Grapevine Phyllosphere Community Analysis in Response to Elicitor Application against Powdery Mildew
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Experimental Set-Up
2.2. Microbial Functional Diversity
2.3. DNA Isolation and Sequencing
2.4. Fungal Metaphylogenomic Analyses and Taxonomic Distributions
2.5. RNA Extraction, Sequencing, and Virome Analysis
2.6. Statistical Analyses
3. Results
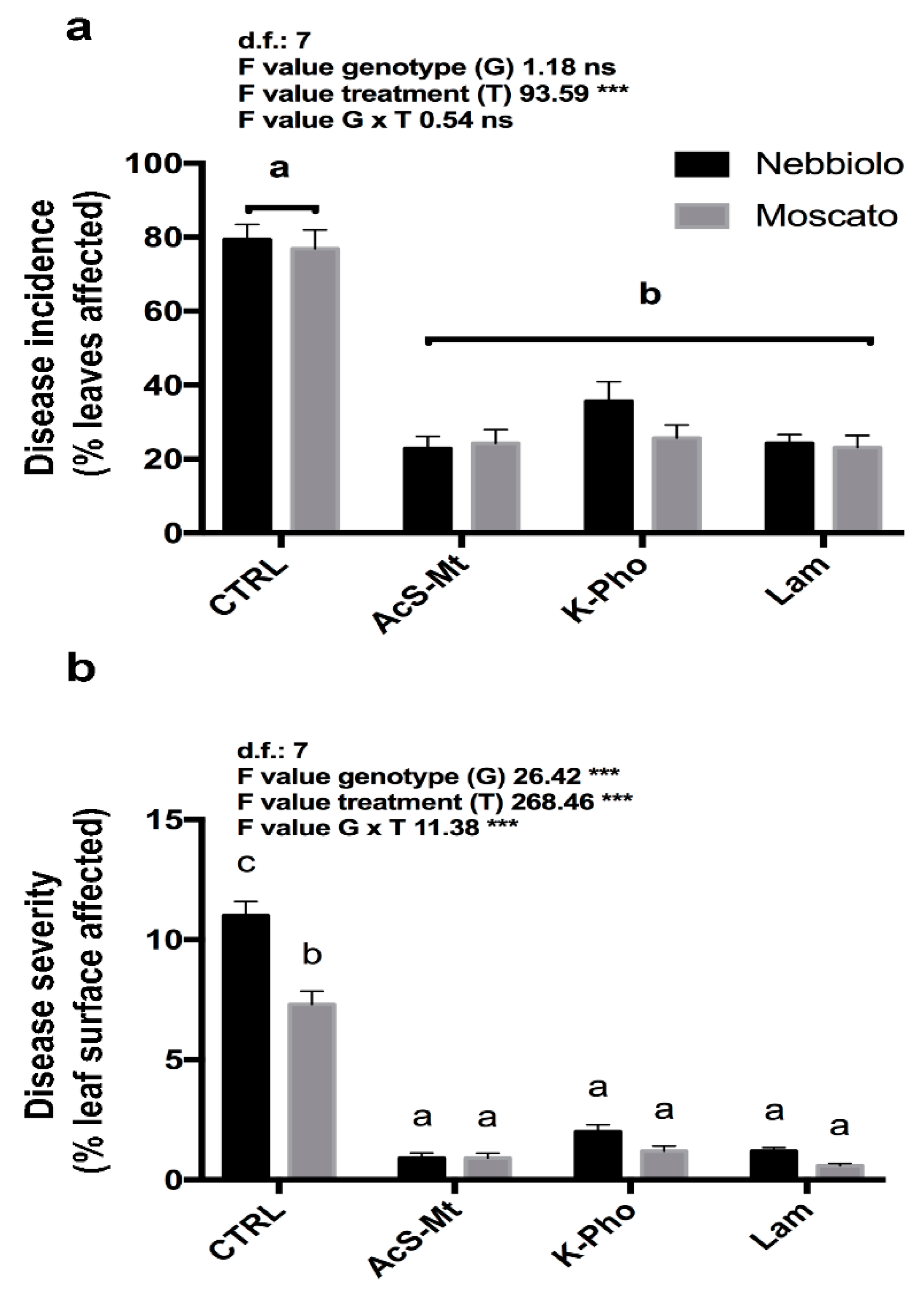
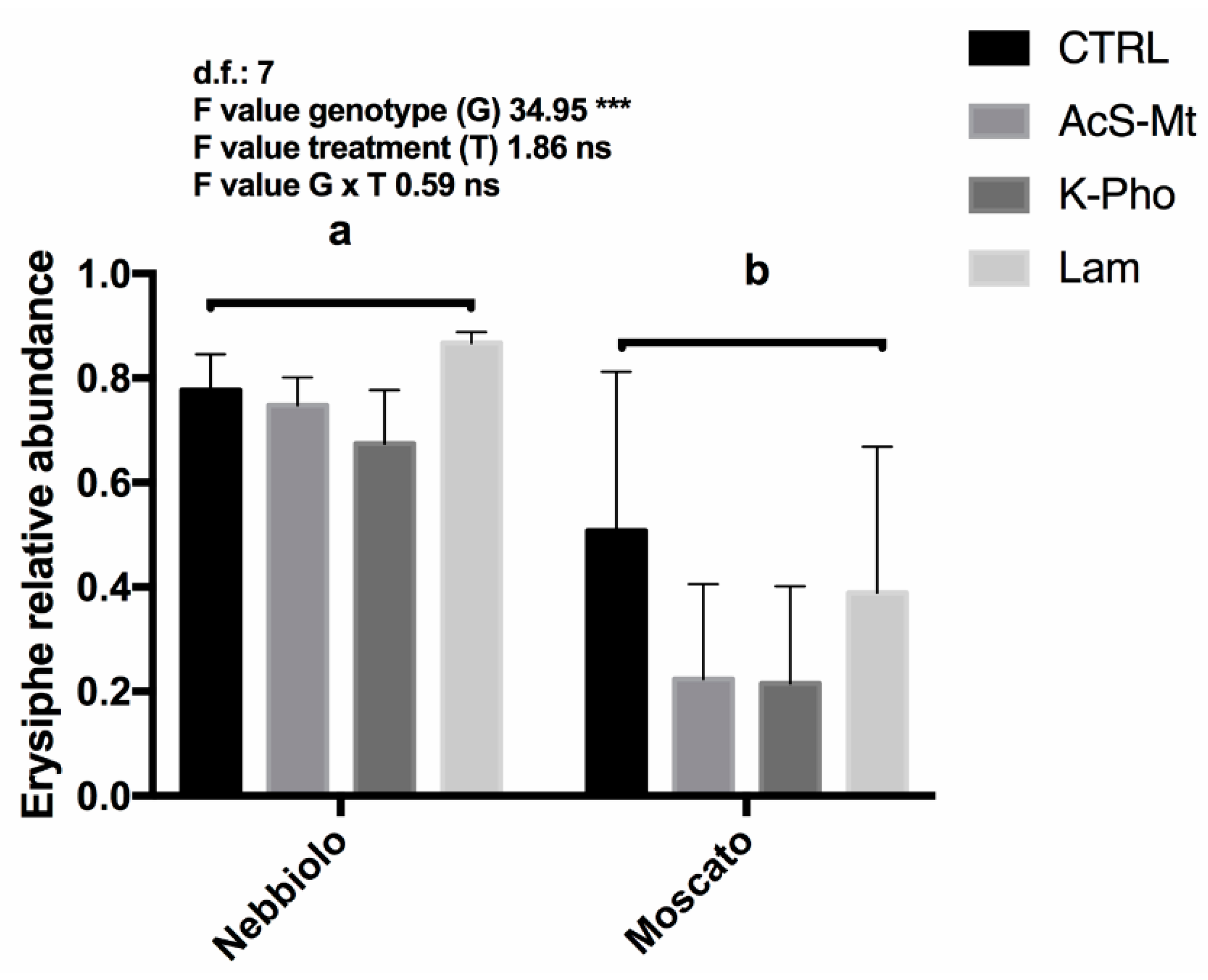
3.1. Disease Development
3.2. Treatment Effects on Microbial Functional Diversity
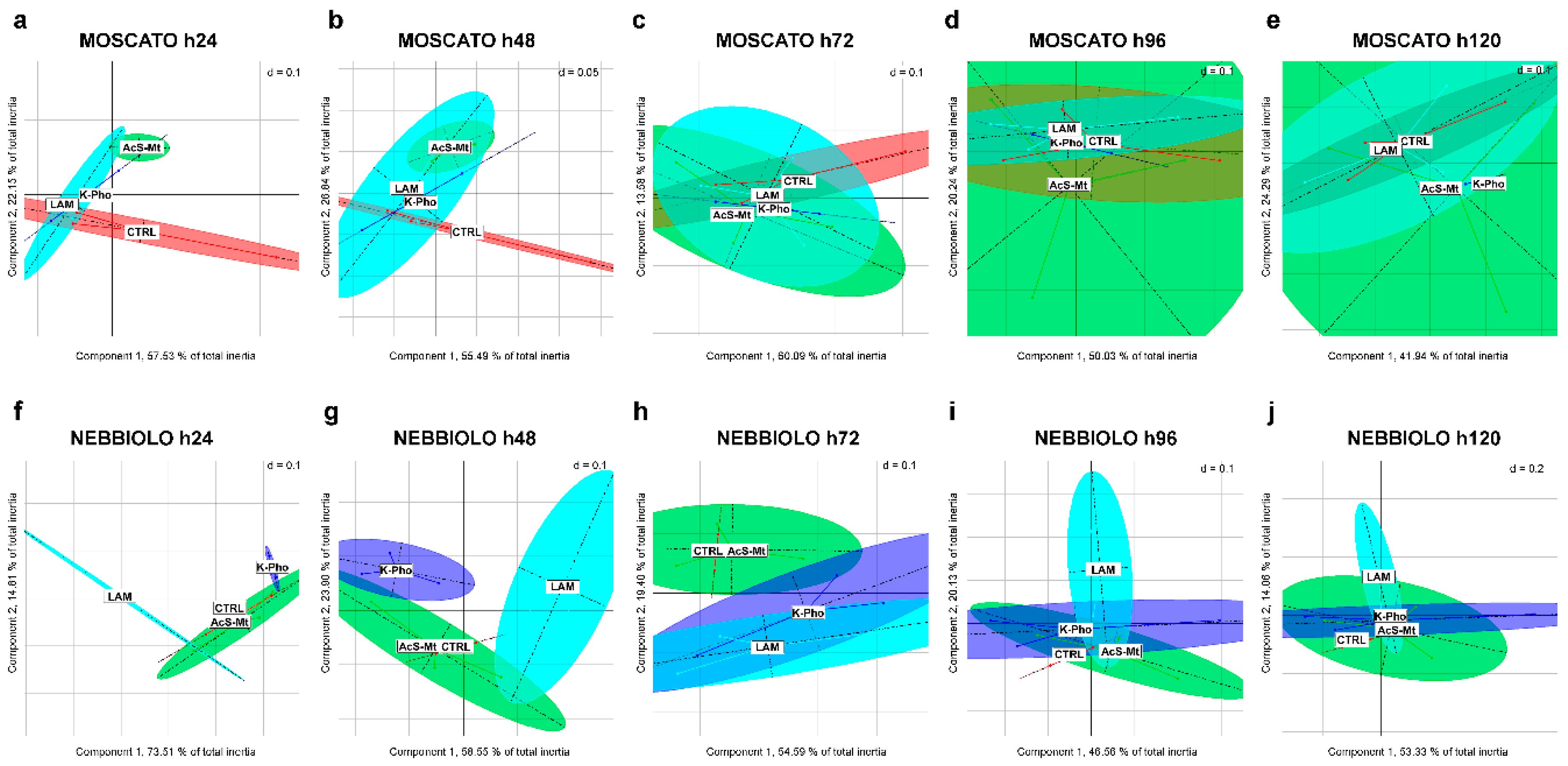
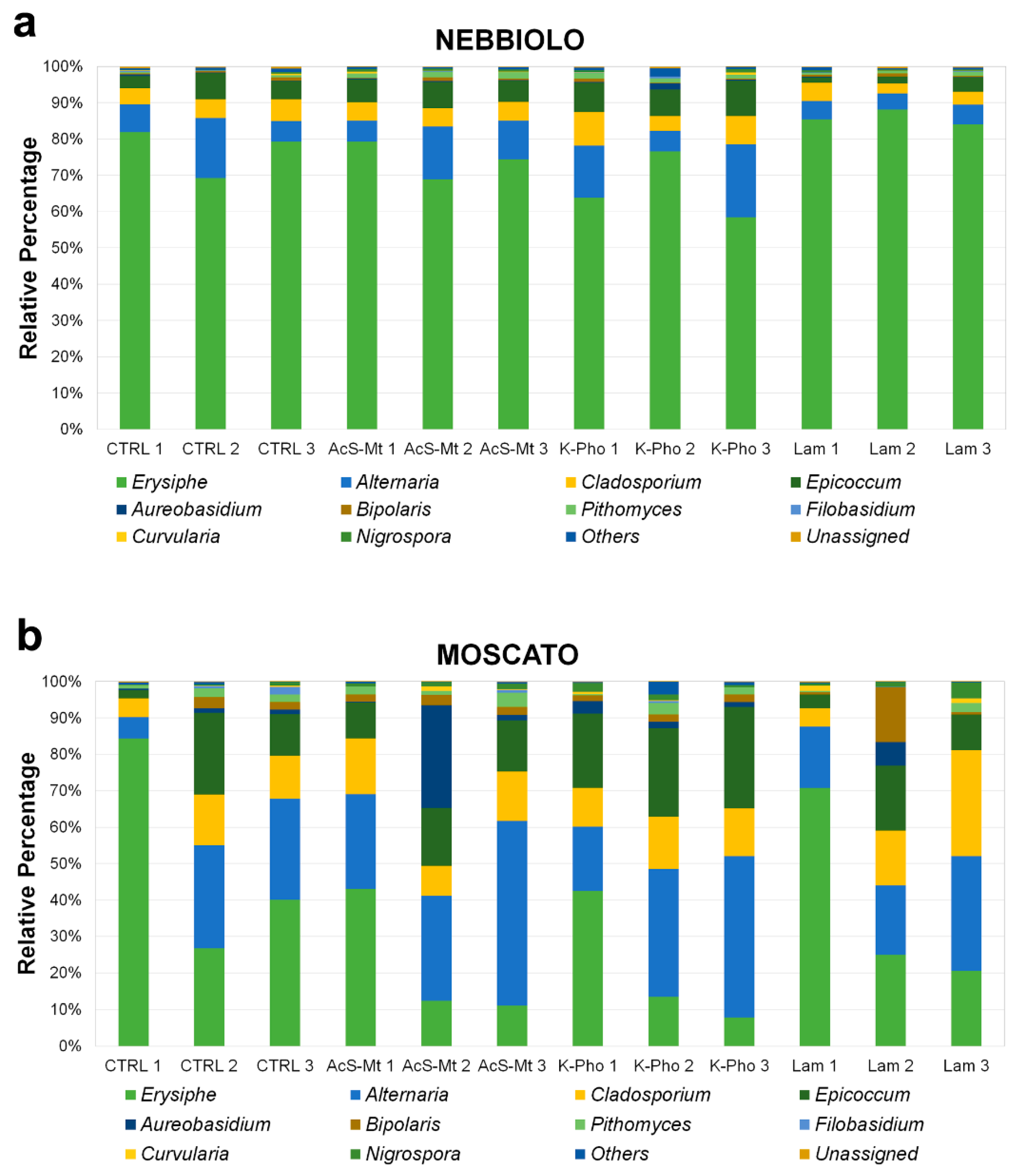
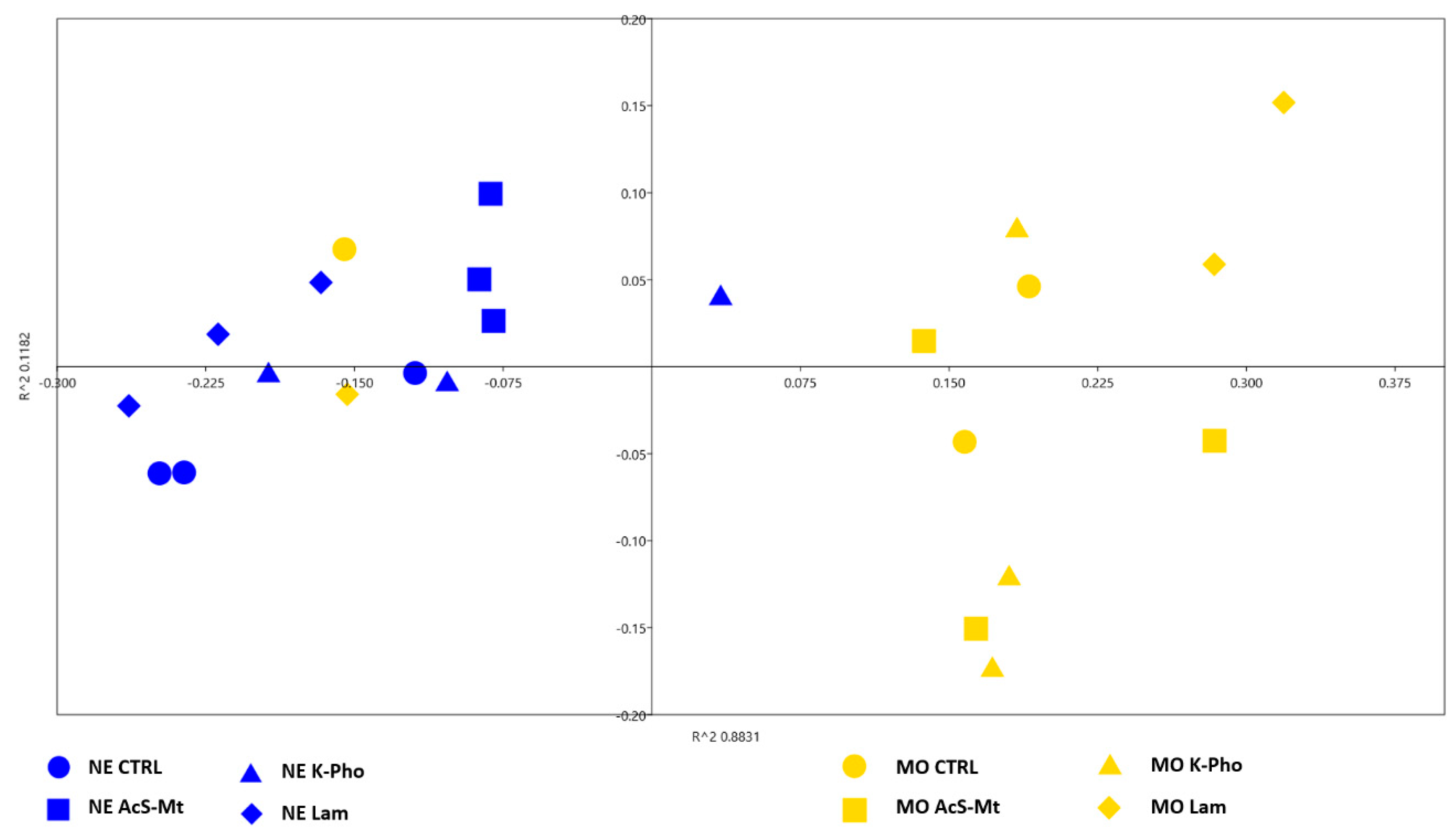
3.3. Treatment Effects on Fungal Community Diversity and Composition
3.4. Grapevine Virome, Viroid, and Phytoplasma Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Provost, C.; Pedneault, K. The organic vineyard as a balanced ecosystem: Improved organic grape management and impacts on wine quality. Sci. Hortic. 2016, 208, 43–56. [Google Scholar] [CrossRef]
- Pertot, I.; Caffi, T.; Rossi, V.; Mugnai, L.; Hoffmann, C.; Grando, M.; Gary, C.; Lafond, D.; Duso, C.; Thiery, D. A critical review of plant protection tools for reducing pesticide use on grapevine and new perspectives for the implementation of IPM in viticulture. Crop Prot. 2017, 97, 70–84. [Google Scholar] [CrossRef]
- Eurostat, Your Key to European Statistics. Available online: https://ec.europa.eu/eurostat/cache/metadata/en/aei_fm_salpest09_esms.htm (accessed on 7 August 2019).
- Pugliese, M.; Gullino, M.; Garibaldi, A. Effect of climate change on infection of grapevine by downy and powdery mildew under controlled environment. Commun. Agric. Appl. Biol. Sci. 2011, 76, 579–582. [Google Scholar] [PubMed]
- Gullino, M.L.; Pugliese, M.; Gilardi, G.; Garibaldi, A. Effect of increased CO2 and temperature on plant diseases: A critical appraisal of results obtained in studies carried out under controlled environment facilities. J. Plant Pathol. 2018, 100, 371–389. [Google Scholar] [CrossRef]
- Large, E.C. The Advance of the Fungi; Henry Holt and Co.: New York, NY, USA, 1940. [Google Scholar]
- Campisano, A.; Antonielli, L.; Pancher, M.; Yousaf, S.; Pindo, M.; Pertot, I. Bacterial endophytic communities in the grapevine depend on pest management. PLoS ONE 2014, 9, e112763. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Bischoff-Schaefer, M.; Bluemel, S.; Dachbrodt-Saaydeh, S.; Dreux, L.; Jansen, J.P.; Kiss, J.; Köhl, J.; Kudsk, P.; Malausa, T. Identifying obstacles and ranking common biological control research priorities for Europe to manage most economically important pests in arable, vegetable and perennial crops. Pest Manag. Sci. 2017, 73, 14–21. [Google Scholar] [CrossRef]
- Töpfer, R.; Hausmann, L.; Harst, M.; Maul, E.; Zyprian, E.; Eibach, R. New horizons for grapevine breeding. Methods Temp. Fruit Breed. Fruit Veg. Cereal Sci. Biotechnol. 2011, 5, 79–100. [Google Scholar]
- Eder, J.; Cosio, E.G. Elicitors of plant defense responses. In International Review of Cytology; Elsevier: Amsterdam, The Netherlands, 1994; Volume 148, pp. 1–36. [Google Scholar]
