Inactivation of Genes Encoding MutL and MutS Proteins Influences Adhesion and Biofilm Formation by Neisseria gonorrhoeae
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Cell Lines and Culture Conditions
2.3. Construction of Neisseria gonorrhoeae (N. gonorrhoeae) Mutants
2.4. Field Emission Scanning Electron Microscopy (FE SEM)
2.5. Scanning Confocal Laser Microscopy (SCLM)
2.6. The Crystal Violet Assay
2.7. RNA Extraction
2.8. Microarray Experiments
2.9. Array Analysis
2.10. RT-qPCR
2.11. Adhesion Assay
2.12. Invasion Assay
2.13. Computer Analysis, In Silico Analysis of Metabolic Pathways, and Determination of Clusters of Orthologous Groups (COG) Category
2.14. Chemicals and Kits
3. Results and Discussion
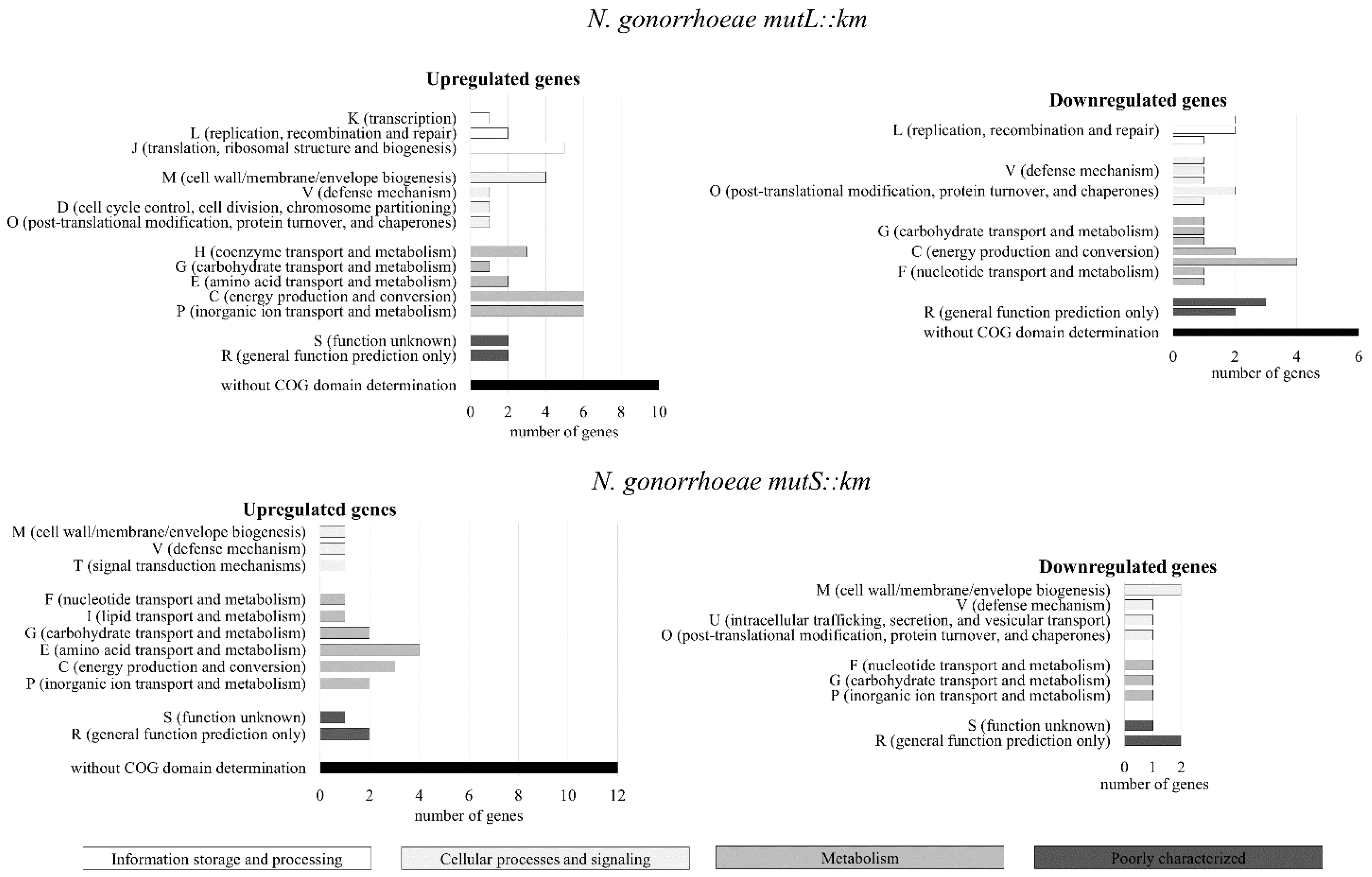
3.1. The Genes Encoding Proteins that can Influence Adhesion and Biofilm Formation Are Differentially Expressed in N. gonorrhoeae mutL::km and N. gonorrhoeae mutS::km Mutants Compared to the Wild-Type Strain
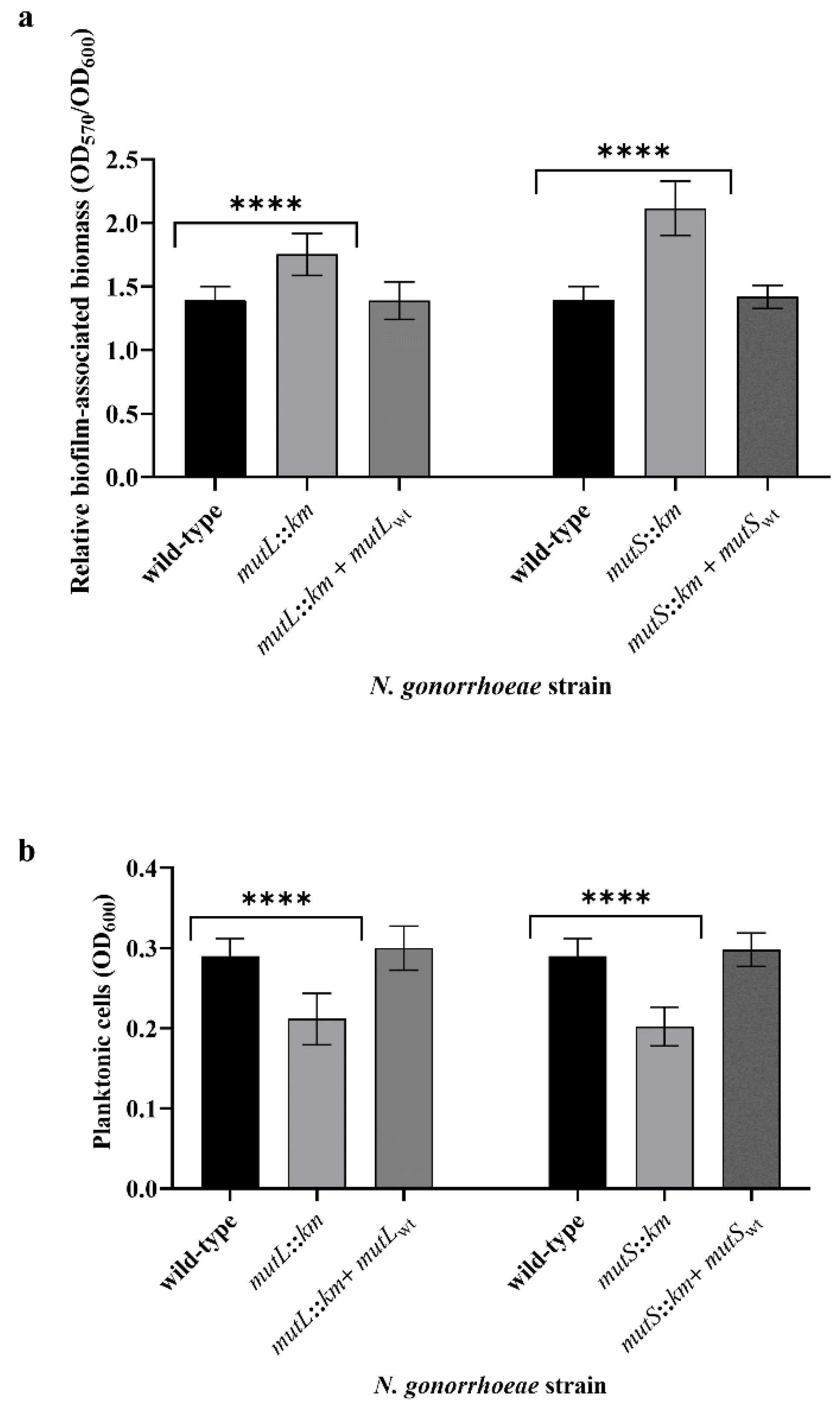
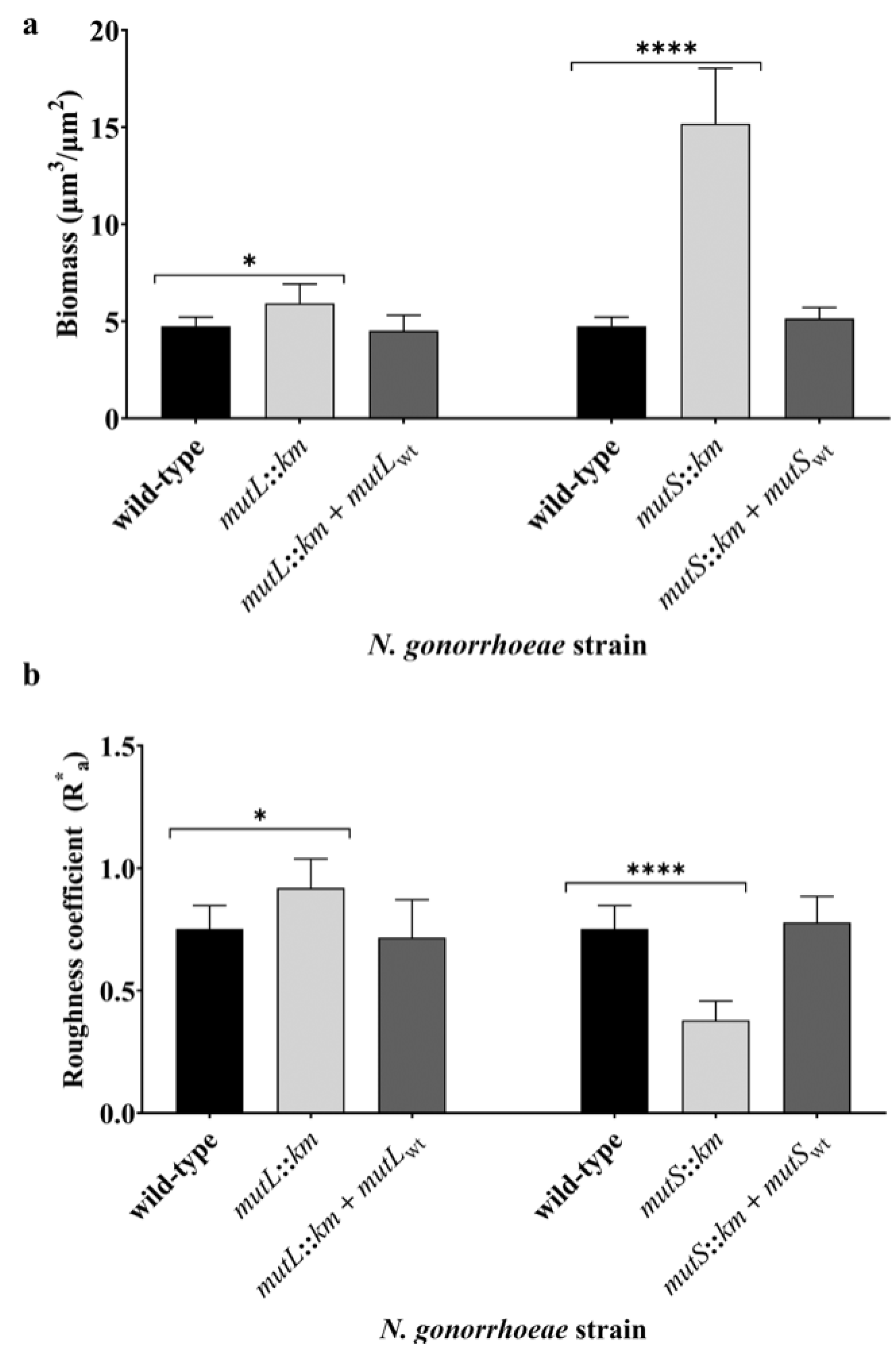
3.2. Inactivation of MutL or MutS Genes Changes the Formation and Structure of the Biofilm by N. gonorrhoeae on Abiotic Surfaces
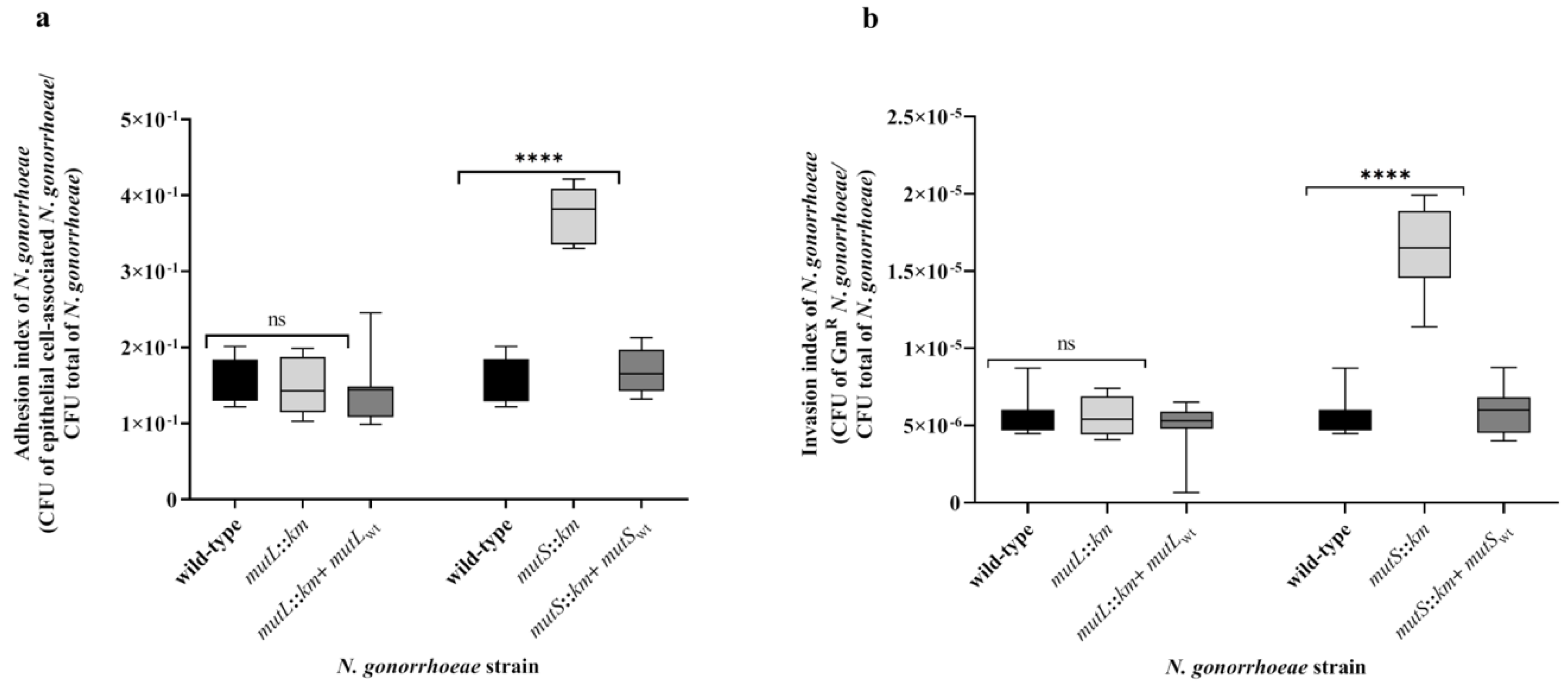
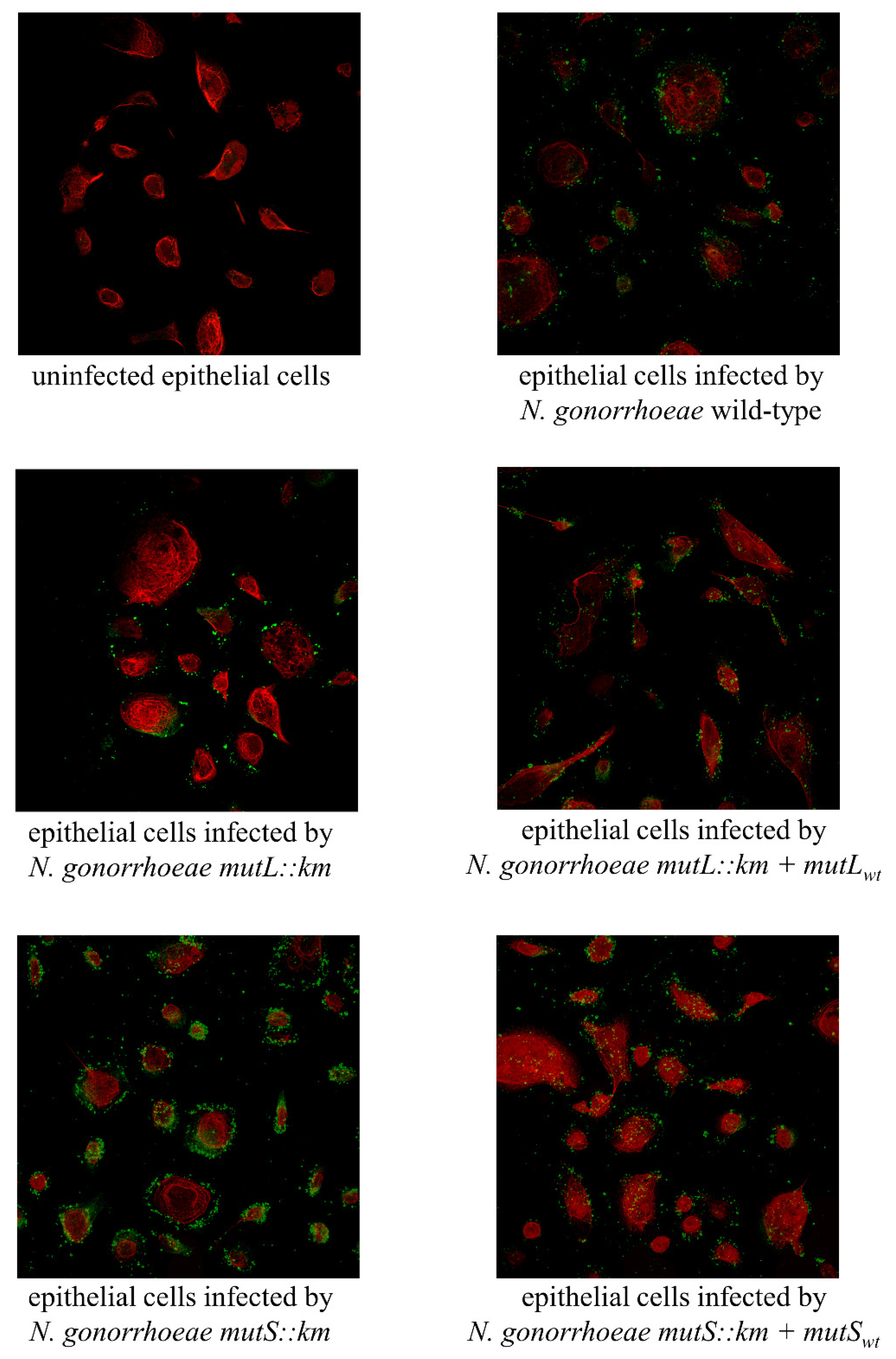
3.3. N. gonorrhoeae mutS::km but not N. gonorrhoeae mutL::km Is Characterized by Increased Adhesion and Invasiveness to Human Epithelial Cells Compared to the Wild-Type Strain
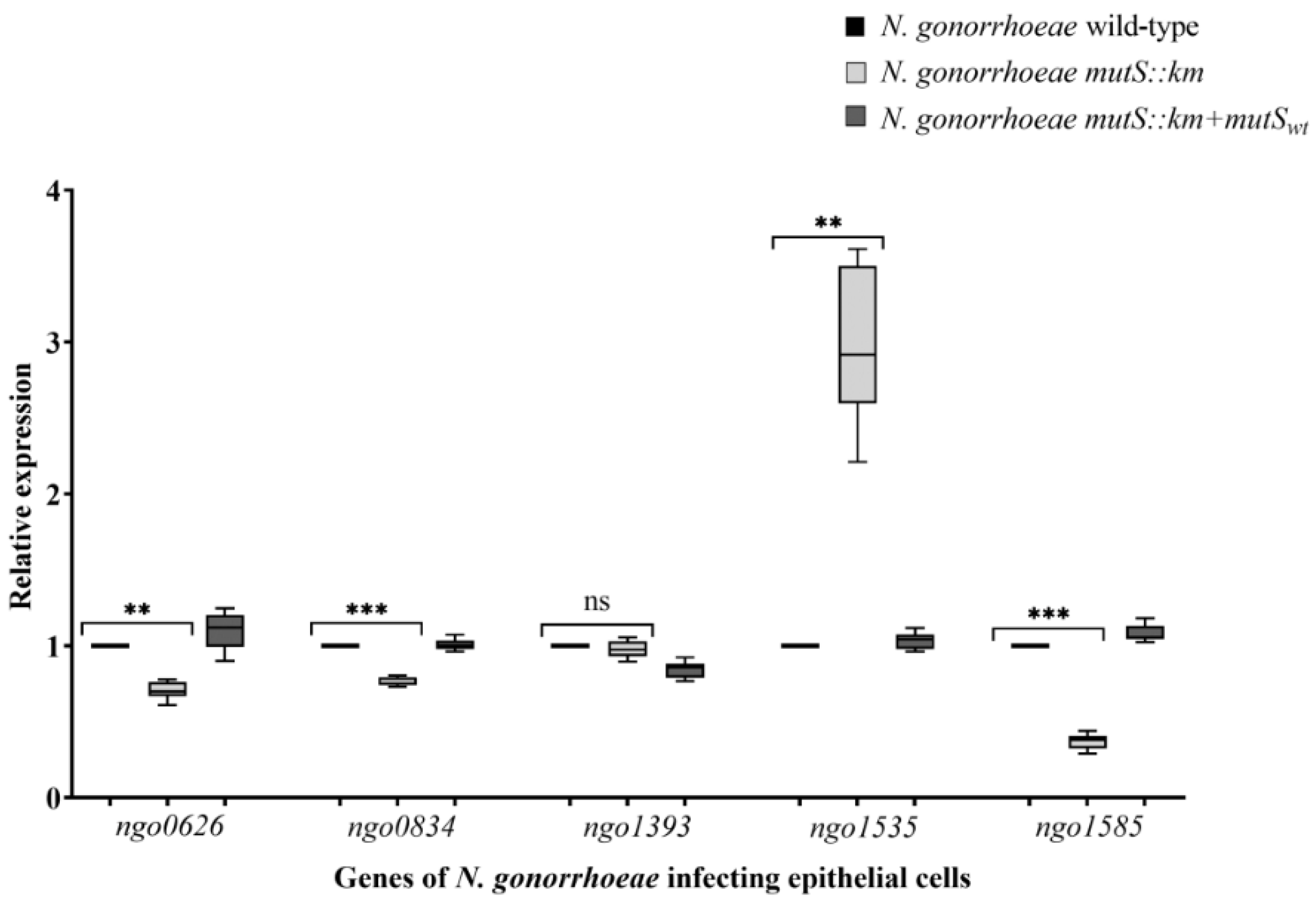
3.4. N. gonorrhoeae mutS::km Has an Altered Expression of Genes Encoding Proteins that can Influence Adhesion to Human Epithelial Cells
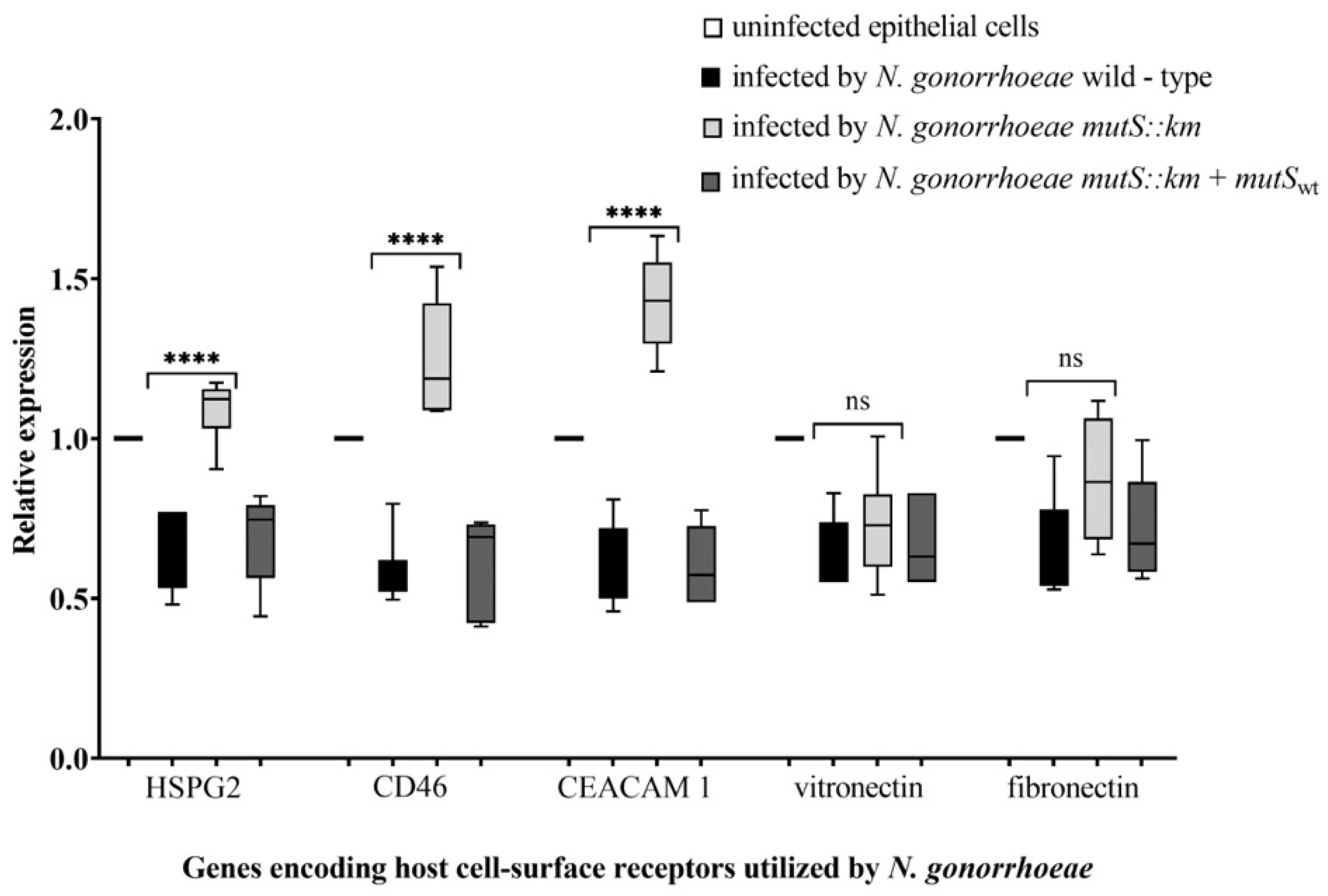
3.5. During Infection of Human Epithelial Cells with N. gonorrhoeae mutS::km, the Expression of Some Host Cell-Surface Receptors is Increased
