Abstract
Reducing acute mortality in aquatic crustaceans using natural alternatives to antibiotics has become a necessity, firstly for its positive impact on the aquaculture industry and, secondly, because the extensive use of antibiotics may lead to increased levels of drug resistance in pathogenic microorganisms. This study aimed to investigate the effect of a mixture of natural antimicrobials on the in vitro and in vivo virulence abilities of Type VI secretion system (T6SS)-positive Vibrio parahaemolyticus (A3 and D4), strains known as having potentially harmful health consequences for aquatic crustaceans and consumers. Herein, we report that a natural antimicrobial mixture (A3009) was capable of significantly reducing the virulence of V. parahaemolyticus strains A3 and D4 in an in vitro infection model, using the fish cell line CHSE-214, an effect which correlates with the bacterial downregulation of hcp1 and hcp2 gene expression and with the ability of the antimicrobial to efficiently retain low cytotoxic levels (p < 0.001). We show for the first time that a natural antimicrobial is able to significantly reduce the mortality of shrimps in a challenge experiment and is able to significantly attenuate H2O2 release during infection (p < 0.001), indicating that it could harbor positive intestinal redox balance effects.
1. Introduction
V. parahaemolyticus is a Gram-negative bacterium, halophilic in nature with a typically rigid curve or straight non-spore forming rod shape [1]. The bacteria of the Vibrio genus are commonly recognised as inhabitants of fresh and marine waters, with over 100 species being described so far [2]. Bacterial pathogens commonly elicit an inflammatory response upon infection; however, V. parahaemolyticus has not been shown to do so [3]. V. parahaemolyticus is a toxin producing bacterium, in which the toxins are controlled by the genes pirA and pirB and together are responsible for acute hepatopancreatic necrosis disease (AHPND) [4]. Individually, the toxins appear to be incapable of inducing AHPND and both genes must be present for induction to take place [5]. This syndrome has proven to be a frequent cause of early mortality syndrome (EMS) in the US and many Asian countries [6]. AHPND induces severe atrophy of aquatic crustaceans [7] and has been responsible for considerable aquaculture production losses [8]. Studies have endeavoured to determine the causative agent, collectively identifying the primary pathogen as V. parahaemolyticus [3].
The Type VI secretion systems (T6SS) are associated with severe forms of gastrointestinal diseases and have been described mostly in Gram-negative bacteria [9]. Bacteria use these secretion systems to eliminate competing microorganisms by transporting toxins into target bacteria or eukaryotic cells, leading to a reduction in growth and ultimately death [10]. Coincidentally, the T6SS was first discovered in Vibrio cholerae [11] and it was characterised by being responsible for the secretion of haemolysin protein (hcp), an important virulence factor in many bacteria, including Pseudomonas aeruginosa [12]. Infecting human gastrointestinal cells lines (HCT-8) with Campylobacter spp. highlighted that the T6SS system is involved via the downregulation of hcp gene expression in the reduction of the bacterium’s ability to adhere and infect in vitro [13]. The same study also showed that the effect was extrapolated in vivo when chicken broilers where challenged with Campylobacter spp. In V. parahaemolyticus, the hcp haemolysin signature gene has two proteins, hcp1 and hcp2, which interestingly are expressed under different growth conditions mainly related to salt concentration (1.5–3% NaCl) [14]. The differential expression of hcp1 and hcp2 and their dependency on the salt concentration seems to be related to the ability of the pathogen to compete for a niche, especially in a marine environment, and to enhance their virulence ability [14].
V. parahaemolyticus detection strategies are continually being developed in an attempt to reduce the risk of infection via seafood consumption [15,16]. Various other control and prevention measures have also been proposed to limit the growth of V. parahaemolyticus in contaminated seafood and prevent potential outbreaks [17]. However, due to its ubiquitous nature in marine environments and relatively low infectious dose, bacteria-free seafood is seemingly unattainable [18]. Antibiotics such as tetracycline are used extensively in aquaculture in an attempt to improve food safety [19]. Despite their efficiency and low cost, antibiotic overuse has become associated with increasing levels of drug resistance and the establishment of resistance determinants [20]. The large-scale disinfection of water prior to stocking is a current practice to remove potential pathogens. This practice causes a considerable ecosystem disturbance by increasing the nutrient availability post-disinfection; destabilising the microbial community and in turn encouraging environmental re-colonisation with fast-growing bacteria, such as the pathogenic Vibrio species [21].
This study aimed to investigate how a mixture of natural antimicrobials could regulate the growth of V. parahaemolyticus strains A3 and D4 in vitro and in vivo and identify the virulence factors involved. We also aimed to investigate their potential to attenuate V. parahaemolyticus virulence by using sub-lethal concentrations of the antimicrobial mixture as there is a constant concern that bacteria could also gain resistance to natural antimicrobials [22].
2. Materials and Methods
2.1. Bacterial and Eukaryotic Cells Lines and Antimicrobials
The Vibrio parahaemolyticus A3 (origin Vietnam) and D4 (origin Mexico) strains were kindly donated by Kim Orth from the Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX, USA. Both strains were routinely cultured in marine Luria–Bertani (MLB) broth (Luria–Bertani broth containing 1.5% or 3.5% NaCl). The CHSE-214 cells were obtained from the European Collection of Authenticated Cell Cultures (ECACC). The CHSE-214 cells (ECACC No. 91041114) are a fish cell line derived from Oncorhync hustshawytscha embryos. They were cultured in minimum essential medium (MEM) (Gibco, Oxford, UK) supplemented with 10% foetal bovine serum (Life Technologies, Oxford, UK), 2 nM l-glutamine (Corning, Oxford, UK), and an antibiotic/antimycotic solution (10,000 IU/mL penicillin, 10,000 μg/mL streptomycin and 25 μg/mL amphotericin B). A mixture of antimicrobials containing lactic acid, E330 citric acid and citrus extract was used. The composition (per kg) of the feed material was sodium chloride (1 g) and maltodextrin. The additives (per kg) were preservatives, including lactic acid and E330 citric acid, and the flavouring was citrus extract. The analytical constituents (%) were crude ash (0.2%), crude protein (0.84%), crude oils and fats (10.83%), crude fibre (0.03%), phosphorus (0.01%), sodium (1.12%), calcium (1.34%), lysine (0%) and methionine (0%). The antimicrobial product (Auranta 3009, Nova UCD, Ireland) was obtained from Envirotech Innovative Products Ltd.
2.2. Determination of Minimum Inhibitory Concentrations
The minimum inhibitory concentration (MIC) and minimum microbicidal concentration (MBC) were determined for two strains of acute hepatopancreatic necrosis disease (AHPND)-causing Vibrio parahaemolyticus, strains A3 and D4. The strains were grown overnight at 37 °C in nutrient broth (Oxoid, Basingstoke, UK) containing 1.5% or 3% NaCl. The resulting stationary phase cultures were diluted using a nutrient broth containing 1.5% or 3% NaCl, to give a suspension containing approximately 6 log CFU/mL A series of Auranta 3009 solutions were prepared in a nutrient broth containing 1.5% or 3% NaCl, to give a range of concentrations from 4% down to 0.015625%. One ml of each Auranta 3009 solution was transferred into separate sterile plastic bijou bottles and 1 mL of either strain of V. parahaemolyticus was added to each bottle. A positive control containing 1 mL of nutrient broth and 1 mL of the 6 log CFU/mL suspension was prepared. The negative control was 2 mL of uninoculated nutrient broth plus salt. The mixtures were incubated aerobically at 37 °C for 24 h. This procedure was repeated on three separate occasions. The MIC value was determined as the lowest concentration of Auranta 3009 that showed no bacterial growth. After 24 h, the bijou bottles were observed for bacterial growth. If the broth was clear, indicating no bacterial growth, then 100 µL was spread-plated onto nutrient agar containing 1.5% or 3% NaCl using a sterile glass spreader. This was repeated for all concentrations of Auranta 3009 that appeared clear in the bijou container. All plates were incubated aerobically at 37 °C for 24 h. This procedure was repeated on three separate occasions for both V. parahaemolyticus strains A3 and D4. The MBC value was determined as the lowest concentration of Auranta 3009 that showed no bacterial growth on the plates.