- Thakur, M.; Sohal, B.S. Role of elicitors in inducing resistance in plants against pathogen infection: A review. ISRN Biochem. 2013, 2013. [Google Scholar] [CrossRef]
- Zaker, M. Natural plant products as eco-friendly fungicides for plant diseases control—A review. Agriculturists 2016, 14, 134–141. [Google Scholar] [CrossRef]
- Gnanamanickam, S.S. Biological Control of Crop Diseases; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Aziz, A.; Poinssot, B.; Daire, X.; Adrian, M.; Bézier, A.; Lambert, B.; Joubert, J.-M.; Pugin, A. Laminarin elicits defense responses in grapevine and induces protection against Botrytis cinerea and Plasmopara viticola. Mol. Plant Microbe Interact. 2003, 16, 1118–1128. [Google Scholar] [CrossRef]
- Gauthier, A.; Trouvelot, S.; Kelloniemi, J.; Frettinger, P.; Wendehenne, D.; Daire, X.; Joubert, J.-M.; Ferrarini, A.; Delledonne, M.; Flors, V. The sulfated laminarin triggers a stress transcriptome before priming the SA-and ROS-dependent defenses during grapevine’s induced resistance against Plasmopara viticola. PLoS ONE 2014, 9, e88145. [Google Scholar] [CrossRef]
- Chalal, M.; Winkler, J.B.; Gourrat, K.; Trouvelot, S.; Adrian, M.; Schnitzler, J.-P.; Jamois, F.; Daire, X. Sesquiterpene volatile organic compounds (VOCs) are markers of elicitation by sulfated laminarine in grapevine. Front. Plant Sci. 2015, 6, 350. [Google Scholar] [CrossRef]
- Romanazzi, G.; Mancini, V.; Feliziani, E.; Servili, A.; Endeshaw, S.; Neri, D. Impact of alternative fungicides on grape downy mildew control and vine growth and development. Plant Dis. 2016, 100, 739–748. [Google Scholar] [CrossRef]
- Pugliese, M.; Monchiero, M.; Gullino, M.L.; Garibaldi, A. Application of laminarin and calcium oxide for the control of grape powdery mildew on Vitis vinifera cv. Moscato. J. Plant Dis. Prot. 2018, 125, 477–482. [Google Scholar] [CrossRef]
- Speiser, B.; Berner, A.; Häseli, A.; Tamm, L. Control of downy mildew of grapevine with potassium phosphonate: Effectivity and phosphonate residues in wine. Biol. Agric. Hortic. 2000, 17, 305–312. [Google Scholar] [CrossRef]
- Dagostin, S.; Schärer, H.-J.; Pertot, I.; Tamm, L. Are there alternatives to copper for controlling grapevine downy mildew in organic viticulture? Crop Prot. 2011, 30, 776–788. [Google Scholar] [CrossRef]
- Lim, S.; Borza, T.; Peters, R.D.; Coffin, R.H.; Al-Mughrabi, K.I.; Pinto, D.M.; Wang-Pruski, G. Proteomics analysis suggests broad functional changes in potato leaves triggered by phosphites and a complex indirect mode of action against Phytophthora infestans. J. Proteom. 2013, 93, 207–223. [Google Scholar] [CrossRef]
- Hollomon, D.W. Fungicide resistance: Facing the challenge-a review. Plant Prot. Sci. 2015, 51, 170–176. [Google Scholar] [CrossRef]
- Pereira, V.F.; Resende, M.L.V.d.; Monteiro, A.C.A.; Ribeiro Júnior, P.M.; Regina, M.d.A.; Medeiros, F.C.L. Alternative products for the protection of vine against downy mildew. Pesqui. Agropecuária Bras. 2010, 45, 25–31. [Google Scholar] [CrossRef]
- Campbell, P.; Latorre, B. Suppression of grapevine powdery mildew (Uncinula necator) by acibenzolar-S-methyl. Vitis 2004, 43, 209–210. [Google Scholar]
