Potential Role for the Gut Microbiota in Modulating Host Circadian Rhythms and Metabolic Health
Abstract
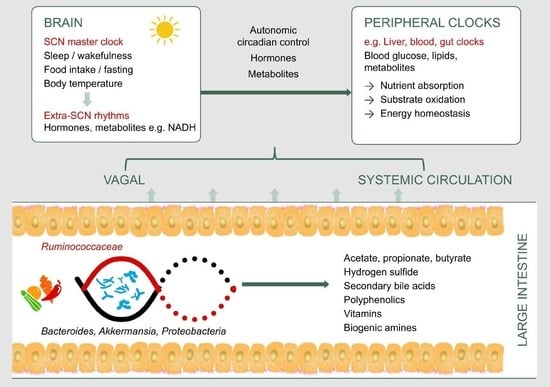
:1. Introduction
2. Factors Implicated in Gut Microbiome–Circadian Rhythm Interactions
2.1. Light/Dark Cycles
2.2. Sunlight Exposure
2.3. Sleep
2.4. Jet Lag
2.5. Diet and Dietary Patterns
3. Gut Microbiota as a Potential Pathway to Restore Circadian Rhythm and Metabolic Health
3.1. Secondary Bile Acids
3.2. Hydrogen Sulfide
3.3. Short Chain Fatty Acids
3.4. Vitamins
3.5. Biogenic Amines
3.6. Probiotics
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bass, J.; Takahashi, J.S. Circadian integration of metabolism and energetics. Science 2010, 330, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Oosterman, J.E.; Kalsbeek, A.; la Fleur, S.E.; Belsham, D.D. Impact of nutrients on circadian rhythmicity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 308, R337–R350. [Google Scholar] [CrossRef] [PubMed]
- Depner, C.M.; Stothard, E.R.; Wright, K.P., Jr. Metabolic consequences of sleep and circadian disorders. Curr. Diabetes Rep. 2014, 14, 507. [Google Scholar] [CrossRef]
- Shi, S.Q.; Ansari, T.S.; McGuinness, O.P.; Wasserman, D.H.; Johnson, C.H. Circadian disruption leads to insulin resistance and obesity. Curr. Biol. 2013, 23, 372–381. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Remely, M.; Aumueller, E.; Merold, C.; Dworzak, S.; Hippe, B.; Zanner, J.; Pointner, A.; Brath, H.; Haslberger, A.G. Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene 2014, 537, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Bhute, S.S.; Suryavanshi, M.V.; Joshi, S.M.; Yajnik, C.S.; Shouche, Y.S.; Ghaskadbi, S.S. Gut microbial diversity assessment of Indian type-2-diabetics reveals alterations in eubacteria, archaea, and eukaryotes. Front. Microbiol. 2017, 8, A214. [Google Scholar] [CrossRef] [PubMed]
- Hippe, B.; Remely, M.; Aumueller, E.; Pointner, A.; Magnet, U.; Haslberger, A. Faecalibacterium prausnitzii phylotypes in type two diabetic, obese, and lean control subjects. Benef. Microbes 2016, 7, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.M.; Pan, X. Circadian regulators of intestinal lipid absorption. J. Lipid Res. 2015, 56, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009, 1, 6ra14. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, P.; Scott, K.P.; Duncan, S.H.; Flint, H.J. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 2007, 102, 1197–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbonero, F.; Benefiel, A.; Alizadeh-Ghamsari, A.; Gaskins, H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012, 3, 448. [Google Scholar] [CrossRef]
- Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dao, M.C.; Everard, A.; Aron-Wisnewsky, J.; Sokolovska, N.; Prifti, E.; Verger, E.O.; Kayser, B.D.; Levenez, F.; Chilloux, J.; Hoyles, L. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut 2016, 65, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Verdam, F.J.; Fuentes, S.; de Jonge, C.; Zoetendal, E.G.; Erbil, R.; Greve, J.W.; Buurman, W.A.; de Vos, W.M.; Rensen, S.S. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity 2013, 21, E607–E615. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Chaix, A.; Yooseph, S.; Panda, S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014, 20, 1006–1017. [Google Scholar] [CrossRef]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [Green Version]
- Marcobal, A.; Southwick, A.M.; Earle, K.A.; Sonnenburg, J.L. A refined palate: Bacterial consumption of host glycans in the gut. Glycobiology 2013, 23, 1038–1046. [Google Scholar] [CrossRef] [Green Version]