2.3. Growth Curves
Growth curves were established for both strains of V. parahaemolyticus; A3 and D4. Broth cultures were prepared in a nutrient broth containing 1.5% or 3% NaCl and incubated overnight at 37 °C. These were diluted using nutrient broth containing 1.5% or 3% NaCl, to give a final concentration of approximately 6 log CFU/mL. The MIC value for both V. parahaemolyticus A3 and V. parahaemolyticus D4 was previously determined to be 0.0625% of Auranta 3009. An Auranta 3009 solution was prepared using a nutrient broth containing 1.5% or 3% NaCl to give a final concentration of 0.0625% when the suspension of bacteria was added. Two-fold dilutions of this solution were prepared in a 96-well plate giving a range of Auranta 3009 concentrations from the MIC (0.0625%) down to 1/8 of the MIC concentration (0.0078125%). The final volume of Auranta 3009 solution in each well was 90 µL. The bacterial suspension (10 µL) was added to each well and thoroughly mixed. Nutrient broth containing 1.5% or 3% NaCl with no Auranta 3009 added was inoculated with the bacterial suspension as a positive control. The negative control was uninoculated nutrient broth containing 1.5% or 3% NaCl. The 96-well plate was sealed with plastic film and the optical density was measured at 600 nm at intervals of 1 h over a 24 h period at 37 °C using a FLUOstar Omega automatic plate reader (BGM Labtech, Aylesbury, UK). This procedure was repeated on three separate occasions for both Vibrio parahaemolyticus A3 and D4.
2.4. Challenge Tests (Counting Living Larvae)
V. parahaemolyticus strains A3 and D4 were selected to be tested for their pathogenicity by a challenge test using healthy Penaeus vannamei post larvae, following a procedure previously described [23]. Twenty-five shrimp post larvae per replicate were plated in sterile petri dishes and exposed to infection with 109/mL bacteria for 10 min. The antimicrobial mixture was applied at the time of infection in concentrations of 0.0625%, 0.031%, 0.015% and 0.007% in 500 mL flasks. Survival was determined by counting the larvae at 30 h after infection. A positive and a negative control (± antimicrobial mixture or ± larvae) was also included in the challenge at 0% of the antimicrobial mixture. The experiment was performed in triplicate.
2.5. H2O2 Production in CHSE-214 Cells Response to Treatment Following Infection
A culture mixture of V. parahaemolyticus strains A3 and D4 was prepared as described above. The CHSE-214 cells were grown in a minimum essential medium (Minimum Essential Media (MEM) -Gibco®, Waltham, MA, USA) until they reached confluence and were supplemented with 10% fetal bovine serum and 2 mL glutamine. The cells were routinely grown in 75 cm2 tissue culture flasks (Sigma-Aldrich, St. Louis, MO, USA), in a humidified incubator at 37 °C with 5% CO2. The H2O2 production from infected and un-infected CHSE-214 in response to treatment with Auranta 3009 at was measured using the PeroxiDetect™ Kit (Sigma-Aldrich, St. Louis, MO, USA), following manufacturer guidelines and as described in [24].
2.6. In Vitro Adhesion and Infection Assay on CHSE-214 Cell Line and qRT-PCR for hcp1 and hcp2 Expression
To confirm the efficacy of Auranta 3009 in reducing the adhesion and invasion abilities of V. parahaeomolyticus strains A3 and D4 the isolates were grown to the OD600 of 0.5. The CHSE-214 were cultured as described above. Prior to the infection of the cells, the cell monolayers were incubated for 3 h with tissue culture media containing 0.031% Auranta 3009 followed by washing three times with the tissue culture media, and then infected for 3 h with 104 CFU/mL V. parahaemolyticus strains A3 and D4. After infection, the infection media was removed and the infected monolayers were washed three times with the tissue culture media. The infected cells were then incubated with tissue culture media containing gentamicin (100 µg/mL) in order to expose the internalised bacteria or to total lysis without gentamicin inclusion, but, instead, 0.1% Triton X was included to reveal the total bacterial adhesion. In order to determine the role of hcp1 in marine conditions (3% NaCl), in V. parahaeomolyticus infection, we have co-cultured the microorganism in nutrient broth containing 1.5% or 3% NaCl in the presence of 0.031% Auranta 3009 for 3 h. The co-cultured microorganisms were then used in the infection assay at an MOI of 100. Similarly, the experiment was also performed to investigate the effect of hcp2 expression, but this time with bacteria grown in low salt concentrations (1.5% NaCl). The bacteria grown in the above conditions was also used to determine the hcp1 and hcp2 gene expression in the above conditions. The primers used to detect hcp1 were: cacgtgacggctcggtgg and ctcttctttcgcgtcttggtcg and for hcp2 cgagtatccactcgaaactttc and ttctgctccctcagtactttctg [14]. The RNA was reverse transcribed using Transcriptor First Strand cDNA Synthesis Kit (Roche, United Kingdom) according to the manufacturer’s protocols. The mRNA levels were determined by quantitative RT-PCR using QuantiNovaSYBR® Green PCR Kit (Qiagen, Manchester, UK) on a LightCycler® 96 (Roche, Buckinghamshire, UK). A total of 5 μL of SYBR Green master mixture was used in each reaction, along with 0.5 μL of 10 μM primer mixture, 3 μL of molecular grade water, and 1 μL of DNA sample. For hcp 1 and 2 (2 min at 95 °C, followed by 40 cycles of 95 °C for 5 s, 60 °C for 10 s, and a final extension at 72 °C for 5 min), a total of 5 μL of SYBR Green master mixture was used in each reaction along with 0.8 μL of 20 μM primer mixture, 7.4 μL of molecular grade water, and 1 μL of DNA sample. The relative quantity of the mRNA was calculated using the ΔCt method. The rARN 16S gene was used as an endogenous control since it was transcribed in equal rates in both the treated and untreated cells.
2.7. Cytotoxicity Assay
The CHSE-214 cells were infected as described above over a 4 h time course with V. parahaemolyticus strains A3 and D4. The direct impact of Auranta 3009 was investigated by pre-treatment of CHSE-214 cells with 0.031% Auranta 3009 for 3 h. After 3 h, the media was removed, the cells were washed 3× with fresh MEM, followed by infection with V. parahaemolyticus strains A3 and D4 at an MOI of 100. The CHSE-214 cells were also infected with pre-treated bacteria. Uninfected cells exposed to 0.031% of Auranta 3009 were used as controls. Lactate dehydrogenase release (LDH) was measured with a cytotoxicity detection kit (Roche, Buckinghamshire, UK) following the manufacturer’s instructions. The results are expressed as cytotoxicity calculated as a percentage of the total lysis of cells lysed in Triton X 100.
2.8. Motility Assay
Motility was assessed as previously described [25]. Briefly, cultures of V. parahaemolyticus strains A3 and D4 were co-cultured in nutrient broth containing 1.5% NaCl or 3% NaCl in the presence of 0.031% Auranta 3009 for 3 h. The co-cultured organisms where then placed (~1 × 106 CFU) at the centre of the agar plate. The diameter of the motility area was measured. Non-co-cultured strains were used as a control. The experiments were performed in triplicate and on three separate occasions.