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Unemo, M.; Fasth, O.; Fredlund, H.; Limnios, A.; Tapsall, J. Phenotypic and genetic characterization of the 2008 WHO Neisseria gonorrhoeae reference strain panel intended for global quality assurance and quality control of gonococcal antimicrobial resistance surveillance for public health purposes. J. Antimicrob. Chemother. 2009, 63, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017; pp. 1–7. [Google Scholar]
- Jarvis, G.A.; Chang, T.L. Modulation of HIV transmission by Neisseria gonorrhoeae: Molecular and immunological aspects. Curr. HIV Res. 2012, 10, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Rapista, A.; Teleshova, N.; Mosoyan, G.; Jarvis, G.A.; Klotman, M.E.; Chang, T.L. Neisseria gonorrhoeae enhances HIV-1 infection of primary resting CD4+ T cells through TLR2 activation. J. Immunol. 2010, 184, 2814–2824. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Boulton, I.C.; Pongoski, J.; Cochrane, A.; Gray-Owen, S.D. Induction of HIV-1 long terminal repeat-mediated transcription by Neisseria gonorrhoeae. AIDS 2003, 17, 625–628. [Google Scholar] [CrossRef] [PubMed]
- Greiner, L.L.; Edwards, J.L.; Shao, J.; Rabinak, C.; Entz, D.; Apicella, M.A. Biofilm Formation by Neisseria gonorrhoeae. Infect. Immun. 2005, 73, 1964–1970. [Google Scholar] [CrossRef] [PubMed]
- Steichen, C.T.; Shao, J.Q.; Ketterer, M.R.; Apicella, M.A. Gonococcal cervicitis: A role for biofilm in pathogenesis. J. Infect. Dis. 2008, 198, 1856–1861. [Google Scholar] [CrossRef]
- Reyes, G.X.; Schmidt, T.T.; Kolodner, R.D.; Hombauer, H. New insights into the mechanism of DNA mismatch repair. Chromosoma 2015, 124, 443–462. [Google Scholar] [CrossRef]
- Criss, A.K.; Bonney, K.M.; Chang, R.A.; Duffin, P.M.; LeCuyer, B.E.; Seifert, H.S. Mismatch correction modulates mutation frequency and pilus phase and antigenic variation in Neisseria gonorrhoeae. J. Bacteriol. 2010, 192, 316–325. [Google Scholar] [CrossRef]