- Thaiss, C.A.; Zeevi, D.; Levy, M.; Zilberman-Schapira, G.; Suez, J.; Tengeler, A.C.; Abramson, L.; Katz, M.N.; Korem, T.; Zmora, N.; et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 2014, 159, 514–529. [Google Scholar] [CrossRef]
- Liang, X.; Bushman, F.D.; FitzGerald, G.A. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci. USA 2015, 112, 10479–10484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thaiss, C.A.; Levy, M.; Korem, T.; Dohnalová, L.; Shapiro, H.; Jaitin, D.A.; David, E.; Winter, D.R.; Gury-BenAri, M.; Tatirovsky, E.; et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 2016, 167, 1495–1510. [Google Scholar] [CrossRef] [PubMed]
- Voigt, R.M.; Summa, K.C.; Forsyth, C.B.; Green, S.J.; Engen, P.; Naqib, A.; Vitaterna, M.H.; Turek, F.W.; Keshavarzian, A. The circadian clock mutation promotes intestinal dysbiosis. Alcohol.-Clin. Exp. Res. 2016, 40, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Bellet, M.M.; Deriu, E.; Liu, J.Z.; Grimaldi, B.; Blaschitz, C.; Zeller, M.; Edwards, R.A.; Sahar, S.; Dandekar, S.; Baldi, P.; et al. Circadian clock regulates the host response to Salmonella. Proc. Natl. Acad. Sci. USA 2013, 110, 9897–9902. [Google Scholar] [CrossRef] [PubMed]
- Marianelli, C.; Cifani, N.; Pasquali, P. Evaluation of antimicrobial activity of probiotic bacteria against Salmonella enterica subsp. enterica serovar typhimurium 1344 in a common medium under different environmental conditions. Res. Microbiol. 2010, 161, 673–680. [Google Scholar]
- Deaver, J.A.; Eum, S.Y.; Toborek, M. Circadian disruption changes gut microbiome taxa and functional gene composition. Front. Microbiol. 2018, 9, 737. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Tang, W.; He, Y.; Hu, J.; Gong, S.; He, Z.; Wei, G.; Lv, L.; Jiang, Y.; Zhou, H.; et al. Light exposure influences the diurnal oscillation of gut microbiota in mice. Biochem. Biophys. Res. Commun. 2018, 501, 16–23. [Google Scholar] [CrossRef]
- Kaczmarek, J.L.; Musaad, S.M.; Holscher, H.D. Time of day and eating behaviors are associated with the composition and function of the human gastrointestinal microbiota. Am. J. Clin. Nutr. 2017, 106, 1220–1231. [Google Scholar] [CrossRef]
- Marquet, P.; Duncan, S.H.; Chassard, C.; Bernalier-Donadille, A.; Flint, H.J. Lactate has the potential to promote hydrogen sulphide formation in the human colon. FEMS Microbiol. Lett. 2009, 299, 128–134. [Google Scholar] [CrossRef] [Green Version]
- Ndiaye, M.A.; Nihal, M.; Wood, G.S.; Ahmad, N. Skin, reactive oxygen species, and circadian clocks. Antioxid. Redox Signal. 2014, 20, 2982–2996. [Google Scholar] [CrossRef]
- Gangloff, A.; Bergeron, J.; Lemieux, I.; Després, J.-P. Changes in circulating vitamin D levels with loss of adipose tissue. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Nie, Y.; Zhu, A.; Chen, Z.; Wu, P.; Zhang, L.; Luo, M.; Sun, Q.; Cai, L.; Lai, Y.; et al. Vitamin D signaling through induction of Paneth cell defensins maintains gut microbiota and improves metabolic disorders and hepatic steatosis in animal models. Front. Physiol. 2016, 7, 498. [Google Scholar] [CrossRef]
- Assa, A.; Vong, L.; Pinnell, L.J.; Rautava, J.; Avitzur, N.; Johnson-Henry, K.C.; Sherman, P.M. Vitamin D deficiency predisposes to adherent-invasive Escherichia coli-induced barrier dysfunction and experimental colonic injury. Inflamm. Bowel Dis. 2015, 21, 297–306. [Google Scholar] [CrossRef]
- Chen, S.W.; Wang, P.Y.; Zhu, J.; Chen, G.W.; Zhang, J.L.; Chen, Z.Y.; Zuo, S.; Liu, Y.C.; Pan, Y.S. Protective effect of 1,25-dihydroxyvitamin D3 on lipopolysaccharide-induced intestinal epithelial tight junction injury in Caco-2 cell monolayers. Inflammation 2015, 38, 375–383. [Google Scholar] [CrossRef]