3. Results
3.1. The Role of Antimicrobial Mixtures in the Growth and Survival of V. parahaemolyticus Strains A3 and D4
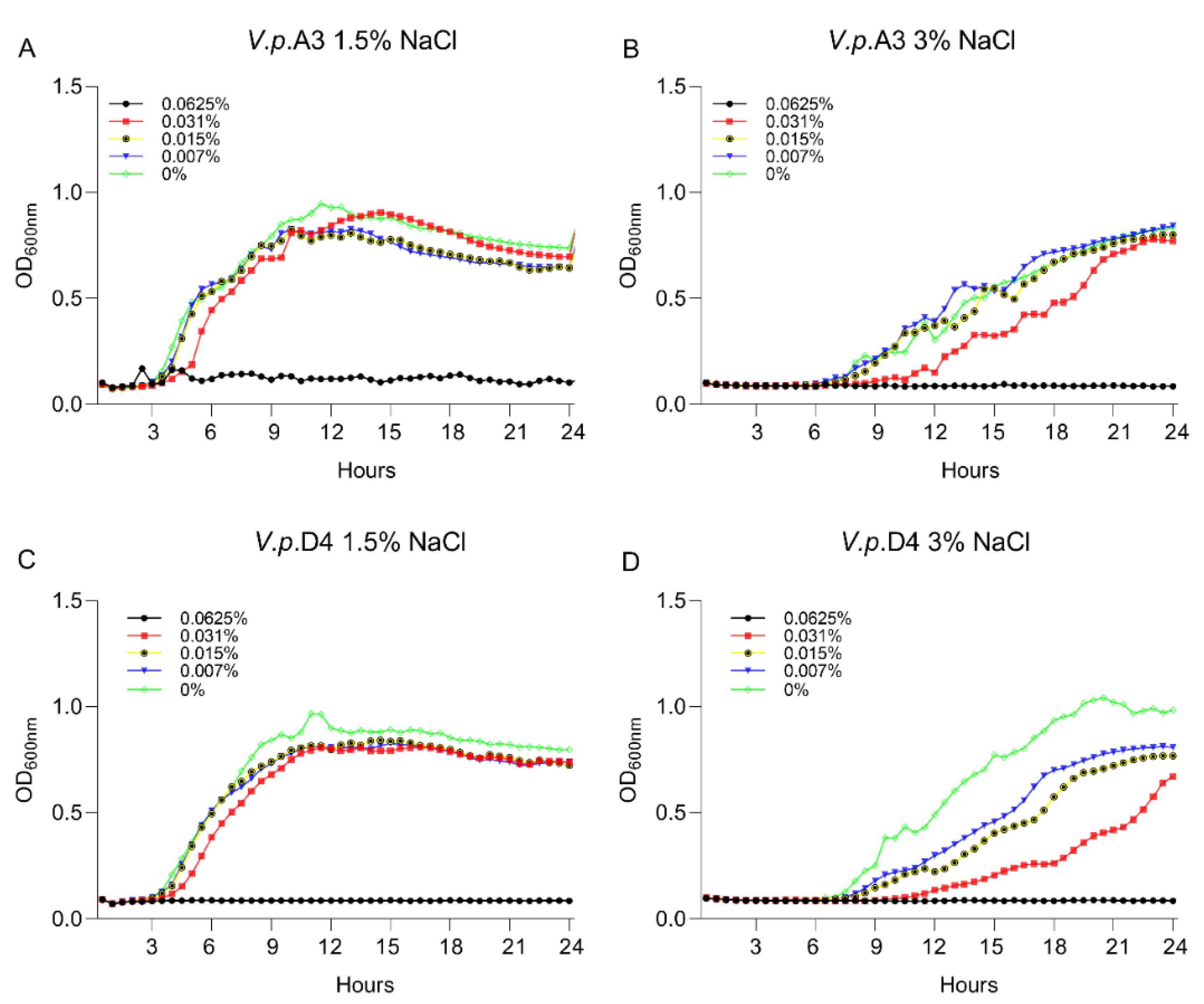
First, we aimed to gain insight into the growth characteristics of V. parahaemolyticus strains A3 and D4 in the presence of 1.5% and 3% NaCl. The experiment was designed to identify the concentration at which the antimicrobial mixture has no lethal effect on the V. parahaemolyticus strains A3 and D4. This concentration was required to allow us to investigate the possible alteration of bacterial virulence factors while the pathogens are still alive. The MIC and MBC values at levels of 1.5% and 3% NaCl against V. parahaemolyticus A3 and V. parahaemolyticus D4 are shown in Table 1. In order to identify the effect of the antimicrobial mixture on the bacterium virulence factors, we used various concentrations and identified that at half ratio (1/2) of the highest MBC (0.031%), the antimicrobial mixture had no lethal effect; however, it disrupted the growth of both strains (Figure 1). Firstly, we have concluded that the salt concentration had no significant effect on the MIC and MBC values and, secondly, that the concentration of 0.031% of the antimicrobial mixture can be used further in our investigations to assess the implications in virulence.

Table 1.
MIC and MBC antimicrobial mixture concentrations required for V. parahaemolyticus A3 and D4 inactivation.
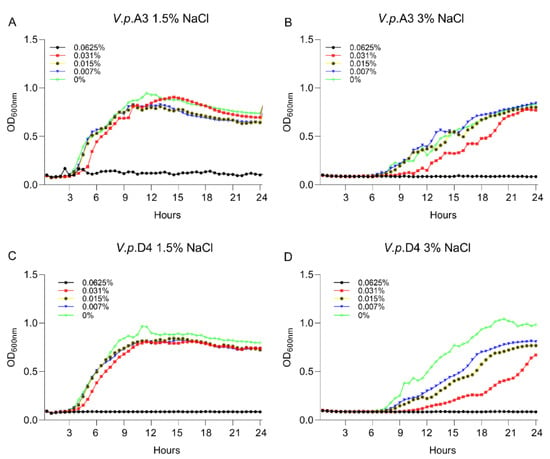
Figure 1.
The impact of A3009 on the growth and survival of Vibrio parahaemolyticus A3 (A)—1.5% NaCl, (B)—3% NaCl and D4 (C)—1.5% NaCl, (D)—3% NaCl. The MIC concentrations are indicated on the graphs. The experiments were performed in triplicate and on three separate occasions. In order to quantify the growth, the absorbance was measured at 600 nm every 0.5 h for 24 h.
3.2. Motility
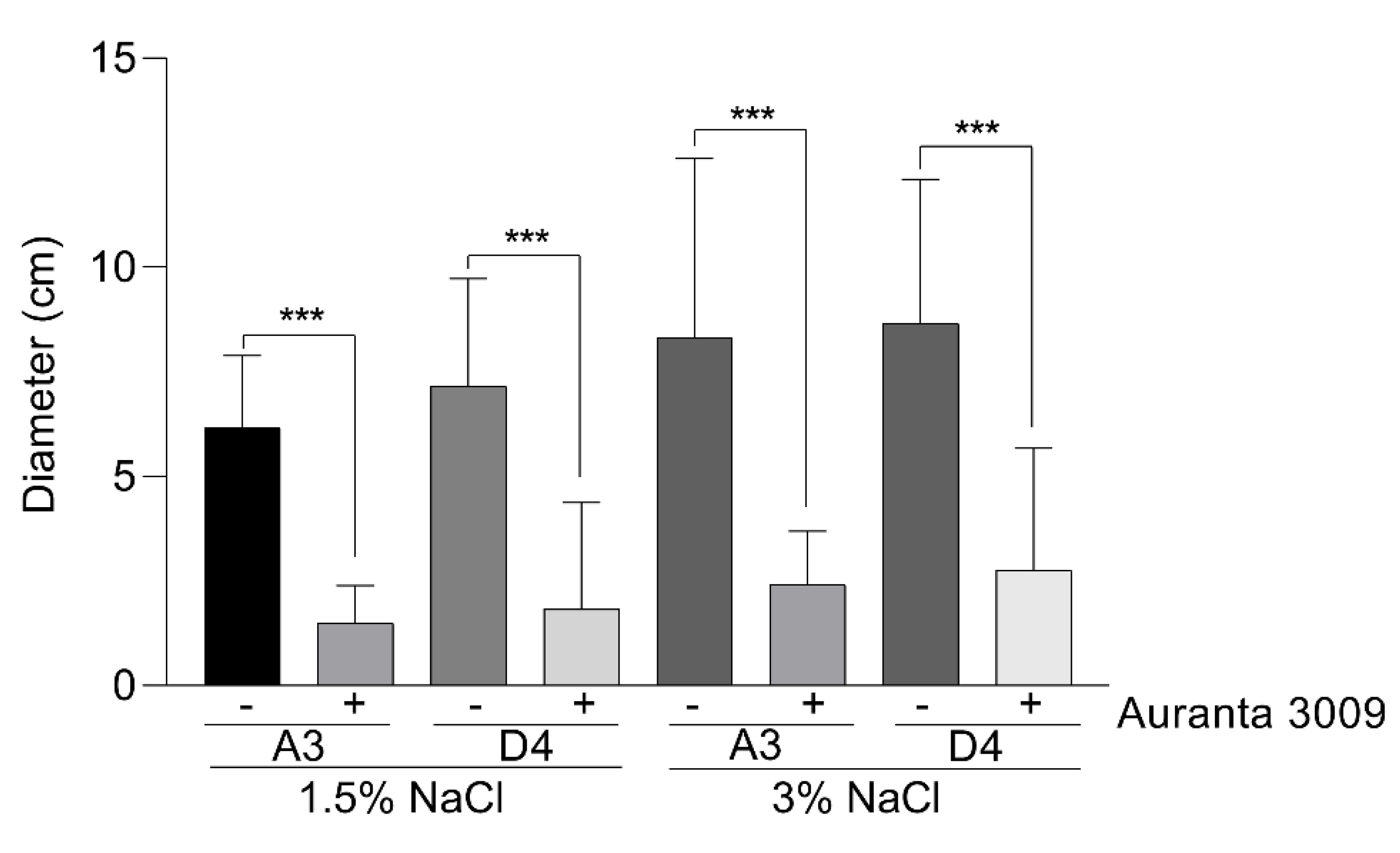
We next investigated the potential impact of the antimicrobial mixture on the V. parahaemolyticus A3 and D4 motility, a major virulence factor, at the sub-inhibitory concentration calculated at half of the MIC (0.031%) in the presence of 1.5% and 3% NaCl (Figure 2). The motility plate assays clearly showed that both strains are generally very motile, but the pre-treatment with 0.031% Auranta 3009 significantly reduced motility, independent of the NaCl2 concentration (∗∗∗ p < 0.001). These results suggested that the antimicrobial mixture reduces the bacterial motility and potentially has an impact on virulence.