- Adamczyk-Popławska, M.; Bandyra, K.; Kwiatek, A. Activity of Vsr endonucleases encoded by Neisseria gonorrhoeae FA1090 is influenced by MutL and MutS proteins. BMC Microbiol. 2018, 18, 95. [Google Scholar] [CrossRef]
- Rotman, E.; Seifert, H.S. Neisseria gonorrhoeae MutS affects pilin antigenic variation through mismatch correction and not by pilE guanine quartet binding. J. Bacteriol. 2015, 197, 1828–1838. [Google Scholar] [CrossRef][Green Version]
- Ambur, O.H.; Davidsen, T.; Frye, S.A.; Balasingham, S.V.; Lagesen, K.; Rognes, T.; Tønjum, T. Genome dynamics in major bacterial pathogens. FEMS Microbiol. Rev. 2009, 33, 453–470. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.A.; Sechman, E.V.; Skaar, E.P.; Seifert, H.S. Recombination, repair and replication in the pathogenic Neisseriae: The 3 R’s of molecular genetics of two human-specific bacterial pathogens. Mol. Microbiol. 2003, 50, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Eisen, J.A.; Hanawalt, P.C. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 1999, 435, 171–213. [Google Scholar] [CrossRef]
- Richardson, A.R.; Stojiljkovic, I. Mismatch repair and the regulation of phase variation in Neisseria meningitidis. Mol. Microbiol. 2001, 40, 645–655. [Google Scholar] [CrossRef]
- Richardson, A.R.; Yu, Z.; Popovic, T.; Stojiljkovic, I. Mutator clones of Neisseria meningitidis in epidemic serogroup a disease. Proc. Natl. Acad. Sci. USA 2002, 99, 6103–6107. [Google Scholar] [CrossRef]
- Bayliss, C.D.; Hoe, J.C.; Makepeace, K.; Martin, P.; Hood, D.W.; Moxon, E.R. Neisseria meningitidis escape from the bactericidal activity of a monoclonal antibody is mediated by phase variation of lgtG and enhanced by a mutator phenotype. Infect. Immun. 2008, 76, 5038–5048. [Google Scholar] [CrossRef]
- LeClerc, J.E.; Li, B.; Payne, W.L.; Cebula, T.A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 1996, 274, 1208–1211. [Google Scholar] [CrossRef]