- Bashir, M.; Prietl, B.; Tauschmann, M.; Mautner, S.I.; Kump, P.K.; Treiber, G.; Wurm, P.; Gorkiewicz, G.; Hogenauer, C.; Pieber, T.R. Effects of high doses of vitamin D-3 on mucosa-associated gut microbiome vary between regions of the human gastrointestinal tract. Eur. J. Nutr. 2016, 55, 1479–1489. [Google Scholar] [CrossRef]
- Archer, S.N.; Oster, H. How sleep and wakefulness influence circadian rhythmicity: Effects of insufficient and mistimed sleep on the animal and human transcriptome. J. Sleep Res. 2015, 24, 476–493. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.; Price, R.J.; King, M.G.; Husband, A.J. Are antibiotic effects on sleep behavior in the rat due to modulation of gut bacteria? Physiol. Behav. 1990, 48, 561–565. [Google Scholar] [CrossRef]
- Nonaka, K.; Nakazawa, Y.; Kotorii, T. Effects of antibiotics, minocycline and ampicillin, on human sleep. Brain Res. 1983, 288, 253–259. [Google Scholar] [CrossRef]
- Brundula, V.; Rewcastle, N.B.; Metz, L.M.; Bernard, C.C.; Yong, V.W. Targeting Leukocyte MMPs and Transmigration: Minocycline as a Potential Therapy for Multiple Sclerosis. Brain 2002, 125, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.L.; Butt, H.; Ball, M.; Lewis, D.P.; Bruck, D. Sleep quality and the treatment of intestinal microbiota imbalance in Chronic Fatigue Syndrome: A pilot study. Sleep Sci. 2015, 8, 124–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quigley, E.M.M.; Quera, R. Small Intestinal Bacterial Overgrowth: Roles of Antibiotics, Prebiotics, and Probiotics. Gastroenterology 2006, 130, S78–S90. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, L.B.; Walters, A.S.; Paueksakon, P. Restless legs syndrome—Theoretical roles of inflammatory and immune mechanisms. Sleep Med. Rev. 2012, 16, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Benedict, C.; Vogel, H.; Jonas, W.; Woting, A.; Blaut, M.; Schurmann, A.; Cedernaes, J. Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals. Mol. Metab. 2016, 5, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.L.; Bai, L.; Goel, N.; Bailey, A.; Jang, C.J.; Bushman, F.D.; Meerlo, P.; Dinges, D.F.; Sehgal, A. Human and rat gut microbiome composition is maintained following sleep restriction. Proc. Natl. Acad. Sci. USA 2017, 114, E1564–E1571. [Google Scholar] [CrossRef] [PubMed]
- Poroyko, V.A.; Carreras, A.; Khalyfa, A.; Khalyfa, A.A.; Leone, V.; Peris, E.; Almendros, I.; Gileles-Hillel, A.; Qiao, Z.; Hubert, N.; et al. Chronic sleep disruption alters gut microbiota, induces systemic and adipose tissue inflammation and insulin resistance in mice. Sci. Rep. 2016, 6, 35405. [Google Scholar] [CrossRef] [PubMed]
- Giskeodegard, G.F.; Davies, S.K.; Revell, V.L.; Keun, H.; Skene, D.J. Diurnal rhythms in the human urine metabolome during sleep and total sleep deprivation. Sci. Rep. 2015, 5, 14843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voigt, R.M.; Forsyth, C.B.; Green, S.J.; Mutlu, E.; Engen, P.; Vitaterna, M.H.; Turek, F.W.; Keshavarzian, A. Circadian disorganization alters intestinal microbiota. PLoS ONE 2014, 9, e97500. [Google Scholar] [CrossRef] [PubMed]
- Bishehsari, F.; Saadalla, A.; Khazaie, K.; Engen, P.A.; Voigt, R.M.; Shetuni, B.B.; Forsyth, C.; Shaikh, M.; Vitaterna, M.H.; Turek, F.; et al. Light/dark shifting promotes alcohol-induced colon carcinogenesis: Possible role of intestinal inflammatory milieu and microbiota. Int. J. Mol. Sci. 2016, 17, 2017. [Google Scholar] [CrossRef]
- Bobinski, R.; Michalik, A. Evaluation of early jet lag symptoms by passengers crossing 7 time zones. In Transport System Telematics; Mikulski, J., Ed.; Springer: Katowice-Ustron, Poland, 2010; Volume 104, pp. 356–363. [Google Scholar]