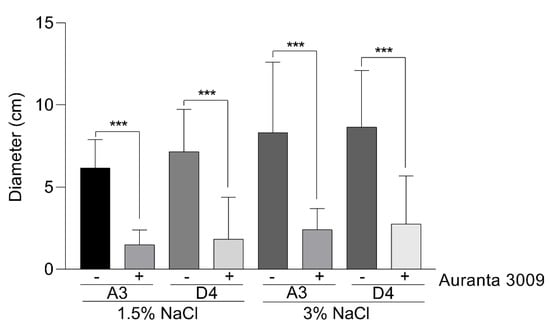
Figure 2.
The effect of 0.031% (concentration) Auranta 3009 on V. parahaemolyticus A3 and D4 motility. Asterisks indicate significant differences (∗∗∗ p < 0.001). Error bars represent the standard deviation of means from three different experiments, each experiment being performed in triplicate.
3.3. The Impact of Auranta 3009 on the hcp1 and hcp2 Gene Expression at 1.5% and 3% NaCl
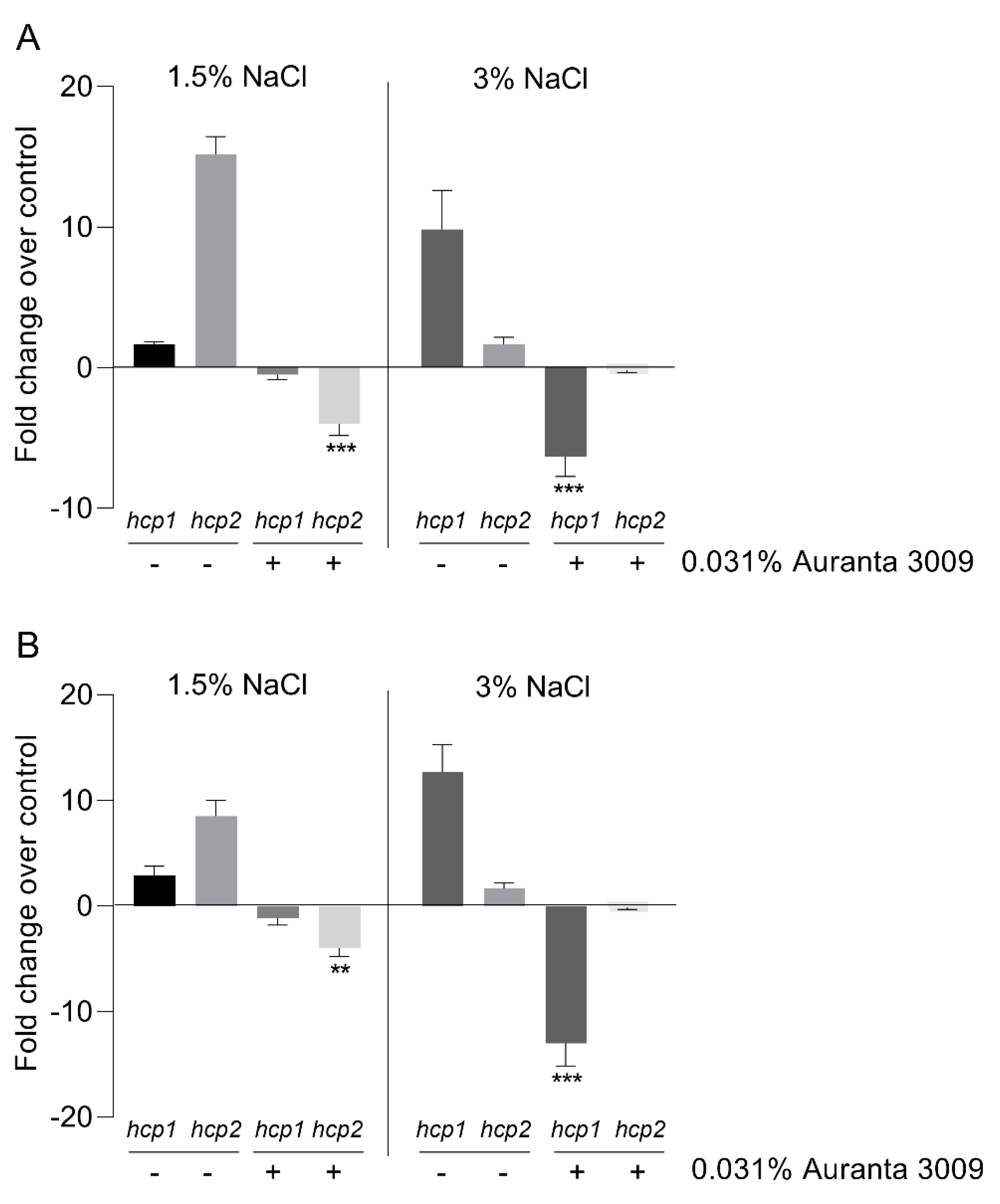
The effect of the antimicrobial mixture on V. parahaemolyticus A3 and D4 hcp gene expression is shown in Figure 3. The qRT-PCR results indicate a significant downregulation (∗∗∗ p < 0.001) in hcp1 gene expression levels in the presence of 3% NaCl in V. parahaemolyticus A3, with no effect on hcp2 at this salt concentration. The downregulation effect on hcp2 was only observed in the presence of 1.5% NaCl, as indicated in Figure 3 (panel A) (∗∗∗ p < 0.001). The results were similar for V. parahaemolyticus D4 in regard to the downregulation effect on hcp1 (∗∗ p < 0.01) and hcp2 (∗∗∗ p < 0.001) gene expression produced by Auranta 3009 at 1.5% and 3% NaCl (Figure 3, panel B).
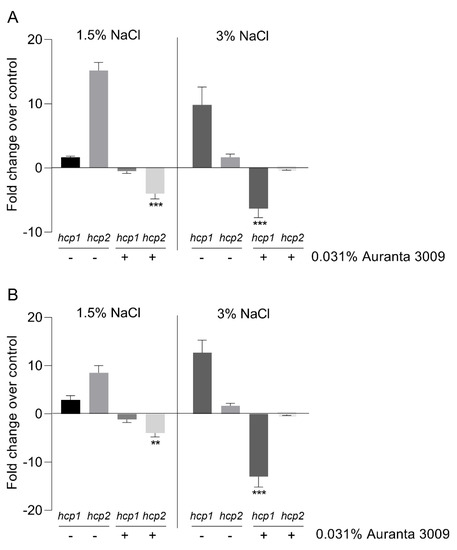
Figure 3.
Effect of the antimicrobial mixture Auranta 3009 on V. parahaemolyticus A3 and D4 hcp1 and hcp2 gene expression in the presence of 1.5% and 3% NaCl. V. parahaemolyticus A3 (A) and D4 (B). Asterisks indicate significant differences (Student’s t-test ∗∗ p < 0.01, ∗∗∗ p < 0.001). Error bars represent the standard deviation of means from three different experiments performed in triplicate.