- Walker, A.P.; McCormack, M.L.; Messier, J.; Myers-Smith, I.H.; Wullschleger, S.D. Trait covariance: The functional warp of plant diversity? New Phytol. 2017, 216, 976–980. [Google Scholar] [CrossRef]
- Chauvin, K.M.; Asner, G.; Martin, R.; Kress, W.; Wright, S.; Field, C. Decoupled dimensions of leaf economic and anti-herbivore defense strategies in a tropical canopy tree community. Oecologia 2018, 186, 765–782. [Google Scholar] [CrossRef]
- Rosado, B.H.; Almeida, L.C.; Alves, L.F.; Lambais, M.R.; Oliveira, R.S. The importance of phyllosphere on plant functional ecology: A phyllo trait manifesto. New Phytol. 2018, 219, 1145–1149. [Google Scholar] [CrossRef]
- Laforest-Lapointe, I.; Paquette, A.; Messier, C.; Kembel, S.W. Leaf bacterial diversity mediates plant diversity and ecosystem function relationships. Nature 2017, 546, 145. [Google Scholar] [CrossRef]
- Hacquard, S. Disentangling the factors shaping microbiota composition across the plant holobiont. New Phytol. 2016, 209, 454–457. [Google Scholar] [CrossRef]
- Perrone, I.; Chitarra, W.; Boccacci, P.; Gambino, G. Grapevine-virus-environment interactions: An intriguing puzzle to solve. New Phytol. 2017, 213, 983–987. [Google Scholar] [CrossRef]
- Müller, T.; Ruppel, S. Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol. Ecol. 2014, 87, 2–17. [Google Scholar] [CrossRef]
- Yang, C.-H.; Crowley, D.E.; Borneman, J.; Keen, N.T. Microbial phyllosphere populations are more complex than previously realized. Proc. Natl. Acad. Sci. USA 2001, 98, 3889–3894. [Google Scholar] [CrossRef]
- Lindow, S.E.; Brandl, M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef]
- Rastogi, G.; Coaker, G.L.; Leveau, J.H. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 2013, 348, 1–10. [Google Scholar] [CrossRef]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828. [Google Scholar] [CrossRef] [PubMed]
- Moulas, C.; Petsoulas, C.; Rousidou, K.; Perruchon, C.; Karas, P.; Karpouzas, D.G. Effects of systemic pesticides imidacloprid and metalaxyl on the phyllosphere of pepper plants. BioMed Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Remus-Emsermann, M.N.; Schlechter, R.O. Phyllosphere microbiology: At the interface between microbial individuals and the plant host. New Phytol. 2018, 218, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Pal, K.K.; Gardener, B.M. Biological control of plant pathogens. Plant Health Instr. 2006, 2, 1117–1142. [Google Scholar] [CrossRef]
- Hacquard, S.; Spaepen, S.; Garrido-Oter, R.; Schulze-Lefert, P. Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 2017, 55, 565–589. [Google Scholar] [CrossRef]
- Gilbert, J.A.; van der Lelie, D.; Zarraonaindia, I. Microbial terroir for wine grapes. Proc. Natl. Acad. Sci. USA 2014, 111, 5–6. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef]
- Martelli, G.P. An Overview on Grapevine Viruses, Viroids, and the Diseases They Cause. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer: Cham, Switzerland, 2017; pp. 31–46. [Google Scholar]