- Roenneberg, T.; Allebrandt, K.V.; Merrow, M.; Vetter, C. Social jetlag and obesity. Curr. Biol. 2012, 22, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.J.; Moffitt, T.E.; Gregory, A.M.; Goldman-Mellor, S.; Nolan, P.M.; Poulton, R.; Caspi, A. Social jetlag, obesity and metabolic disorder: Investigation in a cohort study. Int. J. Obes. 2015, 39, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, A.C.; Paterson, J.L.; Ferguson, S.A.; Stanley, D.; Wright, K.P., Jr.; Dawson, D. The shift work and health research agenda: Considering changes in gut microbiota as a pathway linking shift work, sleep loss and circadian misalignment, and metabolic disease. Sleep Med. Rev. 2017, 34, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Stenvers, D.J.; Jonkers, C.F.; Fliers, E.; Bisschop, P.H.; Kalsbeek, A. Nutrition and the circadian timing system. Prog. Brain Res. 2012, 199, 359–376. [Google Scholar] [PubMed]
- Dallmann, R.; Viola, A.U.; Tarokh, L.; Cajochen, C.; Brown, S.A. The human circadian metabolome. Proc. Natl. Acad. Sci. USA 2012, 109, 2625–2629. [Google Scholar] [CrossRef] [Green Version]
- Kohsaka, A.; Laposky, A.D.; Ramsey, K.M.; Estrada, C.; Joshu, C.; Kobayashi, Y.; Turek, F.W.; Bass, J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007, 6, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Shankar, V.; Gouda, M.; Moncivaiz, J.; Gordon, A.; Reo, N.V.; Hussein, L.; Paliy, O. Differences in gut metabolites and microbial composition and functions between Egyptian and U.S. children are consistent with their diets. mSystems 2017, 2, e00169-16. [Google Scholar] [CrossRef] [PubMed]
- Jumpertz, R.; Duc Son, L.; Turnbaugh, P.J.; Trinidad, C.; Bogardus, C.; Gordon, J.I.; Krakoff, J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 2011, 94, 58–65. [Google Scholar] [CrossRef] [Green Version]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559. [Google Scholar] [CrossRef]
- Walker, A.W.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011, 5, 220–230. [Google Scholar] [CrossRef]
- Tuohy, K.M.; Conterno, L.; Gasperotti, M.; Viola, R. Up-regulating the human intestinal microbiome using whole plant foods, polyphenols, and/or fiber. J. Agric. Food Chem. 2012, 60, 8776–8782. [Google Scholar] [CrossRef]
- Parkar, S.G.; Blatchford, P.A.; Kim, C.C.; Rosendale, D.I.; Ansell, J. New and tailored prebiotics: Established applications. In Probiotics and Prebiotics: Current Research and Future Trends; Venema, K., Ed.; Caister Academic Press: Poole, UK, 2015; pp. 289–314. [Google Scholar]
- Tahara, Y.; Yamazaki, M.; Sukigara, H.; Motohashi, H.; Sasaki, H.; Miyakawa, H.; Haraguchi, A.; Ikeda, Y.; Fukuda, S.; Shibata, S. Gut microbiota-derived short chain fatty acids induce circadian clock entrainment in mouse peripheral tissue. Sci. Rep. 2018, 8, 1395. [Google Scholar] [CrossRef]
- Thompson, R.S.; Roller, R.; Mika, A.; Greenwood, B.N.; Knight, R.; Chichlowski, M.; Berg, B.M.; Fleshner, M. Dietary prebiotics and bioactive milk fractions improve nrem sleep, enhance rem sleep rebound and attenuate the stress-induced decrease in diurnal temperature and gut microbial alpha diversity. Front. Behav. Neurosci. 2017, 10, 240. [Google Scholar] [CrossRef] [PubMed]
- Firrman, J.; Liu, L.; Zhang, L.; Arango Argoty, G.; Wang, M.; Tomasula, P.; Kobori, M.; Pontious, S.; Xiao, W. The effect of quercetin on genetic expression of the commensal gut microbes Bifidobacterium catenulatum, Enterococcus caccae and Ruminococcus gauvreauii. Anaerobe 2016, 42, 130–141. [Google Scholar] [CrossRef]