3.4. Infection Assay
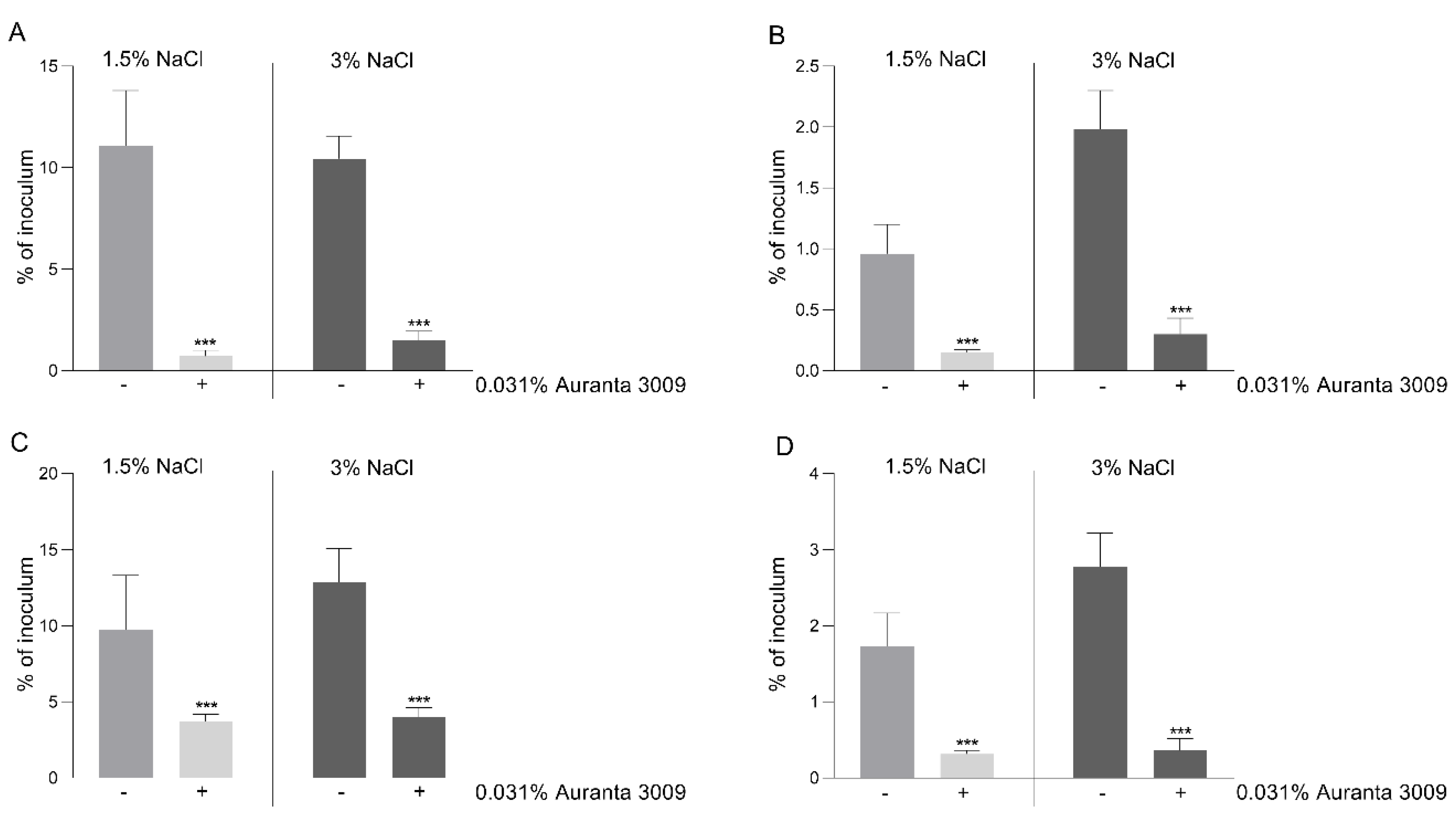
In order to correlate the negative effect on motility and a possible decrease in virulence, we performed in vitro infection assays using the CHSE-214 cell line as described in Materials and Methods. The pre-treatment of V. parahaemolyticus A3 (Figure 4, panel A) and D4 (Figure 4, panel C) with 0.031% of Auranta 3009 significantly reduced the adherence of both strains (p < 0.001). The impact on bacterial internalisation was as significant as for adhesion for both strains, as indicated in Figure 5 panel B for V. parahaemolyticus A3 and panel D for V. parahaemolyticus D4 (p < 0.001). When the antimicrobial mixture was only present in the tissue culture media during the adhesion/invasion assay, the reductions followed the same pattern (data not shown). These results indicate that the reduced in vitro infection abilities of V. parahaemolyticus A3 and D4 follow a similar pattern and in concordance with the reduction in bacterial motility.
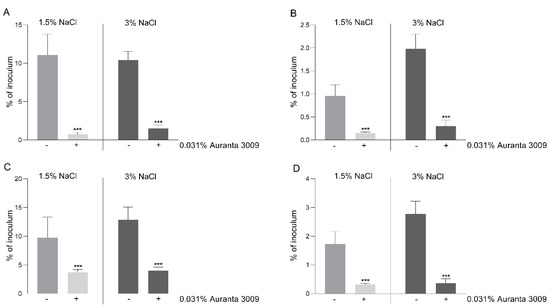
Figure 4.
Adhesion and invasion of CHSE-214 cells by V. parahaemolyticus A3 and D4. The adherence (A) and invasion (B) of V. parahaemolyticus A3 to CHSE214 cells in the presence of Auranta 3009. Panel C shows the adherence of V. parahaemolyticus D4 and panel D the invasion levels of D4 to CHSE-214 cells. The results are expressed as percentages of the initial inoculum. Asterisks indicate significant differences (Student’s t-test ∗∗∗ p < 0.001). Error bars represent the standard deviation of means from three different experiments, each containing triplicate samples.
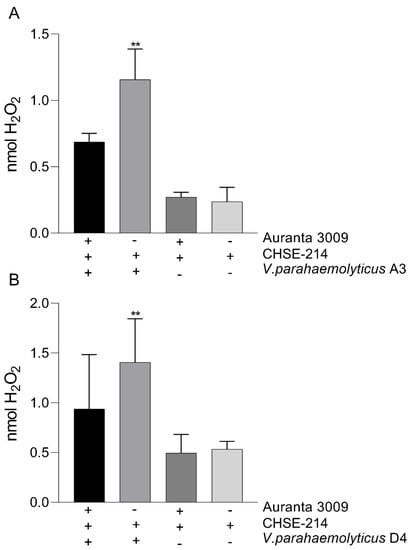
Figure 5.
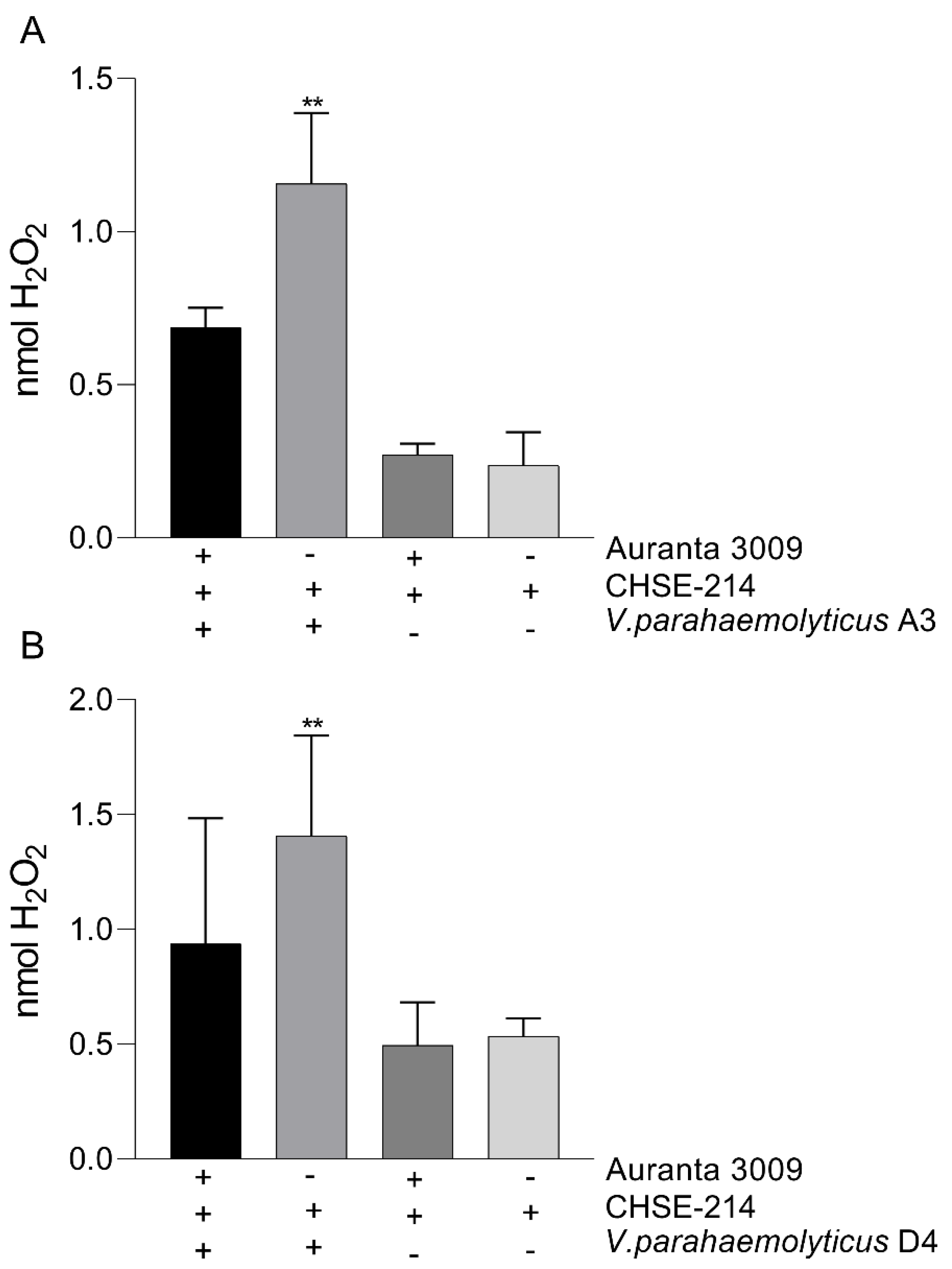
The impact of Auranta 3009 on the H2O2 production during 3 h co-culture of Vibrio parahaemolyticus A3 (A) and D4 (B) with CHSE-214 cells. The experiments were performed in triplicate and on three separate occasions. Student t test was used to analyse the impact of Auranta 3009 on H2O2 release (** p < 0.01).
