. gallinarum (n = 9) | 25.7 | ||
| Other (n = 1) | 2 | Other (n = 1) | 2.9 | ||
| BAF Biomass (n = 1) | E. faecium (n = 0) | 0 | CAS Biomass (n = 10) | E. faecium (n = 0) | 0 |
| E. faecalis (n = 0) | 0 | E. faecalis (n = 8) | 80 | ||
| E. casseliflavus/E. gallinarum (n = 0) | 0 | E. casseliflavus/E. gallinarum (n = 2) | 20 | ||
| Other (n = 1) | 100 | Other (n = 0) | 0 |
| VNCO | TEIC | AMPI | DOXY | ERTH | NTRO | GENT | LNZD | LVFL | QUIN | STEP | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. faecalis | 151 | 0 15 | 30 | 0 | 56 34 | 42 95 | 0 0 | 9 3 | 5 20 | 10 3 | 137 7 | 137 7 |
| E. faecium | 94 | 20 1 | 22 | 26 | 6 3 | 44 41 | 12 23 | 1 0 | 0 3 | 30 7 | 1 5 | 1 5 |
| E. casseliflavus/E. gallinarum | 58 | 0 4 | 1 | 1 | 0 1 | 4 32 | 0 0 | 0 0 | 0 1 | 1 2 | 1 9 | 1 9 |
| Other Enterococcus spp. | 5 | 0 0 | 0 | 0 | 0 1 | 0 2 | 0 2 | 0 0 | 0 0 | 0 0 | 0 0 | 0 0 |
| Total | 308 | 20 20 | 53 | 27 | 62 58 | 90 168 | 12 25 | 10 3 | 5 24 | 41 12 | 139 21 | 25 17 |
| bP values | ||||||||||||
| E. faecalis vs E. faecium | <0.0001 | 0.6187 | <0.0001 | <0.0001 | 0.0073 | <0.0001 | 0.062 | 0.0052 | <0.0001 | <0.0001 | 0.0134 | |
| All Species | <0.0001 | 0.016 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.4059 | 0.0497 | <0.0001 | <0.0001 | 0.1227 | |
| Location | Total | VNCO broth | ABX | VNCO disk | TEIC | AMPI | DOXY | ERTH | NTRO | GENT | LNZD | LVFL | QUIN | STEP | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # | # | R or I | # | # | # | # | # | # | # | # | # | # | # | ||
| Biological Aerated Filter (n = 484) | PE | 270 | 101 | R | 12 | 21 | 15 | 14 | 33 | 2 | 2 | 1 | 21 | 48 | 11 |
| I | 5 | 16 | 51 | 7 | 2 | 7 | 2 | 5 | 11 | ||||||
| FE | 214 | 51 | R | 1 | 8 | 3 | 14 | 14 | 2 | 4 | 2 | 3 | 23 | 5 | |
| I | 4 | 6 | 29 | 4 | 1 | 5 | 4 | 5 | 1 | ||||||
| Conventional Activated Sludge (n = 611) | PE | 375 | 110 | R | 7 | 21 | 9 | 26 | 35 | 3 | 4 | 2 | 15 | 54 | 9 |
| I | 9 | 11 | 59 | 7 | 0 | 5 | 4 | 6 | 5 | ||||||
| FE | 236 | 35 | R | 0 | 0 | 0 | 2 | 5 | 5 | 0 | 0 | 2 | 6 | 0 | |
| I | 2 | 5 | 24 | 7 | 0 | 4 | 2 | 3 | 0 | ||||||
| Total | 1095 | 297 | Total R | 20 | 50 | 27 | 56 | 87 | 12 | 10 | 5 | 41 | 131 | 25 | |
| Total I | 20 | 38 | 163 | 25 | 3 | 21 | 12 | 19 | 17 | ||||||
| p values | |||||||||||||||
| ABX | VNCO | TEIC | AMPI | DOXY | ERTH | NTRO | GENT | LNZD | LVFL | QUIN | STEP | ||||
| (Disk) | |||||||||||||||
| All Four Locations | 0.1467 | 0.034 | 0.04 | 0.1008 | 0.5162 | 0.004 | 0.2447 | 0.5282 | 0.0793 | 0.0382 | 0.0343 | ||||
| WWTP | 0.3924 | 0.3665 | 0.1371 | 0.6744 | 0.7242 | 0.3193 | 0.1968 | 0.7799 | 0.5973 | 0.612 | 0.0875 | ||||
| Effluent | 0.0501 | 0.0409 | 0.0547 | 0.9974 | 0.2058 | 0.0124 | 0.7236 | 0.2886 | 0.0139 | 0.0516 | 0.0474 | ||||
| BAF PE vs FE | 0.1016 | 0.5907 | 0.177 | 0.1198 | 0.7456 | 0.7577 | 0.215 | 0.376 | 0.0177 | 0.5224 | 0.1438 | ||||
| CAS PE vs FE | 0.2623 | 0.0118 | 0.1786 | 0.0618 | 0.128 | 0.0012 | 0.5812 | 0.2545 | 0.4053 | 0.0037 | 0.085 | ||||
| BAF FE vs CAS FE | 0.6509 | 0.0373 | 0.3885 | 0.0389 | 0.347 | 0.0394 | 0.1618 | 0.4874 | 0.9285 | 0.0188 | 0.1093 | ||||
| Chemical and Environmental Parameters | Primary Effluent | Final Effluent | ||||
|---|---|---|---|---|---|---|
| Parameter | Unit | Detection Limit | BAF | CAS | BAF | CAS |