- Leone, V.; Gibbons, S.M.; Martinez, K.; Hutchison, A.L.; Huang, E.Y.; Cham, C.M.; Pierre, J.F.; Heneghan, A.F.; Nadimpalli, A.; Hubert, N.; et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 2015, 17, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Parkar, S.G.; Trower, T.M.; Stevenson, D.E. Fecal microbial metabolism of polyphenols and its effects on human gut microbiota. Anaerobe 2013, 23, 12–19. [Google Scholar] [CrossRef]
- Eeckhaut, E.; Struijs, K.; Possemiers, S.; Vincken, J.P.; De Keukeleire, D.; Verstraete, W. Metabolism of the lignan macromolecule into enterolignans in the gastrointestinal lumen as determined in the simulator of the human intestinal microbial ecosystem. J. Agric. Food Chem. 2008, 56, 4806–4812. [Google Scholar] [CrossRef] [PubMed]
- Clavel, T.; Henderson, G.; Alpert, C.A.; Philippe, C.; Rigottier-Gois, L.; Dore, J.; Blaut, M. Intestinal bacterial communities that produce active estrogen-like compounds enterodiol and enterolactone in humans. Appl. Environ. Microbiol. 2005, 71, 6077–6085. [Google Scholar] [CrossRef]
- Damdimopoulou, P.; Nurmi, T.; Salminen, A.; Damdimopoulos, A.E.; Kotka, M.; van der Saag, P.; Strauss, L.; Poutanen, M.; Pongratz, I.; Makela, S. A single dose of enterolactone activates estrogen signaling and regulates expression of circadian clock genes in mice. J. Nutr. 2011, 141, 1583–1589. [Google Scholar] [CrossRef] [PubMed]
- Chilloux, J.; Neves, A.L.; Boulange, C.L.; Dumas, M.E. The microbial-mammalian metabolic axis: A critical symbiotic relationship. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Bass, J. Circadian topology of metabolism. Nature 2012, 491, 348–356. [Google Scholar] [CrossRef]
- Fiorucci, S.; Distrutti, E. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol. Med. 2015, 21, 702–714. [Google Scholar] [CrossRef]
- Labbe, A.; Ganopolsky, J.G.; Martoni, C.J.; Prakash, S.; Jones, M.L. Bacterial bile metabolising gene abundance in Crohn’s, ulcerative colitis and type 2 diabetes metagenomes. PLoS ONE 2014, 9, e115175. [Google Scholar] [CrossRef] [PubMed]
- Eggink, H.M.; Oosterman, J.E.; de Goede, P.; de Vries, E.M.; Foppen, E.; Koehorst, M.; Groen, A.K.; Boelen, A.; Romijn, J.A.; la Fleur, S.E.; et al. Complex interaction between circadian rhythm and diet on bile acid homeostasis in male rats. Chronobiol. Int. 2017, 34, 1339–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.K.J.; Guo, G.L.; Klaassen, C.D. Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS ONE 2011, 6, e16683. [Google Scholar] [CrossRef]
- Joyce, S.A.; MacSharry, J.; Casey, P.G.; Kinsella, M.; Murphy, E.F.; Shanahan, F.; Hill, C.; Gahan, C.G.M. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. USA 2014, 111, 7421–7426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokota, A.; Fukiya, S.; Islam, K.B.M.S.; Ooka, T.; Ogura, Y.; Hayashi, T.; Hagio, M.; Ishizuka, S. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes 2012, 3, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.; Lucio, M.; Pfitzner, B.; Scheerer, M.F.; Neschen, S.; de Angelis, M.H.; Hartmann, A.; Schmitt-Kopplin, P. Importance of sulfur-containing metabolites in discriminating fecal extracts between normal and type-2 diabetic mice. J. Proteome Res. 2014, 13, 4220–4231. [Google Scholar] [CrossRef]
- Nicholls, P.; Marshall, D.C.; Cooper, C.E.; Wilson, M.T. Sulfide inhibition of and metabolism by cytochrome c oxidase. Biochem. Soc. Trans. 2013, 41, 1312–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frost, G.; Cai, Z.; Raven, M.; Otway, D.T.; Mushtaq, R.; Johnston, J.D. Effect of short chain fatty acids on the expression of free fatty acid receptor 2 (Ffar2), Ffar3 and early-stage adipogenesis. Nutr. Diabetes 2014, 4, e128. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.J.; Peng, L.; Barry, N.A.; Cline, G.W.; Zhang, D.Y.; Cardone, R.L.; Petersen, K.F.; Kibbey, R.G.; Goodman, A.L.; Shulman, G.I. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature 2016, 534, 213–217. [Google Scholar] [CrossRef]