3.5. Challenge Tests
To estimate the efficiency of the antimicrobial mixture we performed challenge studies by infecting P. vannamei shrimps with V. parahaemolyticus strains A3 and D4. Our results show that at the MIC/MBC concentration of the antimicrobial (0.0625%) the lowest shrimp mortality was recorded (Table 2) for both V. parahaemolyticus strains A3 and D4. The challenge test confirmed the results obtained following the MIC/MBC and growth curve experiments, indicating the efficiency of the antimicrobial mixture. The percentage of mortality decreased as the concentration of the antimicrobial mixture increased. These results clearly indicated that the antimicrobial mixture has potential of protecting the shrimp cultures in vivo.

Table 2.
Mortality of Penaeus vannamei after 45 h of challenge after infection with. V. parahaemolyticus A3 and D4.
3.6. In vitro Epithelial H2O2 Release in Response to Treatment
We next investigated the impact of V. parahaemolyticus A3 and D4 infection on H2O2 release by the CHSE-214 in the presence of Auranta 3009. Our results demonstrate that the amount of H2O2 released in the media during infection is significantly reduced (p < 0.001) by the presence of Auranta 3009 (Figure 5). No effect was identified when the antimicrobial mixture was applied to un-infected control as the cells released H2O2 levels similar to the control.
3.7. Cytotoxicity
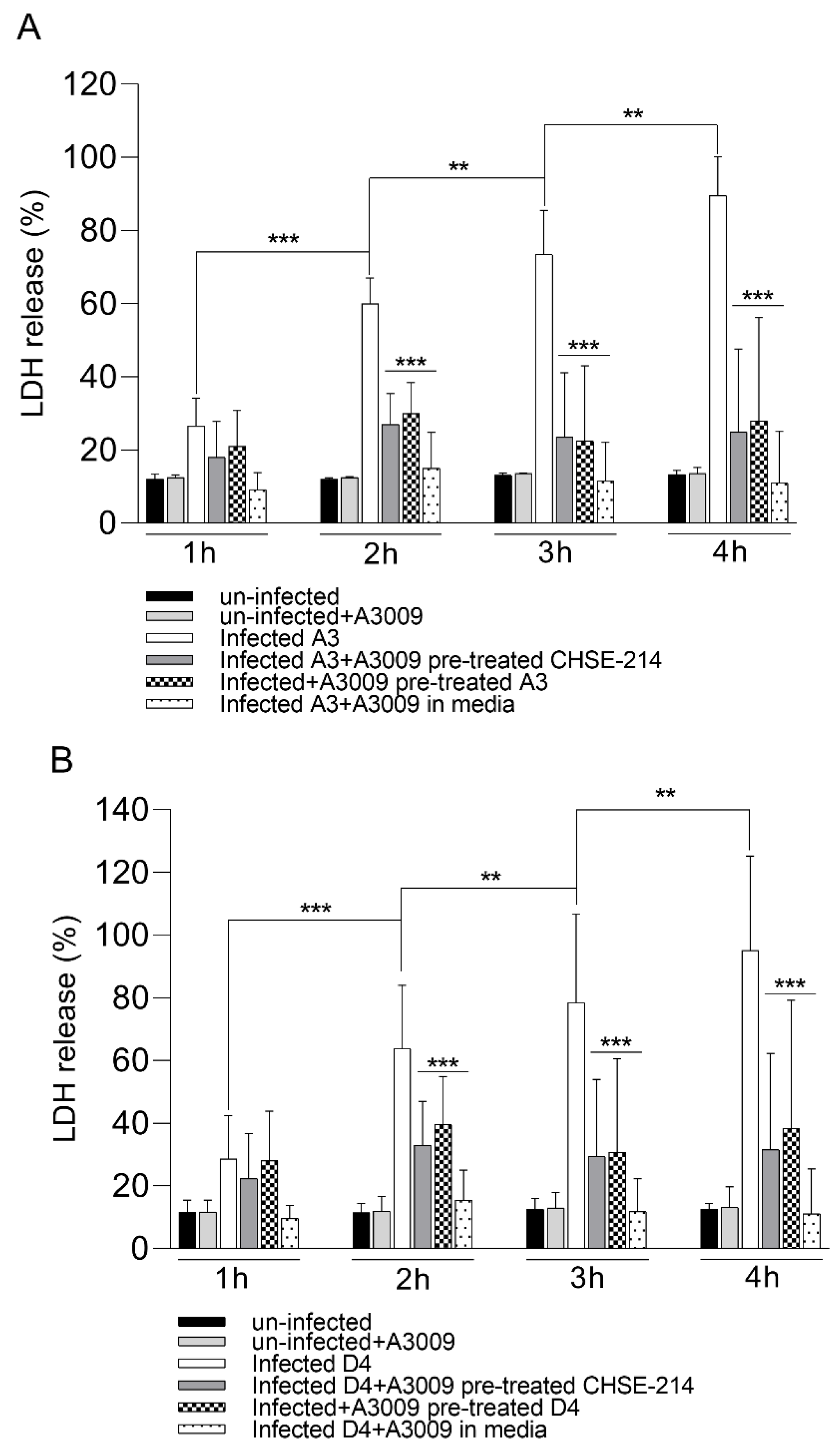
Following exposure to the 0.031% antimicrobial mixture, the infected CHSE-214 cells showed a clear reduction in cytotoxicity during infection with V. parahaemolyticus A3 (Figure 6, panel A) and D4 (Figure 6, panel B). The cytotoxic levels recorded for infected and un-treated cells reached over 80% at 4 h post-infection, compared to a maximum of 25% at 2 h post-infection. The highest anti-cytotoxic effect was recorded when the antimicrobial mixture was present in the culture media at all time points during infection. These results clearly show that the pre-treatment of the bacteria or of the epithelial cells prior to infection will have a significant role in keeping the cytotoxicity levels reduced (∗∗∗ p < 0.001).
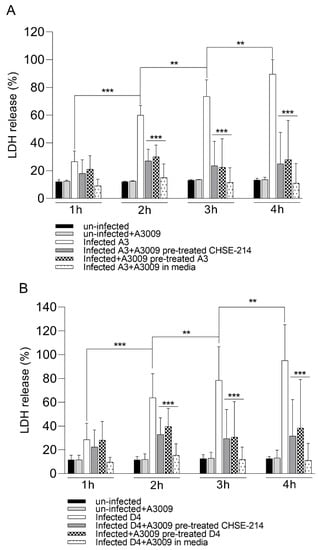
Figure 6.
Cytotoxicity of the antimicrobial mixture to CHSE-214 cells as determined following 4 h of exposure to V. parahaemolyticus A3 (pannel A) and D4 (panel B) by LDH release. Data expressed as a percentage of unexposed controls of three replicates for each exposure concentration. (Student’s t-test ∗∗ p < 0.01, ∗∗∗ p < 0.001). Error bars represent the standard deviation of means from three different experiments performed in triplicate.
4. Discussion
Natural antimicrobials are known to be effective in reducing bacterial pathogenicity via direct [13] or indirect [26] anti-virulence activity. For example, the surface proteins involved in the virulence of Staphylococcus aureus are inhibited by venom peptides from being promising candidates for the treatment of related infections caused by multi-drug resistant bacteria [27] Quorum-sensing molecules originating from marine bacteria can control virulence, with gene expression being described as a novel antimicrobial strategy in aquaculture [28]. Another mechanism by which natural antimicrobials can reduce virulence refers to their involvement in bacterial physiology, as it has been shown that phenolic compounds can disrupt the expression and function of efflux pumps [29]. They can also reduce the ability of bacterial pathogens to adhere to epithelial cells in vitro (e.g., Daphnetin, a coumarin-derivative) by reducing the expression of H. pylori genes involved in colonisation [30]. Antimicrobial mixtures of probiotics, organic acids and essential oils were effective in reducing the devastating impact of Vibrio strain infections in South American farmed shrimps [31]; however, the study did not elucidate the biological mechanism responsible for the effect.