- LeClerc, J.E.; Payne, W.L.; Kupchella, E.; Cebula, T.A. Detection of mutator subpopulations in Salmonella typhimurium LT2 by reversion of his alleles. Mutat. Res. 1998, 400, 89–97. [Google Scholar] [CrossRef]
- Oliver, A.; Baquero, F.; Blázquez, J. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: Molecular characterization of naturally occurring mutants. Mol. Microbiol. 2002, 43, 1641–1650. [Google Scholar] [CrossRef]
- Nachamkin, I.; Cannon, J.G.; Mittler, R.S. Monoclonal antibodies against Neisseria gonorrhoeae: Production of antibodies directed against a strain-specific cell surface antigen. Infect. Immun. 1981, 32, 641–648. [Google Scholar]
- Dillard, J.P. Genetic Manipulation of Neisseria gonorrhoeae. Curr. Protoc. Microbiol. 2011, 23, Unit4A.2.1–Unit4A.2.24. [Google Scholar] [CrossRef]
- Shaw, J.H.; Falkow, S. Model for invasion of human tissue culture cells by Neisseria gonorrhoeae. Infect. Immun. 1988, 56, 1625–1632. [Google Scholar] [PubMed]
- Ilver, D.; Källström, H.; Normark, S.; Jonsson, A.B. Transcellular passage of Neisseria gonorrhoeae involves pilus phase variation. Infect. Immun. 1998, 66, 469–473. [Google Scholar] [PubMed]
- Merz, A.J.; So, M. Attachment of piliated, Opa- and Opc- gonococci and meningococci to epithelial cells elicits cortical actin rearrangements and clustering of tyrosine-phosphorylated proteins. Infect. Immun. 1997, 65, 4341–4349. [Google Scholar]
- Naumann, M.; Wessler, S.; Bartsch, C.; Wieland, B.; Meyer, T.F. Neisseria gonorrhoeae epithelial cell interaction leads to the activation of the transcription factors nuclear factor kappaB and activator protein 1 and the induction of inflammatory cytokines. J. Exp. Med. 1997, 186, 247–258. [Google Scholar] [CrossRef]
- Kwiatek, A.; Bacal, P.; Wasiluk, A.; Trybunko, A.; Adamczyk-Poplawska, M. The dam replacing gene product enhances Neisseria gonorrhoeae FA1090 viability and biofilm formation. Front. Microbiol. 2014, 5, 712. [Google Scholar] [CrossRef]