- Perazzolli, M.; Antonielli, L.; Storari, M.; Puopolo, G.; Pancher, M.; Giovannini, O.; Pindo, M.; Pertot, I. Resilience of the natural phyllosphere microbiota of the grapevine to chemical and biological pesticides. Appl. Environ. Microbiol. 2014, 80, 3585–3596. [Google Scholar] [CrossRef]
- Cappelletti, M.; Perazzolli, M.; Antonielli, L.; Nesler, A.; Torboli, E.; Bianchedi, P.L.; Pindo, M.; Puopolo, G.; Pertot, I. Leaf treatments with a protein-based resistance inducer partially modify phyllosphere microbial communities of grapevine. Front. Plant Sci. 2016, 7, 1053. [Google Scholar] [CrossRef]
- Knight, S.; Klaere, S.; Fedrizzi, B.; Goddard, M.R. Regional microbial signatures positively correlate with differential wine phenotypes: Evidence for a microbial aspect to terroir. Sci. Rep. 2015, 5, 14233. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. MBio 2016, 7, e00631-16. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Santoni, S.; This, P.; Péros, J.-P. Genotype-Environment Interaction Shapes the Microbial Assemblage in Grapevine’s Phyllosphere and Carposphere: An NGS Approach. Microorganisms 2018, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, Y.; Zhang, X.; Wang, J.; Gao, M. Effects of chlortetracycline on soil microbial communities: Comparisons of enzyme activities to the functional diversity via Biolog EcoPlates™. Eur. J. Soil Biol. 2015, 68, 69–76. [Google Scholar] [CrossRef]
- Palaghianu, C. A tool for computing diversity and consideration on differences between diversity indices. arXiv 2016, arXiv:1602.04005. [Google Scholar]
- Zak, J.C.; Willig, M.R.; Moorhead, D.L.; Wildman, H.G. Functional diversity of microbial communities: A quantitative approach. Soil Biol. Biochem. 1994, 26, 1101–1108. [Google Scholar] [CrossRef]
- Janniche, G.S.; Spliid, H.; Albrechtsen, H.-J. Microbial Community-Level Physiological Profiles (CLPP) and herbicide mineralization potential in groundwater affected by agricultural land use. J. Contam. Hydrol. 2012, 140, 45–55. [Google Scholar] [CrossRef]
- Lindahl, B.D.; Nilsson, R.H.; Tedersoo, L.; Abarenkov, K.; Carlsen, T.; Kjøller, R.; Kõljalg, U.; Pennanen, T.; Rosendahl, S.; Stenlid, J. Fungal community analysis by high-throughput sequencing of amplified markers—A user’s guide. New Phytol. 2013, 199, 288–299. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press, Inc.: Cambridge, MA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef]
- Rivers, A.R.; Weber, K.C.; Gardner, T.G.; Liu, S.; Armstrong, S.D. ITSxpress: Software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Research 2018, 7, 1418. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Henrik Nilsson, R.; Larsson, K.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T. The UNITE database for molecular identification of fungi—Recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data, P. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Gambino, G. Multiplex RT-PCR method for the simultaneous detection of nine grapevine viruses. In Plant Virology Protocols; Springer: Berlin/Heidelberg, Germany, 2015; pp. 39–47. [Google Scholar]
- Hajizadeh, M.; Navarro, B.; Bashir, N.S.; Torchetti, E.M.; Di Serio, F. Development and validation of a multiplex RT-PCR method for the simultaneous detection of five grapevine viroids. J. Virol. Methods 2012, 179, 62–69. [Google Scholar] [CrossRef]