V. parahaemolyticus is often found in estuaries and coastal waters and because it causes severe disease is also of significant importance as a human pathogen [32]. Phage and probiotic treatments were proposed as alternative method to control infections in humans and aquaculture [23,33]. Constant and un-controlled application of increased concentrations of antibiotics in aquatic environments lead to increased resistance in the bacterial populations (e.g., V. parahaemolyticus), posing a threat to human and animal health and is of major concern to fish and shellfish farmers [34,35]. Early mortality syndrome is caused by infection with V. parahaemolyticus and strains A3 and D4 [36] were previously reported to affect shrimp farming by causing the acute hepatopancreatic necrosis disease (AHPND) [37]. V. parahaemolyticus strains A3 and D4 [3] acquired the type VI secretions system (T6SS), offering them a competitive advantage in survival and virulence over other bacteria in the shrimp ecosystem [7]. This development increases the need for effective control measures to prevent infection of fish and shellfish farms, especially by preventing the multiplication and virulence of V. parahaemolyticus in such environments [38].
Natural antimicrobials offer such an opportunity by being known to affect bacterial cell membranes [39]. One such example is lavender extract, which has previously been shown to reduce the ability of V. parahaemolyticus to produce and release its toxin [40]. The anti-virulent effect of natural antimicrobial mixtures was previously described in other pathogens e.g., Campylobacter jejuni, Campylobacter coli and Shiga Toxin Producing E. Coli (STEC) [13,41]. This anti-virulent effect was exhibited by decreasing the activity of major virulence factors including motility, and by reducing the in vitro and in vivo infection abilities, T6SS and biofilm formation [13,42].
According to our results, such mixtures can be used efficiently in both fresh and marine water environments, as we have observed similar results in low and high salt conditions. Similar antimicrobial mixtures have been previously used to prevent the bacterial infection of epithelial cells, both in vitro and in vivo [13], through the inactivation of virulence factors. The link between T6SS inactivation and decreased virulence has been previously shown, as it has been indicated that its presence enhances the adhesion to cultured cell monolayers [43]. This proposed hypothesis has been previously described for C. jejuni RC039, where the downregulation of the hcp gene was associated with reduced virulence both in vitro and in vivo [13].
In addition, V. parahaemolyticus A3 and D4 appear to stimulate H2O2 release from human intestinal epithelial cells, possibly by altering the redox balance within the host’s lumen or by triggering the host NADPH oxidases. Microbiota-induced alterations to the lumen redox balance could have a modulatory effect upon pathogenicity, favoring the intestinal pathogen [24]. Therefore, our results indicate that a suitable antimicrobial mixture (e.g., A3009) could be used to maintain the intestinal redox balance by restricting H2O2 release. However, the presence of hydrogen peroxide during the interaction between V. parahaemolyticus and the host epithelium does not seem to be critical, since it is able to significantly express catalase enzymes and induce a detoxification process [44].
5. Conclusions
The data presented herein describe the effect of a mixture of antimicrobials (A3009) on the virulence and pathogenicity of T6SS V. parahaemolyticus A3 and D4 strains. The antimicrobials reduced the ability of V. parahaemolyticus A3 and D4 to infect the CHSE-214 fish cell line by inhibiting the expression of the T6SS hcp1 and hcp2 genes and through the reduction of bacterial motility in both low and high salt environments. These results corroborated with the antioxidant effect, low tissue cytotoxicity and the low mortality recorded in the in vivo challenge test, suggesting that this antimicrobial mixture (A3009) can be efficiently used to improve the survival of shrimp populations when subject to V. parahaemolyticus infections.
Author Contributions
Conceptualization, N.C.; Data curation, P.W., T.A. and N.C.; Formal analysis, L.P., M.L., C.K., G.G.P., I.P., L.S. and F.S.; Funding acquisition, N.C.; Investigation, O.G.; Methodology, M.L., C.K. and G.G.P.; Project administration, P.W. and N.C.; Resources, L.S.; Writing—original draft, O.G. and N.C.; Writing—review & editing, M.L., O.G. and N.C.
Funding
This study was supported by a grant awarded to NC by Auranta, Dublin, Ireland.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Letchumanan, V.; Chan, K.G.; Lee, L.H. Vibrio parahaemolyticus: A review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 2014, 5, 705. [Google Scholar] [CrossRef] [PubMed]
- Osunla, C.A.; Okoh, A.I. Vibrio Pathogens: A Public Health Concern in Rural Water Resources in Sub-Saharan Africa. Int. J. Environ. Res. Public Health 2017, 14, 1188. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.; Nunan, L.; Redman, R.M.; Mohney, L.L.; Pantoja, C.R.; Fitzsimmons, K.; Lightner, D.V. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Org. 2013, 105, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Kanrar, S.; Dhar, A.K. Complete Genome Sequence of a Novel Mutant Strain of Vibrio parahaemolyticus from Pacific White Shrimp (Penaeus vannamei). Genome Announc. 2018, 6. [Google Scholar] [CrossRef]
- Lee, C.T.; Chen, I.T.; Yang, Y.T.; Ko, T.P.; Huang, Y.T.; Huang, J.Y.; Huang, M.F.; Lin, S.J.; Chen, C.Y.; Lin, S.S.; et al. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. USA 2015, 112, 10798–10803. [Google Scholar] [CrossRef]
- Lai, H.C.; Ng, T.H.; Ando, M.; Lee, C.T.; Chen, I.T.; Chuang, J.C.; Mavichak, R.; Chang, S.H.; Yeh, M.D.; Chiang, Y.A.; et al. Pathogenesis of acute hepatopancreatic necrosis disease (AHPND) in shrimp. Fish Shellfish Immunol. 2015, 47, 1006–1014. [Google Scholar] [CrossRef]
- Li, P.; Kinch, L.N.; Ray, A.; Dalia, A.B.; Cong, Q.; Nunan, L.M.; Camilli, A.; Grishin, N.V.; Salomon, D.; Orth, K. Acute Hepatopancreatic Necrosis Disease-Causing Vibrio parahaemolyticus Strains Maintain an Antibacterial Type VI Secretion System with Versatile Effector Repertoires. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Flegel, T.W. Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 2012, 110, 166–173. [Google Scholar] [CrossRef]
- Corcionivoschi, N.; Gundogdu, O.; Moran, L.; Kelly, C.; Scates, P.; Stef, L.; Cean, A.; Wren, B.; Dorrell, N.; Madden, R.H. Virulence characteristics of hcp (+) Campylobacter jejuni and Campylobacter coli isolates from retail chicken. Gut Pathog. 2015, 7, 20. [Google Scholar] [CrossRef]
- Jana, B.; Salomon, D. Type VI secretion system: A modular toolkit for bacterial dominance. Future Microbiol. 2019. [Google Scholar] [CrossRef]
- Pukatzki, S.; Ma, A.T.; Sturtevant, D.; Krastins, B.; Sarracino, D.; Nelson, W.C.; Heidelberg, J.F.; Mekalanos, J.J. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 2006, 103, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Mougous, J.D.; Cuff, M.E.; Raunser, S.; Shen, A.; Zhou, M.; Gifford, C.A.; Goodman, A.L.; Joachimiak, G.; Ordonez, C.L.; Lory, S.; et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 2006, 312, 1526–1530. [Google Scholar] [CrossRef] [PubMed]
- Sima, F.; Stratakos, A.C.; Ward, P.; Linton, M.; Kelly, C.; Pinkerton, L.; Stef, L.; Gundogdu, O.; Lazar, V.; Corcionivoschi, N. A Novel Natural Antimicrobial Can Reduce the in vitro and in vivo Pathogenicity of T6SS Positive Campylobacter jejuni and Campylobacter coli Chicken Isolates. Front. Microbiol. 2018, 9, 2139. [Google Scholar] [CrossRef] [PubMed]