- Kwiatek, A.; Mrozek, A.; Bacal, P.; Piekarowicz, A.; Adamczyk-Popławska, M. Type III Methyltransferase M.NgoAX from Neisseria gonorrhoeae FA1090 Regulates Biofilm Formation and Interactions with Human Cells. Front. Microbiol. 2015, 6, 1426. [Google Scholar] [CrossRef]
- Bhoopalan, S.V.; Piekarowicz, A.; Lenz, J.D.; Dillard, J.P.; Stein, D.C. nagZ Triggers Gonococcal Biofilm Disassembly. Sci. Rep. 2016, 6, 22372. [Google Scholar] [CrossRef]
- Stepanovic, S.; Vukovic, D.; Dakic, I.; Savic, B.; Svabic-Vlahovic, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Hopper, S.; Wilbur, J.S.; Vasquez, B.L.; Larson, J.; Clary, S.; Mehr, I.J.; Seifert, H.S.; So, M. Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infect. Immun. 2000, 68, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Natale, D.A.; Galperin, M.Y.; Tatusov, R.L.; Koonin, E.V. Using the COG database to improve gene recognition in complete genomes. Genetica 2000, 108, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef] [PubMed]
- Jamet, A.; Jousset, A.B.; Euphrasie, D.; Mukorako, P.; Boucharlat, A.; Ducousso, A.; Charbit, A.; Nassif, X. A new family of secreted toxins in pathogenic Neisseria species. PLoS Pathog. 2015, 11, e1004592. [Google Scholar] [CrossRef] [PubMed]
- Jamet, A.; Nassif, X. Characterization of the Maf family of polymorphic toxins in pathogenic. Microb. Cell 2015, 2, 88–90. [Google Scholar] [CrossRef] [PubMed]
- Arenas, J.; de Maat, V.; Catón, L.; Krekorian, M.; Herrero, J.C.; Ferrara, F.; Tommassen, J. Fratricide activity of MafB protein of N. meningitidis strain B16B6. BMC Microbiol. 2015, 15, 156. [Google Scholar] [CrossRef]
- Paruchuri, D.K.; Seifert, H.S.; Ajioka, R.S.; Karlsson, K.A.; So, M. Identification and characterization of a Neisseria gonorrhoeae gene encoding a glycolipid-binding adhesin. Proc. Natl. Acad. Sci. USA 1990, 87, 333–337. [Google Scholar] [CrossRef]
