Insights into Antagonistic Interactions of Multidrug Resistant Bacteria in Mangrove Sediments from the South Indian State of Kerala
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling, Enrichment, and Isolation of Antibiotic-Resistant Bacteria
2.2. Multidrug-Resistance Profiling
2.3. Molecular Identification and Phylogenetic Analysis
2.4. Biofilm Assay
2.5. Antagonism
3. Results
3.1. Distribution of Antibiotic-Resistant Bacteria, MDR Profiles, and Phylogenetic Analysis
3.2. Biofilm Formation in Environmental Isolates
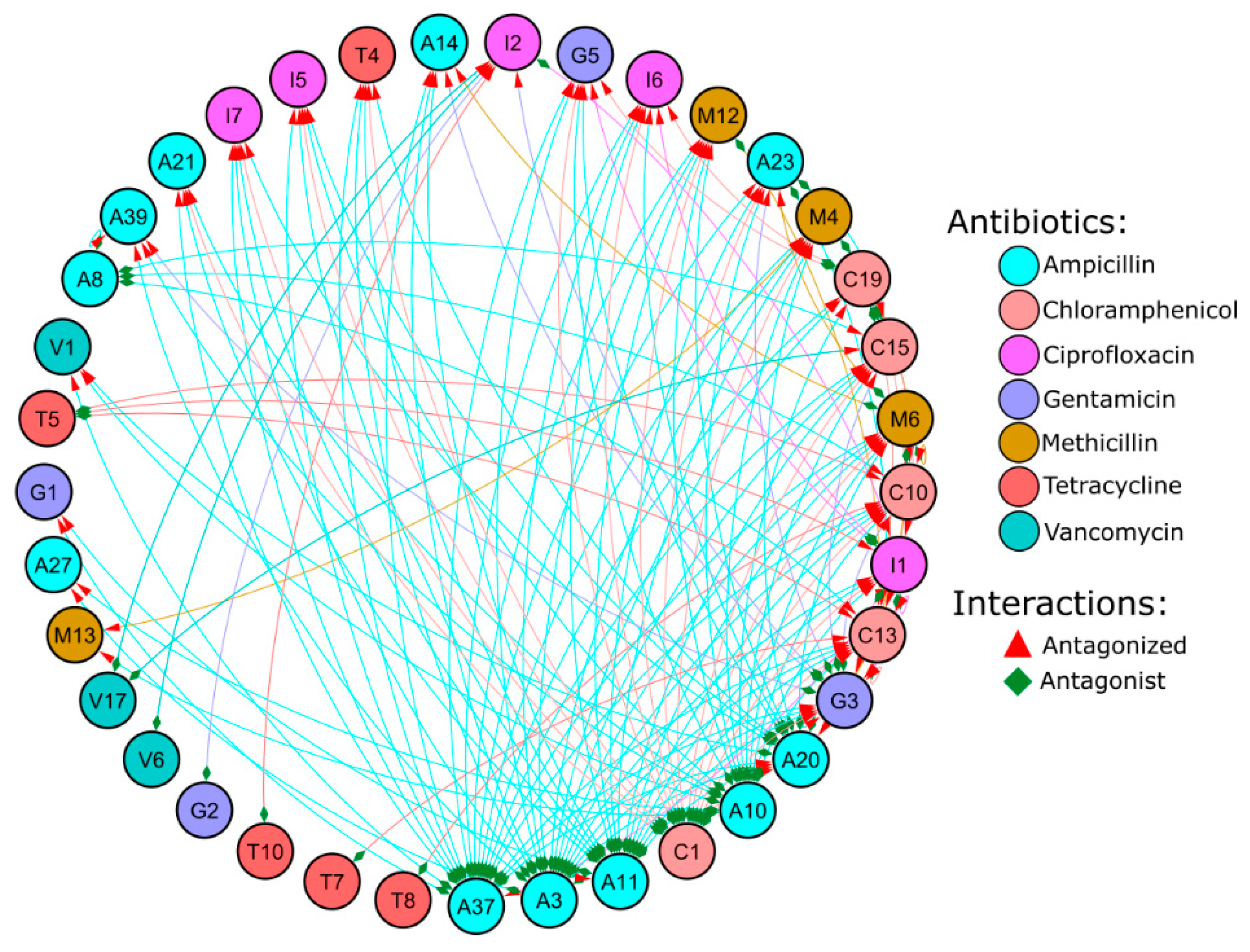
3.3. Antagonist Interactions
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Walsh, F.; Duffy, B. The culturable soil antibiotic resistome: A community of multi-drug resistant bacteria. PLoS ONE 2013, 8, e65567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wikler, M.A. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: Approved standard. CLSI (NCCLS) 2006, 26, M7-A7. [Google Scholar]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the antibiotic resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; Koteva, K.; Banks, E.D.; Johnston, M.D.; Barton, H.A.; Wright, G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 2012, 7, e34953. [Google Scholar] [CrossRef]
- Li, Y.; Li, Q.; Zhou, K.; Sun, X.L.; Zhao, L.R.; Zhang, Y.B. Occurrence and distribution of the environmental pollutant antibiotics in Gaoqiao mangrove area, China. Chemosphere 2016, 147, 25–35. [Google Scholar] [CrossRef]
- Zhao, H.; Yan, B.; Mo, X.; Li, P.; Li, B.; Li, Q.; Li, N.; Mo, S.; Ou, Q.; Shen, P.; et al. Prevalence and proliferation of antibiotic resistance genes in the subtropical mangrove wetland ecosystem of South China Sea. MicrobiologyOpen 2019, 8, e871. [Google Scholar] [CrossRef] [Green Version]
- Lee, L.H.; Zainal, N.; Azman, A.S.; Eng, S.K.; Goh, B.H.; Yin, W.F.; Ab Mutalib, N.S.; Chan, K.G. Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.B.; Ye, W.W.; Han, Y.; Deng, Z.X.; Hong, K. Natural products from mangrove actinomycetes. Mar. Drugs 2014, 12, 2590–2613. [Google Scholar] [CrossRef] [Green Version]
- Dhayanithi, N.B.; Kumar, T.A.; Murthy, R.G.; Kathiresan, K. Isolation of antibacterials from the mangrove, Avicennia marina and their activity against multi drug resistant Staphylococcus aureus. Asian Pac. J. Trop. Biomed. 2012, 2, S1892–S1895. [Google Scholar] [CrossRef]
- Imchen, M.; Kumavath, R.; Barh, D.; Azevedo, V.; Ghosh, P.; Viana, M.; Wattam, A.R. Searching for signatures across microbial communities: Metagenomic analysis of soil samples from mangrove and other ecosystems. Sci. Rep. 2017, 7, 8859. [Google Scholar] [CrossRef] [Green Version]
- Imchen, M.; Kumavath, R.; Barh, D.; Vaz, A.; Góes-Neto, A.; Tiwari, S.; Ghosh, P.; Wattam, A.R.; Azevedo, V. Comparative mangrove metagenome reveals global prevalence of heavy metals and antibiotic resistome across different ecosystems. Sci. Rep. 2018, 8, 11187. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A. Microtiter dish biofilm formation assay. JoVE (J. Vis. Exp.) 2011, e2437. [Google Scholar] [CrossRef]