- Pant, K.; Yadav, A.K.; Gupta, P.; Islam, R.; Jain, A.; Venugopal, S.K. Butyrate induces ROS-mediated apoptosis by modulating miR-22/SIRT-1 pathway in hepatic cancer cells. Redox Biol. 2017, 12, 340–349. [Google Scholar] [CrossRef]
- Rada-Iglesias, A.; Enroth, S.; Ameur, A.; Koch, C.M.; Clelland, G.K.; Respuela-Alonso, P.; Wilcox, S.; Dovey, O.M.; Ellis, P.D.; Langford, C.F.; et al. Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007, 17, 708–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakahata, Y.; Kaluzova, M.; Grimaldi, B.; Sahar, S.; Hirayama, J.; Chen, D.; Guarente, L.P.; Sassone-Corsi, P. The NAD(+)-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008, 134, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Gillum, M.P.; Erion, D.M.; Shulman, G.I. Sirtuin-1 regulation of mammalian metabolism. Trends Mol. Med. 2011, 17, 8–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogelauer, M.; Krall, A.S.; McBrian, M.A.; Li, J.Y.; Kurdistani, S.K. Stimulation of histone deacetylase activity by metabolites of intermediary metabolism. J. Biol. Chem. 2012, 287, 32006–32016. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef]
- Wolf, G. Three vitamins are involved in regulation of the circadian rhythm. Nutr. Rev. 2002, 60, 257–260. [Google Scholar]
- Beydoun, M.A.; Gamaldo, A.A.; Canas, J.A.; Beydoun, H.A.; Shah, M.T.; McNeely, J.M.; Zonderman, A.B. Serum nutritional biomarkers and their associations with sleep among us adults in recent national surveys. PLoS ONE 2014, 9, e103490. [Google Scholar] [CrossRef]
- Magnusdottir, S.; Ravcheev, D.; de Crecy-Lagard, V.; Thiele, I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet. 2015, 6, 148. [Google Scholar] [CrossRef]
- Degnan, P.H.; Taga, M.E.; Goodman, A.L. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 2014, 20, 769–778. [Google Scholar] [CrossRef]
- Langlais, P.J.; Hall, T. Thiamine deficiency-induced disruptions in the diurnal rhythm and regulation of body temperature in the rat. Metab. Brain Dis. 1998, 13, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Guy LeBlanc, J.; Milani, C.; Savoy de Giori, G.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Hirano, A.; Braas, D.; Fu, Y.H.; Ptacek, L.J. FAD regulates cryptochrome protein stability and circadian clock in mice. Cell Rep. 2017, 19, 255–266. [Google Scholar] [CrossRef]
- Wakade, C.; Chong, R.; Bradley, E.; Thomas, B.; Morgan, J. Upregulation of GPR109A in Parkinson’s Disease. PLoS ONE 2014, 9, e109818. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.W.; Yang, Y.; Li, X.; He, B.; Jia, Y.M.; Zhang, N.N.; Zhao, R.Q. Folate deprivation modulates the expression of autophagy- and circadian-related genes in HT-22 hippocampal neuron cells through GR-mediated pathway. Steroids 2016, 112, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Chan, A.; Ali, S.; Saha, A.; Haushalter, K.J.; Lam, W.-L.M.; Glasheen, M.; Parker, J.; Brenner, M.; Mahon, S.B.; et al. Hydrogen sulfide—Mechanisms of toxicity and development of an antidote. Sci. Rep. 2016, 6, 20831. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.K.; Ang, J.E.; Revell, V.L.; Holmes, B.; Mann, A.; Robertson, F.P.; Cui, N.; Middleton, B.; Ackermann, K.; Kayser, M.; et al. Effect of sleep deprivation on the human metabolome. Proc. Natl. Acad. Sci. USA 2014, 111, 10761–10766. [Google Scholar] [CrossRef] [Green Version]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [Green Version]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef]