- Zielke, R.A.; Wierzbicki, I.H.; Weber, J.V.; Gafken, P.R.; Sikora, A.E. Quantitative proteomics of the Neisseria gonorrhoeae cell envelope and membrane vesicles for the discovery of potential therapeutic targets. Mol. Cell Proteom. 2014, 13, 1299–1317. [Google Scholar] [CrossRef]
- Dove, J.E.; Yasukawa, K.; Tinsley, C.R.; Nassif, X. Production of the signalling molecule, autoinducer-2, by Neisseria meningitidis: Lack of evidence for a concerted transcriptional response. Microbiology 2003, 149, 1859–1869. [Google Scholar] [CrossRef]
- Falsetta, M.L.; Bair, T.B.; Ku, S.C.; Vanden Hoven, R.N.; Steichen, C.T.; McEwan, A.G.; Jennings, M.P.; Apicella, M.A. Transcriptional profiling identifies the metabolic phenotype of gonococcal biofilms. Infect. Immun. 2009, 77, 3522–3532. [Google Scholar] [CrossRef] [PubMed]
- Bandara, M.; Skehel, J.M.; Kadioglu, A.; Collinson, I.; Nobbs, A.H.; Blocker, A.J.; Jenkinson, H.F. The accessory Sec system (SecY2A2) in Streptococcus pneumoniae is involved in export of pneumolysin toxin, adhesion and biofilm formation. Microbes. Infect. 2017, 19, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Robinson, L.S.; Ashman, E.M.; Hultgren, S.J.; Chapman, M.R. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 2006, 59, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Zhao, Y.; Kou, Y.; Ni, D.; Zhang, X.C.; Huang, Y. Structure of the nonameric bacterial amyloid secretion channel. Proc. Natl. Acad. Sci. USA 2014, 111, E5439–E5444. [Google Scholar] [CrossRef]
- Goyal, P.; Krasteva, P.V.; Van Gerven, N.; Gubellini, F.; Van den Broeck, I.; Troupiotis-Tsaïlaki, A.; Jonckheere, W.; Péhau-Arnaudet, G.; Pinkner, J.S.; Chapman, M.R.; et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 2014, 516, 250–253. [Google Scholar] [CrossRef]
- Berne, C.; Ducret, A.; Hardy, G.G.; Brun, Y.V. Adhesins Involved in Attachment to Abiotic Surfaces by Gram-Negative Bacteria. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Chagnot, C.; Zorgani, M.A.; Astruc, T.; Desvaux, M. Proteinaceous determinants of surface colonization in bacteria: Bacterial adhesion and biofilm formation from a protein secretion perspective. Front. Microbiol. 2013, 4, 303. [Google Scholar] [CrossRef]