- Li, Y.; Li, Q.; Sun, X.L.; Zhao, L.R.; Fan, H.Q. Occurrence and Distribution of Antibiotic-Resistant Bacteria Isolated in Gaoqiao Mangrove Wetland. China J. Antibiot. Res. 2017, 1, 202. [Google Scholar]
- Sayd, M.; Dev, A.; Sahr, Z.; Sajisha, S.; Safna, M.; Anu Ahmmad, B. Prevalence of high multiple antibiotic resistance index (mar) bacterial isolates from mangroves with reference to Calicut zones, kerala. Int. J. Adv. Sci. Eng. Technol. 2017, 5, 1–6. [Google Scholar]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [Green Version]
- Zaman, S.B.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A review on antibiotic resistance: Alarm bells are ringing. Cureus 2017, 9, e1403. [Google Scholar] [CrossRef] [Green Version]
- Allen, H.K.; Moe, L.A.; Rodbumrer, J.; Gaarder, A.; Handelsman, J. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. ISME J. 2009, 3, 243. [Google Scholar] [CrossRef]
- Udikovic-Kolic, N.; Wichmann, F.; Broderick, N.A.; Handelsman, J. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc. Natl. Acad. Sci. USA 2014, 111, 15202–15207. [Google Scholar] [CrossRef] [Green Version]
- Laroche, E.; Pawlak, B.; Berthe, T.; Skurnik, D.; Petit, F. Occurrence of antibiotic resistance and class 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France). FEMS Microbiol. Ecol. 2009, 68, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Ghaderpour, A.; Ho, W.S.; Chew, L.L.; Bong, C.W.; Chong, V.C.; Thong, K.L.; Chai, L.C. Diverse and abundant multi-drug resistant E. coli in Matang mangrove estuaries, Malaysia. Front. Microbiol. 2015, 6, 977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, T.X.; Munekage, Y.; Kato, S.I. Antibiotic resistance in bacteria from shrimp farming in mangrove areas. Sci. Total Environ. 2005, 349, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Mao, D.; Luo, Y.; Wang, L.; Xu, B.; Xu, L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 2012, 46, 2355–2364. [Google Scholar] [CrossRef] [PubMed]
- De WBlackburn, C.; McClure, P.J. (Eds.) Foodborne Pathogens: Hazards, Risk Analysis and Control; Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Taylor, J.M.; Sutherland, A.D.; Aidoo, K.E.; Logan, N.A. Heat-stable toxin production by strains of Bacillus cereus, Bacillus firmus, Bacillus megaterium, Bacillus simplex and Bacillus licheniformis. FEMS Microbiol. Lett. 2005, 242, 313–317. [Google Scholar] [CrossRef] [Green Version]
- Sanyal, S.; Karmaker, M.; Sultana, M.; Hossain, M. Association of Bacillus circulans with non-diabetic foot infection in Bangladeshi patient. Indian J. Med. Microbiol. 2015, 33. [Google Scholar] [CrossRef]
- Ko, K.S.; Oh, W.S.; Lee, M.Y.; Lee, J.H.; Lee, H.; Peck, K.R.; Lee, N.Y.; Song, J.H. Bacillus infantis sp. nov. and Bacillus idriensis sp. nov., isolated from a patient with neonatal sepsis. Int. J. Syst. Evol. Microbiol. 2006, 56, 2541–2544. [Google Scholar] [CrossRef]
- Krishnan, B.; Massilamany, C.; Smith, T.; Loy, J.; Barletta, R.; Reddy, J. Bacillus infantis NRRL B−14911-induced myocarditis: A novel disease model to study the autoimmune mechanisms of myocardial injuries in the myocarditis-susceptible, A/J mice (BA15P. 221). J. Immunol. 2014, 192. [Google Scholar]