| Max Temperature | °C | N/A | 22.2 ± 5.69 | |||
| Min Temperature | °C | N/A | 9.30 ± 6.70 | |||
| Accumulative Precipitation | mm | N/A | 3.77 ± 7.70 | |||
| Alkalinity (CaCO3, pH 4.5) | mg/L | 3 | 172 ± 16.1 | 253 ± 17.5 | 73.6 ± 16.9 | 143 ± 33.5 |
| pH (25 °C) | pH units | N/A | 7.62 ± 0.162 | 7.45 ± 9.24 × 10−2 | 7.40 ± 0.111 | 7.61 ± 0.138 |
| CBOD5 | mg/L | 2 | 57.8 ± 35.7 | 138 ± 39.2 | <2 | 3.26 ± 2.28 |
| TSS ** | mg/L | 3 | 81.9 ± 53.9 | 165 ± 56.5 | 33.2 ± 21.7 | 67.3 ± 68.4 |
| Total P | mg/L | 0.01 | 2.37 ± 0.99 | 4.09 ± 0.99 | 0.460 ± 0.15 | 0.677 ± 0.31 |
| Total Ammonia | mg/L | 0.01 | 13.0 ± 3.58 | 25.9 ± 6.47 | 0.820 ± 0.636 | 14.3 ± 5.87 |
| Ammonia (unionized) | mg/L | 0.01 | 0.150 ± 7.45 × 10−2 | 0.328 ± 0.129 | <0.01 | 0.320 ± 0.264 |
| TKN | mg/L | 0.1 | 17.9 ± 5.88 | 39.3 ± 8.61 | 2.27 ± 0.862 | 16.8 ± 7.89 |
| Nitrite | mg/L | 0.1 | <0.1 | <0.1 | 0.220 ± 0.237 | 1.88 ± 0.801 |
| Nitrate | mg/L | 0.1 | <0.1 | <0.1 | 15.9 ± 3.38 | 14.0 ± 4.85 |
| Aluminum *** | mg/L | 0.01 | 0.388 ± 0.292 | NM | 0.982 ± 7.03 × 10−2 | NM |
| Arsenic *** | mg/L | 5.00 × 10−4 | <5.00 × 10−4 | <5.00 × 10−4 | <5.00 × 10−4 | <5.00 × 10−4 |
| Cadmium *** | mg/L | 0.005 | <0.005 | <0.005 | <0.005 | <0.005 |
| Chromium *** | mg/L | 0.002 | 1.00 × 10−2 ± 9.00 × 10−3 | <0.002 | <0.002 | <0.002 |
| Cobalt *** | mg/L | 0.005 | <0.005 | 2.00 × 10−2 ± 3.40 × 10−2 | <0.005 | <0.005 |
| Copper *** | mg/L | 0.002 | 3.98 × 10−2 ± 2.01 × 10−2 | 3.13 × 10−2 ± 2.02 × 10−2 | 0.01 ± 9.00 × 10−3 | <0.002 |
| Lead *** | mg/L | 0.02 | <0.02 | <0.02 | <0.02 | <0.02 |
| Mercury *** | mg/L | 2.00 × 10−5 | <2.00 × 10−5 | <2.00 × 10−5 | <2.00 × 10−5 | <2.00 × 10−5 |
| Molybdenum *** | mg/L | 0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Nickel *** | mg/L | 0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Selenium *** | mg/L | 0.005 | <0.005 | <0.005 | <0.005 | <0.005 |
| Potassium *** | mg/L | 0.1 | 7.09 ± 0.202 | 20.8 ± 21.7 | 7.01 ± 0.501 | 9.30 ± 2.14 |
| Zinc *** | mg/L | 0.005 | 7.06 × 10−2 ± 3.17 × 10−2 | 0.107 ± 3.31 × 10−2 | 2.23 × 10−2 ± 7.32 × 10−3 | 2.89 × 10−2 ± 4.59 × 10−3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanderson, H.; Ortega-Polo, R.; McDermott, K.; Hall, G.; Zaheer, R.; Brown, R.S.; Majury, A.; McAllister, T.A.; Liss, S.N. Quantification and Multidrug Resistance Profiles of Vancomycin-Resistant Enterococci Isolated from Two Wastewater Treatment Plants in the Same Municipality. Microorganisms 2019, 7, 626. https://doi.org/10.3390/microorganisms7120626
Sanderson H, Ortega-Polo R, McDermott K, Hall G, Zaheer R, Brown RS, Majury A, McAllister TA, Liss SN. Quantification and Multidrug Resistance Profiles of Vancomycin-Resistant Enterococci Isolated from Two Wastewater Treatment Plants in the Same Municipality. Microorganisms. 2019; 7(12):626. https://doi.org/10.3390/microorganisms7120626
Chicago/Turabian StyleSanderson, Haley, Rodrigo Ortega-Polo, Kevin McDermott, Geoffrey Hall, Rahat Zaheer, R. Stephen Brown, Anna Majury, Tim A. McAllister, and Steven N. Liss. 2019. "Quantification and Multidrug Resistance Profiles of Vancomycin-Resistant Enterococci Isolated from Two Wastewater Treatment Plants in the Same Municipality" Microorganisms 7, no. 12: 626. https://doi.org/10.3390/microorganisms7120626