- Fronzes, R.; Remaut, H.; Waksman, G. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 2008, 27, 2271–2280. [Google Scholar] [CrossRef]
- Kikuchi, T.; Mizunoe, Y.; Takade, A.; Naito, S.; Yoshida, S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 2005, 49, 875–884. [Google Scholar] [CrossRef]
- Loferer, H.; Hammar, M.; Normark, S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol. Microbiol. 1997, 26, 11–23. [Google Scholar] [CrossRef]
- Kurtz, H.D.; Smith, J. Analysis of a Caulobacter crescentus gene cluster involved in attachment of the holdfast to the cell. J. Bacteriol. 1992, 174, 687–694. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ragland, S.A.; Schaub, R.E.; Hackett, K.T.; Dillard, J.P.; Criss, A.K. Two lytic transglycosylases in Neisseria gonorrhoeae impart resistance to killing by lysozyme and human neutrophils. Cell. Microbiol. 2017, 19. [Google Scholar] [CrossRef] [PubMed]
- Knilans, K.J.; Hackett, K.T.; Anderson, J.E.; Weng, C.; Dillard, J.P.; Duncan, J.A. Neisseria gonorrhoeae Lytic Transglycosylases LtgA and LtgD Reduce Host Innate Immune Signaling through TLR2 and NOD2. ACS Infect. Dis. 2017, 3, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, C.; Fang, X.; Ahmad, I.; Gomelsky, M.; Römling, U. Regulation of biofilm components in Salmonella enterica serovar Typhimurium by lytic transglycosylases involved in cell wall turnover. J. Bacteriol. 2011, 193, 6443–6451. [Google Scholar] [CrossRef]
- Martin, P.; Sun, L.; Hood, D.W.; Moxon, E.R. Involvement of genes of genome maintenance in the regulation of phase variation frequencies in Neisseria meningitidis. Microbiology 2004, 150, 3001–3012. [Google Scholar] [CrossRef]