- Pinheiro, L.; Brito, C.; de Oliveira, A.; Martins, P.; Pereira, V.; da Cunha, M. Staphylococcus epidermidis and Staphylococcus haemolyticus: Molecular detection of cytotoxin and enterotoxin genes. Toxins 2015, 7, 3688–3699. [Google Scholar] [CrossRef]
- Czekaj, T.; Ciszewski, M.; Szewczyk, E.M. Staphylococcus haemolyticus—An emerging threat in the twilight of the antibiotics age. Microbiology 2015, 161, 2061–2068. [Google Scholar] [CrossRef]
- Mendis, H.C.; Thomas, V.P.; Schwientek, P.; Salamzade, R.; Chien, J.T.; Waidyarathne, P.; Kloepper, J.; De La Fuente, L. Strain-specific quantification of root colonization by plant growth promoting rhizobacteria Bacillus firmus I–1582 and Bacillus amyloliquefaciens QST713 in non-sterile soil and field conditions. PLoS ONE 2018, 13, e0193119. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, A.R.; Kiewnick, S.; Sikora, R.A. In vitro activity of Bacillus firmus against the burrowing nematode Radopholus similis, the root-knot nematode Meloidogyne incognita and the stem nematode Ditylenchus dipsaci. Biocontrol Sci. Technol. 2008, 18, 377–389. [Google Scholar] [CrossRef]
- Geng, C.; Nie, X.; Tang, Z.; Zhang, Y.; Lin, J.; Sun, M.; Peng, D. A novel serine protease, Sep1, from Bacillus firmus DS–1 has nematicidal activity and degrades multiple intestinal-associated nematode proteins. Sci. Rep. 2016, 6, 25012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rafikova, G.F.; Korshunova, T.Y.; Minnebaev, L.F.; Chetverikov, S.P.; Loginov, O.N. A new bacterial strain, Pseudomonas koreensis IB−4, as a promising agent for plant pathogen biological control. Microbiology 2016, 85, 333–341. [Google Scholar] [CrossRef]
- Kantas, D.; Papatsiros, V.G.; Tassis, P.D.; Giavasis, I.; Bouki, P.; Tzika, E.D. A feed additive containing Bacillus toyonensis (Toyocerin®) protects against enteric pathogens in postweaning piglets. J. Appl. Microbiol. 2015, 118, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Imperial, I.C.; Ibana, J.A. Addressing the antibiotic resistance problem with probiotics: Reducing the risk of its double-edged sword effect. Front. Microbiol. 2016, 7, 1983. [Google Scholar] [CrossRef]
- Wong, A.; Saint Ngu, D.Y.; Dan, L.A.; Ooi, A.; Lim, R.L. Detection of antibiotic resistance in probiotics of dietary supplements. Nutr. J. 2015, 14, 95. [Google Scholar] [CrossRef]
- Reisner, A.; Krogfelt, K.A.; Klein, B.M.; Zechner, E.L.; Molin, S. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: Impact of environmental and genetic factors. J. Bacteriol. 2006, 188, 3572–3581. [Google Scholar] [CrossRef] [Green Version]
- Parker, R.E.; Laut, C.; Gaddy, J.A.; Zadoks, R.N.; Davies, H.D.; Manning, S.D. Association between genotypic diversity and biofilm production in group B Streptococcus. BMC Microbiol. 2016, 16, 86. [Google Scholar] [CrossRef] [Green Version]
- Avila-Novoa, M.G.; Iñíguez-Moreno, M.; Solís-Velázquez, O.A.; González-Gómez, J.P.; Guerrero-Medina, P.J.; Gutiérrez-Lomelí, M. Biofilm Formation by Staphylococcus aureus Isolated from Food Contact Surfaces in the Dairy Industry of Jalisco, Mexico. J. Food Qual. 2018, 2018. [Google Scholar] [CrossRef] [Green Version]