- Glasa, M.; Predajňa, L.; Komínek, P.; Nagyová, A.; Candresse, T.; Olmos, A. Molecular characterization of divergent Grapevine Pinot gris virus isolates and their detection in Slovak and Czech grapevines. Arch. Virol. 2014, 159, 2103–2107. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar]
- Dray, S.; Dufour, A.-B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Julien-Laferriere, A.; Siberchicot, A.; Dray, S. The adegraphics package. 2018. Available online: https://cran.r-project.org/web/packages/adegraphics/vignettes/adegraphics.html (accessed on 25 November 2019).
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST-Palaeontological Statistics. Available online: www. uv. es/~ pardomv/pe/2001_1/past/pastprog/past.pdf (accessed on 25 November 2019).
- Rutgers, M.; Wouterse, M.; Drost, S.M.; Breure, A.M.; Mulder, C.; Stone, D.; Creamer, R.E.; Winding, A.; Bloem, J. Monitoring soil bacteria with community-level physiological profiles using Biolog™ ECO-plates in the Netherlands and Europe. Appl. Soil Ecol. 2016, 97, 23–35. [Google Scholar] [CrossRef]
- Bruez, E.; Haidar, R.; Alou, M.T.; Vallance, J.; Bertsch, C.; Mazet, F.; Fermaud, M.; Deschamps, A.; Guerin-Dubrana, L.; Compant, S. Bacteria in a wood fungal disease: Characterization of bacterial communities in wood tissues of esca-foliar symptomatic and asymptomatic grapevines. Front. Microbiol. 2015, 6, 1137. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Li, F.; Yang, Q.; Wang, R.; Song, Y.; Tao, Y. The influence of different types of urban land use on soil microbial biomass and functional diversity in Beijing, China. Soil Use Manag. 2013, 29, 230–239. [Google Scholar] [CrossRef]
- Martelli, P.G. Where grapevine virology is heading to. In Proceedings of the 19th Congress of International Council for the Study of Viruses and Virus-Lile Diseases of the Grapevine, Santiago, Chile, 9–12 April 2018; pp. 10–15. [Google Scholar]
- Verginer, M.; Leitner, E.; Berg, G. Production of volatile metabolites by grape-associated microorganisms. J. Agric. Food Chem. 2010, 58, 8344–8350. [Google Scholar] [CrossRef]
- Stefanini, I.; Cavalieri, D. Metagenomic Approaches to Investigate the Contribution of the Vineyard Environment to the Quality of Wine Fermentation: Potentials and Difficulties. Front. Microbiol. 2018, 9, 991. [Google Scholar] [CrossRef]
- Berg, G. Plant—microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef]
- Delaunois, B.; Farace, G.; Jeandet, P.; Clément, C.; Baillieul, F.; Dorey, S.; Cordelier, S. Elicitors as alternative strategy to pesticides in grapevine? Current knowledge on their mode of action from controlled conditions to vineyard. Environ. Sci. Pollut. Res. 2014, 21, 4837–4846. [Google Scholar] [CrossRef]
- Melis, P.; Stoffels, K.; Vervoort, M.; Van Delm, T. Integrated approach of powdery mildew control on strawberry cultivar ’Elsanta’ in Belgium. In Proceedings of the 8th International Strawberry Symposium, Québec, QC, Canada, 13–17 August 2016; Volume 1156, pp. 709–714. [Google Scholar]