- Salomon, D.; Gonzalez, H.; Updegraff, B.L.; Orth, K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS ONE 2013, 8, e61086. [Google Scholar] [CrossRef]
- Jones, J.L.; Hara-Kudo, Y.; Krantz, J.A.; Benner, R.A., Jr.; Smith, A.B.; Dambaugh, T.R.; Bowers, J.C.; Depaola, A. Comparison of molecular detection methods for Vibrio parahaemolyticus and Vibrio vulnificus. Food Microbiol. 2012, 30, 105–111. [Google Scholar] [CrossRef]
- Jung, S.W. A foodborne outbreak of gastroenteritis caused by Vibrio parahaemolyticus associated with cross-contamination from squid in Korea. Epidemiol. Health 2018, 40, e2018056. [Google Scholar] [CrossRef]
- Su, Y.C.; Liu, C. Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol. 2007, 24, 549–558. [Google Scholar] [CrossRef]
- DePaola, A.; Kaysner, C.A.; Bowers, J.; Cook, D.W. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 2000, 66, 4649–4654. [Google Scholar] [CrossRef]
- Neela, F.A.; Nonaka, L.; Suzuki, S. The diversity of multi-drug resistance profiles in tetracycline-resistant Vibrio species isolated from coastal sediments and seawater. J. Microbiol. 2007, 45, 64–68. [Google Scholar]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- Attramadal, K.J.K.; Salvesen, I.; Xue, R.; Øie, G.; Størseth, T.R.; Vadstein, O.; Olsen, Y. Recirculation as a possible microbial control strategy in the production of marine larvae. Aquac. Eng. 2012, 46, 27–39. [Google Scholar] [CrossRef]
- Lu, M.; Dai, T.; Murray, C.K.; Wu, M.X. Bactericidal Property of Oregano Oil Against Multidrug-Resistant Clinical Isolates. Front. Microbiol. 2018, 9, 2329. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T.; Boon, N.; Sorgeloos, P.; Verstraete, W.; Bossier, P. Alternatives to antibiotics to control bacterial infections: Luminescent vibriosis in aquaculture as an example. Trends Biotechnol. 2007, 25, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Corcionivoschi, N.; Alvarez, L.A.; Sharp, T.H.; Strengert, M.; Alemka, A.; Mantell, J.; Verkade, P.; Knaus, U.G.; Bourke, B. Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe 2012, 12, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Packiavathy, I.A.; Priya, S.; Pandian, S.K.; Ravi, A.V. Inhibition of biofilm development of uropathogens by curcumin—An anti-quorum sensing agent from Curcuma longa. Food Chem. 2014, 148, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Tabashsum, Z.; Peng, M.; Bernhardt, C.; Patel, P.; Carrion, M.; Biswas, D. Synbiotic-like effect of linoleic acid overproducing Lactobacillus casei with berry phenolic extracts against pathogenesis of enterohemorrhagic Escherichia coli. Gut Pathog. 2019, 11, 41. [Google Scholar] [CrossRef]
- Younis, S.; Taj, S.; Rashid, S. Structural studies of Staphylococcus aureus Sortase inhibiton via Conus venom peptides. Arch. Biochem. Biophys. 2019, 671, 87–102. [Google Scholar] [CrossRef]
- Zhao, J.; Li, X.; Hou, X.; Quan, C.; Chen, M. Widespread Existence of Quorum Sensing Inhibitors in Marine Bacteria: Potential Drugs to Combat Pathogens with Novel Strategies. Mar. Drugs 2019, 17, 275. [Google Scholar] [CrossRef]
- Lima, M.C.; Paiva de Sousa, C.; Fernandez-Prada, C.; Harel, J.; Dubreuil, J.D.; de Souza, E.L. A review of the current evidence of fruit phenolic compounds as potential antimicrobials against pathogenic bacteria. Microb. Pathog. 2019, 130, 259–270. [Google Scholar] [CrossRef]
- Wang, G.; Pang, J.; Hu, X.; Nie, T.; Lu, X.; Li, X.; Wang, X.; Lu, Y.; Yang, X.; Jiang, J.; et al. Daphnetin: A Novel Anti-Helicobacter pylori Agent. Int. J. Mol. Sci. 2019, 20, 850. [Google Scholar] [CrossRef]
- Sotomayor, M.A.; Reyes, J.K.; Restrepo, L.; Dominguez-Borbor, C.; Maldonado, M.; Bayot, B. Efficacy assessment of commercially available natural products and antibiotics, commonly used for mitigation of pathogenic Vibrio outbreaks in Ecuadorian Penaeus (Litopenaeus) vannamei hatcheries. PLoS ONE 2019, 14, e0210478. [Google Scholar] [CrossRef] [PubMed]
- Raszl, S.M.; Froelich, B.A.; Vieira, C.R.; Blackwood, A.D.; Noble, R.T. Vibrio parahaemolyticus and Vibrio vulnificus in South America: Water, seafood and human infections. J. Appl. Microbiol. 2016, 121, 1201–1222. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.W.; Shin, T.H.; Kim, J.H.; Shin, S.P.; Han, J.E.; Heo, G.J.; De Zoysa, M.; Shin, G.W.; Chai, J.Y.; Park, S.C. Bacteriophage therapy of a Vibrio parahaemolyticus infection caused by a multiple-antibiotic-resistant O3:K6 pandemic clinical strain. J. Infect. Dis. 2014, 210, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Elmahdi, S.; DaSilva, L.V.; Parveen, S. Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: A review. Food Microbiol. 2016, 57, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wei, W.; Fu, G.; Zhou, J.; Wang, Y.; Li, X.; Ma, L.; Fang, W. Application of biofertilizers increases fluoroquinolone resistance in Vibrio parahaemolyticus isolated from aquaculture environments. Mar. Pollut. Bull. 2019. [Google Scholar] [CrossRef]
- Han, J.E.; Tang, K.F.; Tran, L.H.; Lightner, D.V. Photorhabdus insect-related (Pir) toxin-like genes in a plasmid of Vibrio parahaemolyticus, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp. Dis. Aquat. Org. 2015, 113, 33–40. [Google Scholar] [CrossRef]
- Velazquez-Lizarraga, A.E.; Juarez-Morales, J.L.; Racotta, I.S.; Villarreal-Colmenares, H.; Valdes-Lopez, O.; Luna-Gonzalez, A.; Rodriguez-Jaramillo, C.; Estrada, N.; Ascencio, F. Transcriptomic analysis of Pacific white shrimp (Litopenaeus vannamei, Boone 1931) in response to acute hepatopancreatic necrosis disease caused by Vibrio parahaemolyticus. PLoS ONE 2019, 14, e0220993. [Google Scholar] [CrossRef]
- Nishina, T.; Wada, M.; Ozawa, H.; Hara-Kudo, Y.; Konuma, H.; Hasegawa, J.; Kumagai, S. Growth kinetics of Vibrio parahaemolyticus O3:K6 under varying conditions of pH, NaCl concentration and temperature. Shokuhin Eiseigaku Zasshi 2004, 45, 35–37. [Google Scholar] [CrossRef][Green Version]
- Gonelimali, F.D.; Lin, J.; Miao, W.; Xuan, J.; Charles, F.; Chen, M.; Hatab, S.R. Antimicrobial Properties and Mechanism of Action of Some Plant Extracts Against Food Pathogens and Spoilage Microorganisms. Front. Microbiol. 2018, 9, 1639. [Google Scholar] [CrossRef]
- Maghsoudi, O.; Zeraatkar, M.; Dolatabadi, M.; Johari, A.; Barati Karizno, M.; Ranjbar, R. The Effect of Lavandula Stoechas on Toxigenesis and the Growth of Vibrio Parahaemolyticus. Iran. J. Pathol. 2018, 13, 245–255. [Google Scholar] [CrossRef]
- Stratakos, A.C.; Linton, M.; Ward, P.; Campbell, M.; Kelly, C.; Pinkerton, L.; Stef, L.; Pet, I.; Stef, D.; Iancu, T.; et al. The Antimicrobial Effect of a Commercial Mixture of Natural Antimicrobials Against Escherichia coli O157:H7. Foodborne Pathog. Dis. 2019, 16, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Stratakos, A.C.; Grant, I.R. Evaluation of the efficacy of multiple physical, biological and natural antimicrobial interventions for control of pathogenic Escherichia coli on beef. Food Microbiol. 2018, 76, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yang, H.; Li, J.; Zhang, P.; Wu, B.; Zhu, B.; Zhang, Y.; Fang, W. Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Arch. Microbiol. 2012, 194, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Fen, S.Y.; Chung, C.H.; Yu, S.C.; Chien, C.L.; Wong, H.C. Functions of VPA1418 and VPA0305 Catalase Genes in Growth of Vibrio parahaemolyticus under Oxidative Stress. Appl. Environ. Microbiol. 2016, 82, 1859–1867. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).