- Chavez-Dozal, A.; Nishiguchi, M.K. Variation in biofilm formation among symbiotic and free-living strains of Vibrio fischeri. J. Basic Microbiol. 2011, 51, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yan, Z.; Xu, J. Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology 2003, 149, 353–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, S.; Devi, K.M.; Damrolien, S.; Devi, K.S.; Krossnunpuii, K.T. Biofilm production and its correlation with antibiotic resistance pattern among clinical isolates of Pseudomonas aeruginosa in a tertiary care hospital in north-east India. Int. J. Adv. Med. 2018, 5, 964. [Google Scholar] [CrossRef] [Green Version]
- Namuq, A.O.; Ali, K.O.; Al-Ani, A.H. Correlation between Biofilm Formation, Multi-Drug Resistance and AlgD Gene among Pseudomonas aeruginosa Clinical Isolates. J. Univ. Babylon Pure Appl. Sci. 2019, 27, 143–150. [Google Scholar]
- Roshani-Asl, P.; Rashidi, N.; Shokoohizadeh, L.; Zarei, J. Relationship Among Antibiotic Resistance, Biofilm Formation and lasB Gene in Pseudomonas Aeruginosa Isolated from Burn Patients. Clin. Lab. 2018, 64, 1477–1484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabatabaei, M.; Sohrabi, N. Comparison of biofilm formation and antibiotic resistance pattern of Pseudomonas aeruginosa in human and environmental isolates. Microb. Pathog. 2017, 109, 94–98. [Google Scholar]
- Rao, R.S.; Karthika, R.U.; Singh, S.P.; Shashikala, P.; Kanungo, R.; Jayachandran, S.; Prashanth, K. Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J. Med. Microbiol. 2008, 26, 333. [Google Scholar]
- Karigoudar, R.M.; Karigoudar, M.H.; Wavare, S.M.; Mangalgi, S.S. Detection of biofilm among uropathogenic Escherichia coli and its correlation with antibiotic resistance pattern. J. Lab. Physicians 2019, 11, 17. [Google Scholar] [CrossRef]
- Shahidul, K.M.; Farahnaaz, F.; Sunjukta, A. Determination of antibiotic resistance pattern of biofilm producing pathogenic bacteria associated with UTI. Int. J. Drug Dev. Res. 2013, 5, 312–319. [Google Scholar]
- Cepas, V.; López, Y.; Munoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship between Biofilm Formation and Antimicrobial Resistance in Gram-Negative Bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Türkel, İ.; Yıldırım, T.; Yazgan, B.; Bilgin, M.; Başbulut, E. Relationship between antibiotic resistance, efflux pumps, and biofilm formation in extended-spectrum β-lactamase producing Klebsiella pneumoniae. J. Chemother. 2018, 30, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Balcázar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, P.; Maira-Litran, T.; McBain, A.J.; Rickard, A.H.; Whyte, F. The Physiology and Collective Recalcitrance of Microbial Biofilm Communities–2 Changes in susceptibility associated with cellular aggregates. Adv. Microb. Physiol. 2002, 46, 213–216. [Google Scholar]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Burns, A.; Ryder, D.S. Potential for biofilms as biological indicators in Australian riverine systems. Ecol. Manag. Restor. 2001, 2, 53–64. [Google Scholar] [CrossRef]