- Muniz, S.; Lacarta, J.; Pata, M.P.; Jimenez, J.J.; Navarro, E. Analysis of the diversity of substrate utilisation of soil bacteria exposed to Cd and earthworm activity using generalised additive models. PLoS ONE 2014, 9, e85057. [Google Scholar] [CrossRef]
- Sylla, J.; Alsanius, B.W.; Krüger, E.; Reineke, A.; Strohmeier, S.; Wohanka, W. Leaf microbiota of strawberries as affected by biological control agents. Phytopathology 2013, 103, 1001–1011. [Google Scholar] [CrossRef]
- Russell, D.; Chard, J.; McKinlay, R. Effect of Bacillus thuringiensis and a pyrethroid insecticide on the leaf microflora of Brassica oleracea. Lett. Appl. Microbiol. 1999, 28, 359–362. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Unravelling the diversity of grapevine microbiome. PLoS ONE 2014, 9, e85622. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, A.J.; Purahong, W.; Wubet, T.; Hyde, K.D.; Zhang, W.; Xu, H.; Zhang, G.; Fu, C.; Liu, M.; Xing, Q. Direct comparison of culture-dependent and culture-independent molecular approaches reveal the diversity of fungal endophytic communities in stems of grapevine (Vitis vinifera). Fungal Divers. 2018, 90, 85–107. [Google Scholar] [CrossRef]
- Whipps, J.; Hand, P.; Pink, D.; Bending, G.D. Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 2008, 105, 1744–1755. [Google Scholar] [CrossRef]
- Adams, P.D.; Kloepper, J.W. Effect of host genotype on indigenous bacterial endophytes of cotton (Gossypium hirsutum L.). Plant Soil 2002, 240, 181–189. [Google Scholar] [CrossRef]
- Rasche, F.; Trondl, R.; Naglreiter, C.; Reichenauer, T.G.; Sessitsch, A. Chilling and cultivar type affect the diversity of bacterial endophytes colonizing sweet pepper (Capsicum anuum L.). Can. J. Microbiol. 2006, 52, 1036–1045. [Google Scholar] [CrossRef]
- Correa, O.; Romero, A.; Montecchia, M.; Soria, M. Tomato genotype and Azospirillum inoculation modulate the changes in bacterial communities associated with roots and leaves. J. Appl. Microbiol. 2007, 102, 781–786. [Google Scholar] [CrossRef]
- Rasche, F.; Velvis, H.; Zachow, C.; Berg, G.; Van Elsas, J.D.; Sessitsch, A. Impact of transgenic potatoes expressing anti-bacterial agents on bacterial endophytes is comparable with the effects of plant genotype, soil type and pathogen infection. J. Appl. Ecol. 2006, 43, 555–566. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Bazán, E.R. Symbiosis: Viruses as intimate partners. Annu. Rev. Virol. 2017, 4, 123–139. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 2013, 3, 546–557. [Google Scholar] [CrossRef]
- Paez-Espino, D.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Thomas, A.D.; Huntemann, M.; Mikhailova, N.; Rubin, E.; Ivanova, N.N.; Kyrpides, N.C. Uncovering Earth’s virome. Nature 2016, 536, 425. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Ciuffo, M.; Vallino, M.; Margaria, P.; Varese, G.C.; Gnavi, G.; Turina, M. Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res. 2016, 219, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Vigani, G.; Di Silvestro, D.; Ciuffo, M.; Forgia, M.; Chitarra, W.; Turina, M. Biological and Molecular Characterization of Chenopodium quinoa Mitovirus 1 Reveals a Distinct Small RNA Response Compared to Those of Cytoplasmic RNA Viruses. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J. A new look at plant viruses and their potential beneficial roles in crops. Mol. Plant Pathol. 2015, 16, 331–333. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Chen, F.; Mannas, J.P.; Feldman, T.; Sumner, L.W.; Roossinck, M.J. Virus infection improves drought tolerance. New Phytol. 2008, 180, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Cuozzo, D.; Fasoli, M.; Pagliarani, C.; Vitali, M.; Boccacci, P.; Pezzotti, M.; Mannini, F. Co-evolution between Grapevine rupestris stem pitting-associated virus and Vitis vinifera L. leads to decreased defence responses and increased transcription of genes related to photosynthesis. J. Exp. Bot. 2012, 63, 5919–5933. [Google Scholar] [CrossRef]
- Pantaleo, V.; Vitali, M.; Boccacci, P.; Miozzi, L.; Cuozzo, D.; Chitarra, W.; Mannini, F.; Lovisolo, C.; Gambino, G. Novel functional microRNAs from virus-free and infected Vitis vinifera plants under water stress. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Repetto, O.; Bertazzon, N.; De Rosso, M.; Miotti, L.; Flamini, R.; Angelini, E.; Borgo, M. Low susceptibility of grapevine infected by GLRaV-3 to late Plasmopara viticola infections: Towards understanding the phenomenon. Physiol. Mol. Plant Pathol. 2012, 79, 55–63. [Google Scholar] [CrossRef]
- Chitarra, W.; Cuozzo, D.; Ferrandino, A.; Secchi, F.; Palmano, S.; Perrone, I.; Boccacci, P.; Pagliarani, C.; Gribaudo, I.; Mannini, F.; et al. Dissecting interplays between Vitis vinifera L. and grapevine virus B (GVB) under field conditions. Mol. Plant Pathol. 2018, 19, 2651–2666. [Google Scholar] [CrossRef]
| Active Ingredient (a.i.) | Commercial Product (c.p.) | a.i. Concentration | Dose of a.i. (g/ha) | Dose of c.p. (g/100 L) |
|---|---|---|---|---|
| Acibenzolar-S-methyl (AcS-Mt) | BION | 50% | 100 | 20 |
| Potassium phosphonate (K-Pho) | CENTURY SL | 755 g/L | 3020 | 600 |
| Laminarin (Lam) | VACCIPLANT | 45 g/L | 90 | 20 |
| Treatments | 10 August 2017 | 17 August 2017 | 17 August 2017 | 26 August 2017 | 4 September 2017 | 5 September 2017 | 11 September 2017 | 19 September 2017 | 22 September 2017 |
|---|---|---|---|---|---|---|---|---|---|
| Inoculated untreated control (CTRL) | - | - | E. necator (1 × 105 conidia/mL) | - | - | E. necator (1 × 105 conidia/mL) | - | - | Disease scoring and Sample collection |
| Acibenzolar-S-methyl (AcS-Mt) | Bion | Bion | E. necator (1 × 105 conidia/mL) | Bion | Bion | E. necator (1 × 105 conidia/mL) | Bion | Bion | |
| Potassium phosphonate (K-Pho) | Century Sl | Century Sl | E. necator (1 × 105 conidia/mL) | Century Sl | Century Sl | E. necator (1 × 105 conidia/mL) | Century Sl | Century Sl | |
| Laminarin (Lam) | Vacciplant | Vacciplant | E. necator (1 × 105 conidia/mL) | Vacciplant | Vacciplant | E. necator (1 × 105 conidia/mL) | Vacciplant | Vacciplant |
| Cultivar | Index | Treatment | h72 |
|---|---|---|---|
| MOSCATO | Simpson index (D) | CTRL | 0.97 ± 0.03 |
| AcS-Mt | 0.96 ± 0.04 | ||
| K-Pho | 0.99 ± 0.00 | ||
| Lam | 0.94 ± 0.04 | ||
| Shannon index (H’) | CTRL | 1.50 ± 1.27 | |