- Fagnocchi, L.; Pigozzi, E.; Scarlato, V.; Delany, I. In the NadR regulon, adhesins and diverse meningococcal functions are regulated in response to signals in human saliva. J. Bacteriol. 2012, 194, 460–474. [Google Scholar] [CrossRef]
- Nassif, X.; Pujol, C.; Morand, P.; Eugène, E. Interactions of pathogenic Neisseria with host cells. Is it possible to assemble the puzzle? Mol. Microbiol. 1999, 32, 1124–1132. [Google Scholar] [CrossRef]
- Dehio, C.; Gray-Owen, S.D.; Meyer, T.F. Host cell invasion by pathogenic Neisseriae. In Subcellular Biochemistry; Springer: Boston, MA, USA, 2000; pp. 61–96. [Google Scholar]
- Schofield, M.J.; Hsieh, P. DNA mismatch repair: Molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003, 57, 579–608. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Płaczkiewicz, J.; Adamczyk-Popławska, M.; Lasek, R.; Bącal, P.; Kwiatek, A. Inactivation of Genes Encoding MutL and MutS Proteins Influences Adhesion and Biofilm Formation by Neisseria gonorrhoeae. Microorganisms 2019, 7, 647. https://doi.org/10.3390/microorganisms7120647
Płaczkiewicz J, Adamczyk-Popławska M, Lasek R, Bącal P, Kwiatek A. Inactivation of Genes Encoding MutL and MutS Proteins Influences Adhesion and Biofilm Formation by Neisseria gonorrhoeae. Microorganisms. 2019; 7(12):647. https://doi.org/10.3390/microorganisms7120647
Chicago/Turabian StylePłaczkiewicz, Jagoda, Monika Adamczyk-Popławska, Robert Lasek, Pawel Bącal, and Agnieszka Kwiatek. 2019. "Inactivation of Genes Encoding MutL and MutS Proteins Influences Adhesion and Biofilm Formation by Neisseria gonorrhoeae" Microorganisms 7, no. 12: 647. https://doi.org/10.3390/microorganisms7120647
APA StylePłaczkiewicz, J., Adamczyk-Popławska, M., Lasek, R., Bącal, P., & Kwiatek, A. (2019). Inactivation of Genes Encoding MutL and MutS Proteins Influences Adhesion and Biofilm Formation by Neisseria gonorrhoeae. Microorganisms, 7(12), 647. https://doi.org/10.3390/microorganisms7120647