- Sabater, S.; Guasch, H.; Ricart, M.; Romaní, A.; Vidal, G.; Klünder, C.; Schmitt-Jansen, M. Monitoring the effect of chemicals on biological communities. The biofilm as an interface. Anal. Bioanal. Chem. 2007, 387, 1425–1434. [Google Scholar] [CrossRef]
- Laport, M.S.; Santos-Gandelman, J.F.; Muricy, G.; Giambiade-deMarval, M.; George, I. Antagonistic interactions among bacteria isolated from either the same or from different sponges native to the Brazilian coast. J. Mar. Sci. Res. Dev. 2016, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Russel, J.; Røder, H.L.; Madsen, J.S.; Burmølle, M.; Sørensen, S.J. Antagonism correlates with metabolic similarity in diverse bacteria. Proc. Natl. Acad. Sci. USA 2017, 114, 10684–10688. [Google Scholar] [CrossRef] [Green Version]
- Mangano, S.; Michaud, L.; Caruso, C.; Brilli, M.; Bruni, V.; Fani, R.; Giudice, A.L. Antagonistic interactions between psychrotrophic cultivable bacteria isolated from Antarctic sponges: A preliminary analysis. Res. Microbiol. 2009, 160, 27–37. [Google Scholar] [CrossRef]
- Long, R.A.; Azam, F. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 2001, 67, 4975–4983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, S.H.; Simidu, U.S. Distribution and significance of heterotrophic marine bacteria with antibacterial activity. Appl. Environ. Microbiol. 1987, 53, 2957–2962. [Google Scholar] [PubMed]
- Lo Giudice, A.; Brilli, M.; Bruni, V.; De Domenico, M.; Fani, R.; Michaud, L. Bacterium–bacterium inhibitory interactions among psychrotrophic bacteria isolated from Antarctic seawater (Terra Nova Bay, Ross Sea). FEMS Microbiol. Ecol. 2007, 60, 383–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanagasabhapathy, M.; Nagata, S. Cross-species induction of antibacterial activity produced by epibiotic bacteria isolated from Indian marine sponge Pseudoceratina purpurea. World J. Microbiol. Biotechnol. 2008, 24, 687–691. [Google Scholar] [CrossRef]
- Hentschel, U.; Schmid, M.; Wagner, M.; Fieseler, L.; Gernert, C.; Hacker, J. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 2001, 35, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Grossart, H.P.; Schlingloff, A.; Bernhard, M.; Simon, M.; Brinkhoff, T. Antagonistic activity of bacteria isolated from organic aggregates of the German Wadden Sea. FEMS Microbiol. Ecol. 2004, 47, 387–396. [Google Scholar] [CrossRef]
- Vetsigian, K.; Jajoo, R.; Kishony, R. Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol. 2011, 9, e1001184. [Google Scholar] [CrossRef]
- Cordero, O.X.; Wildschutte, H.; Kirkup, B.; Proehl, S.; Ngo, L.; Hussain, F.; Le Roux, F.; Mincer, T.; Polz, M.F. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science 2012, 337, 1228–1231. [Google Scholar] [CrossRef]
- Deng, Y.J.; Wang, S.Y. Complex carbohydrates reduce the frequency of antagonistic interactions among bacteria degrading cellulose and xylan. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, M.; Lovett, B.; Angoshtari, R.; Nirenberg, P.; Loch, T.P.; Scribner, K.T.; Marsh, T.L. Antagonistic Interactions and Biofilm Forming Capabilities Among Bacterial Strains Isolated from the Egg Surfaces of Lake Sturgeon (Acipenser fulvescens). Microb. Ecol. 2018, 75, 22–37. [Google Scholar] [CrossRef]