| AcS-Mt | 1.36 ± 1.20 | ||
| K-Pho | 0.92 ± 0.56 | ||
| Lam | 2.16 ± 1.07 | ||
| NEBBIOLO | Simpson index (D) | CTRL | 0.99 ± 0.00 |
| AcS-Mt | 0.96 ± 0.02 | ||
| K-Pho | 0.83 ± 0.15 | ||
| Lam | 0.95 ± 0.04 | ||
| Shannon index (H’) | CTRL | 1.56 ± 0.00 | |
| AcS-Mt | 2.51 ± 0.88 | ||
| K-Pho | 3.16 ± 2.44 | ||
| Lam | 2.29 ± 1.5 |
| Nebbiolo | Moscato | |||||||
|---|---|---|---|---|---|---|---|---|
| Virus/Viroid Name | CTRL | AcS-Mt | K-Pho | Lam | CTRL | AcS-Mt | K-Pho | Lam |
| Grapevine leafroll associated virus 1 | −−− | −−− | −−− | −−− | +++ | +++ | +++ | +++ |
| Grapevine leafroll associated virus 3 | −−− | −−− | −−− | −−− | ++− | +−− | +−− | −−− |
| Grapevine ruspestris stem pitting associated virus | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Grapevine pinot gris virus | ++− | +−− | ++− | −−− | −−− | −−− | −−− | −−− |
| Grapevine virus A | −−− | −−− | −−− | −−− | +++ | +++ | +++ | +++ |
| Grapevine virus B | −−− | −−− | −−− | −−− | −−− | −−− | ++− | −−− |
| Grapevine virus D | −−− | −−− | −−− | −−− | +−− | −−− | −−− | −−− |
| Grapevine virus F | −−− | −−− | −−− | −−− | ++− | −−− | −−− | −−− |
| Grapevine deformation virus | −−− | −−− | −−− | −−− | ++− | +−− | ++− | −−− |
| Grapevine fanleaf virus | −−− | −−− | −−− | −−− | +++ | +++ | ++− | −−− |
| Grapevine syrah virus | −−− | −−− | −−− | −−− | +−− | −−− | +−− | +−− |
| Grapevine rupestris vein feathering virus | −−− | −−− | −−− | −−− | +++ | ++− | +−− | −−− |
| Grapevine fleck virus | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Grapevine virus T | −−− | −−− | −−− | −−− | ++− | ++− | +−− | −−− |
| Grapevine yellow speckle viroid 1 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Grapevine yellow speckle viroid 2 | −−− | −−− | −−− | −−− | +−− | −−− | −−− | −−− |
| Hop stunt viroid | +++ | +++ | +++ | +++ | ++− | +++ | +++ | +++ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nerva, L.; Pagliarani, C.; Pugliese, M.; Monchiero, M.; Gonthier, S.; Gullino, M.L.; Gambino, G.; Chitarra, W. Grapevine Phyllosphere Community Analysis in Response to Elicitor Application against Powdery Mildew. Microorganisms 2019, 7, 662. https://doi.org/10.3390/microorganisms7120662
Nerva L, Pagliarani C, Pugliese M, Monchiero M, Gonthier S, Gullino ML, Gambino G, Chitarra W. Grapevine Phyllosphere Community Analysis in Response to Elicitor Application against Powdery Mildew. Microorganisms. 2019; 7(12):662. https://doi.org/10.3390/microorganisms7120662
Chicago/Turabian StyleNerva, Luca, Chiara Pagliarani, Massimo Pugliese, Matteo Monchiero, Solène Gonthier, Maria Lodovica Gullino, Giorgio Gambino, and Walter Chitarra. 2019. "Grapevine Phyllosphere Community Analysis in Response to Elicitor Application against Powdery Mildew" Microorganisms 7, no. 12: 662. https://doi.org/10.3390/microorganisms7120662
APA StyleNerva, L., Pagliarani, C., Pugliese, M., Monchiero, M., Gonthier, S., Gullino, M. L., Gambino, G., & Chitarra, W. (2019). Grapevine Phyllosphere Community Analysis in Response to Elicitor Application against Powdery Mildew. Microorganisms, 7(12), 662. https://doi.org/10.3390/microorganisms7120662