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 2005, 436, 1171. [Google Scholar] [CrossRef] [PubMed]



| Location | Co-Ordinates (Latitude, Longitude) | Sample ID |
|---|---|---|
| Payannur | 12.1050687, 75.2058 | PYN |
| Bangramanjeshwar | 12.708333, 74.900754 | BNH |
| Kumbla | 12.594195, 74.946623 | KMA |
| Kavvayi | 12.088286, 75.176029 | KVY |
| Valapattanam | 9.996566, 76.247189 | VPM |
| Panangod | 9.8959941, 76.326094 | PGD |
| Madakal | 9.9091896, 76.30629 | MAL |
| Vallarpadam | 9.9994138, 76.253705 | VPDM |
| Antibiotics | Working Concentration (µg/mL) |
|---|---|
| Ampicillin | 100 |
| Gentamicin | 10 |
| Chloramphenicol | 25 |
| Ciprofloxacin | 10 |
| Tetracycline | 10 |
| Vancomycin | 50 |
| Methicillin | 1 |
| Isolate Sample ID | Species | Strain | Similarity (%) | Phylum | Family | Genus |
|---|---|---|---|---|---|---|
| V6 | Klebsiella aerogenes | KCTC 2190 | 100 | Proteobacteria | Enterobacteriaceae | Klebsiella |
| V4 | Enterobacter bugandensis | EB–247(T) | 100 | Proteobacteria | Enterobacteriaceae | Enterobacter |
| V1 | K. aerogenes | KCTC 2190 | 100 | Proteobacteria | Enterobacteriaceae | Klebsiella |
| V17 | Shewanella algae | JCM 21037 | 100 | Proteobacteria | Shewanellaceae | Shewanella |
| T8 | Bacillus circulans | ATCC 4513(T) | 99.15 | Firmicutes | Bacillaceae | Bacillus |
| T7 | Bacillus cereus | ATCC 14579(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| T5 | Lysinibacillus macroides | DSM 54(T) | 100 | Firmicutes | Planococcaceae | Lysinibacillus |
| T4 | Pseudomonas aeruginosa | JCM 5962 | 100 | Proteobacteria | Pseudomonadaceae | Pseudomonas |
| T10 | L. macroides | DSM 54(T) | 100 | Firmicutes | Planococcaceae | Lysinibacillus |
| M6 | Lysinibacillus fusiformis | NBRC 15717(T) | 99.88 | Firmicutes | Planococcaceae | Lysinibacillus |
| M4 | Staphylococcus epidermidis | NCTC 11047(T) | 99.88 | Firmicutes | Staphylococcaceae | Staphylococcus |
| M13 | L. fusiformis | NBRC 15717(T) | 99.84 | Firmicutes | Planococcaceae | Lysinibacillus |
| M12 | Bacillus koreensis | DSM 16467 | 100 | Firmicutes | Bacillaceae | Bacillus |
| I7 | Sporosarcina luteola | Y1 | 99.75 | Firmicutes | Planococcaceae | Sporosarcina |
| I6 | S. luteola | Y1 | 99.72 | Firmicutes | Planococcaceae | Sporosarcina |
| I5 | S. luteola | Y1 | 99.75 | Firmicutes | Planococcaceae | Sporosarcina |
| I2 | S. luteola | Y1 | 99.75 | Firmicutes | Planococcaceae | Sporosarcina |
| I1 | S. luteola | Y1 | 99.74 | Firmicutes | Planococcaceae | Sporosarcina |
| G5 | B. circulans | ATCC 4513(T) | 99.66 | Firmicutes | Bacillaceae | Bacillus |
| G3 | Gracilibacillus marinus | HB09003 | 100 | Firmicutes | Bacillaceae | Gracilibacillus |
| G2 | Bacillus infantis | NRRL B–14911 | 99.73 | Firmicutes | Bacillaceae | Bacillus |
| G1 | Staphylococcus haemolyticus | MTCC 3383 | 100 | Firmicutes | Staphylococcaceae | Staphylococcus |
| C1 | P. aeruginosa | JCM 5962 | 100 | Proteobacteria | Pseudomonadaceae | Pseudomonas |
| C19 | B. infantis | NRRL B–14911 | 99.75 | Firmicutes | Bacillaceae | Bacillus |
| C16 | B. infantis | NRRL B–14911 | 99.74 | Firmicutes | Bacillaceae | Bacillus |
| C15 | Bacillus firmus | NBRC 15306 | 100 | Firmicutes | Bacillaceae | Bacillus |
| C13 | Bacillus oceanisediminis | H2(T) | 99.3 | Firmicutes | Bacillaceae | Bacillus |
| C10 | Bacillus enclensis | SGD–1123(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| A8 | B. cereus | ATCC 14579 | 100 | Firmicutes | Bacillaceae | Bacillus |
| A3 | Shigella flexneri | ATCC 29903 | 100 | Proteobacteria | Enterobacteriaceae | Escherichia |
| A39 | P. aeruginosa | JCM 5962 | 100 | Proteobacteria | Pseudomonadaceae | Pseudomonas |
| A37 | S. flexneri | ATCC 29903(T) | 100 | Proteobacteria | Enterobacteriaceae | Escherichia |
| A27 | Bacillus toyonensis | BCT–7112 | 99.85 | Firmicutes | Bacillaceae | Bacillus |
| A23 | B. cereus | ATCC 14579(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| A21 | B. cereus | ATCC 14579(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| A20 | B. cereus | ATCC 14579(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| A1 | S. flexneri | ATCC 29903(T) | 100 | Proteobacteria | Enterobacteriaceae | Escherichia |
| A14 | B. cereus | ATCC 14579(T) | 100 | Firmicutes | Bacillaceae | Bacillus |
| A11 | B. cereus | ATCC 14579(T) | 99.88 | Firmicutes | Bacillaceae | Bacillus |
| A10 | S. flexneri | ATCC 29903(T) | 100 | Proteobacteria | Enterobacteriaceae | Escherichia |
| Antagonized (%) | Antagonist (%) | ||
|---|---|---|---|
| Isolates | Non-Biofilm | 68.75 | 68.75 |
| Biofilm | 62.5 | 50 | |
| Interactions | Non-Biofilm | 53.28 | 57.66 |
| Biofilm | 46.71 | 42.33 | |
| Isolates | Gram negative | 36.36 | 54.54 |
| Gram positive | 75.86 | 58.62 | |
| Interactions | Gram negative | 9.42 | 52.17 |
| Gram positive | 89.85 | 47.10 |
| Interactions (Antagonist vs. Antagonized) | Antagonist (%) |
|---|---|
| Gram negative vs. gram positive | 47.45 |
| Gram negative vs. gram negative | 5.11 |
| Gram positive vs. gram negative | 4.38 |
| Gram positive vs. gram positive | 43.07 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imchen, M.; Vennapu, R.K.; Ghosh, P.; Kumavath, R. Insights into Antagonistic Interactions of Multidrug Resistant Bacteria in Mangrove Sediments from the South Indian State of Kerala. Microorganisms 2019, 7, 678. https://doi.org/10.3390/microorganisms7120678
Imchen M, Vennapu RK, Ghosh P, Kumavath R. Insights into Antagonistic Interactions of Multidrug Resistant Bacteria in Mangrove Sediments from the South Indian State of Kerala. Microorganisms. 2019; 7(12):678. https://doi.org/10.3390/microorganisms7120678
Chicago/Turabian StyleImchen, Madangchanok, Ravali Krishna Vennapu, Preetam Ghosh, and Ranjith Kumavath. 2019. "Insights into Antagonistic Interactions of Multidrug Resistant Bacteria in Mangrove Sediments from the South Indian State of Kerala" Microorganisms 7, no. 12: 678. https://doi.org/10.3390/microorganisms7120678
APA StyleImchen, M., Vennapu, R. K., Ghosh, P., & Kumavath, R. (2019). Insights into Antagonistic Interactions of Multidrug Resistant Bacteria in Mangrove Sediments from the South Indian State of Kerala. Microorganisms, 7(12), 678. https://doi.org/10.3390/microorganisms7120678