- Versteeg, R.I.; Serlie, M.J.; Kalsbeek, A.; la Fleur, S.E. Serotonin, a possible intermediate between disturbed circadian rhythms and metabolic disease. Neuroscience 2015, 301, 155–167. [Google Scholar] [CrossRef]
- Hardeland, R.; Cardinali, D.P.; Srinivasan, V.; Spence, D.W.; Brown, G.M.; Pandi-Perumal, S.R. Melatonin-A pleiotropic, orchestrating regulator molecule. Prog. Neurobiol. 2011, 93, 350–384. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.K.; Yang, C.; Song, G.H.; Wong, J.; Ho, K.Y. Melatonin regulation as a possible mechanism for probiotic (VSL#3) in irritable bowel syndrome: A randomized double-blinded placebo study. Dig. Dis. Sci. 2015, 60, 186–194. [Google Scholar] [PubMed]
- Paulose, J.K.; Wright, J.M.; Patel, A.G.; Cassone, V.M. Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PLoS ONE 2016, 11, e0146643. [Google Scholar] [CrossRef] [PubMed]
- Daulatzai, M.A. Chronic functional bowel syndrome enhances gut-brain axis dysfunction, neuroinflammation, cognitive impairment, and vulnerability to dementia. Neurochem. Res. 2014, 39, 624–644. [Google Scholar] [CrossRef] [PubMed]
- Malinen, E.; Mättö, J.; Salmitie, M.; Alander, M.; Saarela, M.; Palva, A. PCR-ELISA: II: Analysis of Bifidobacterium populations in human faecal samples from a consumption trial with Bifidobacterium lactis Bb-12 and a galacto-oligosaccharide preparation. Syst. Appl. Microbiol. 2002, 25, 249–258. [Google Scholar] [PubMed]


| Query | Conditions | Examples of Evidences |
|---|---|---|
| How does circadian rhythm affect gut microbiota? | Light/dark cycles | Effect of diurnal variation and altered light/dark cycles on gut microbiome |
| Sleep status | Gut microbiome in sleep deprivation/sleep fragmentation | |
| Diet | High fat diet, night eating | |
| How does gut microbiota affect circadian rhythm? | Antibiotic-induced gut microbial alterations | Do antibiotics affect sleep or circadian-controlled metabolic states? |
| Microbial dysbiosis due to intestinal disease | Chronic gut disorders with altered microbiota showing co-morbidities in sleep | |
| Microbial metabolites | Short chain fatty acids, Secondary bile acids | |
| Vitamins | ||
| Biogenic amines | ||
| Hydrogen sulfide |
| Microorganisms | Microbial Function | Interactions with Host Pathway |
|---|---|---|
| Firmicutes (Lachnospiraceae, Clostridiaceae, Erysepelotrichaceae, Ruminococcaceae, Lactobacillus), Bacteroidetes (Bacteroides), and Bifidobacterium [72,73]. | Microbial bile salt hydrolases deconjugate bile deoxycholic acid and lithocholic acid [76]. | Microbial bile salt hydrolase associated with modulation of canonical clock genes, genes related to lipid metabolism and immune homeostasis [76]. |
| Desulfovibrio, Desulfotobacter, Desulfobulbus, Bilophila wadsworthia [13]. | Act on sulfated compounds to generate hydrogen sulfide in the colon. | Hydrogen sulfide phase-delays hepatic Bmal1 expression, which is also associated with suppressed substrate oxidation and elevated systemic glucose [65]. |
| Lachnospiraceae (Roseburia), Eubacteriaceae (Eubacterium rectale), and Ruminococcaceae (Ruminococcus bromii, Faecalibacterium prausnitzii) [11,12] | Break down dietary fiber to generate butyrate in the colon [58]. | Butyrate is a key metabolic fuel for colonic epithelial cells [11,12]; regulates plasma glucose by multiple mechanisms including activation of receptors that signal the secretion of satiety hormones, stimulation of insulin secretion and suppression of pancreatic glucagon [6,80]; modulates canonical clock genes in peripheral tissue [65]; acts as a histone deacetylase inhibitor [82,83]. |
| Clostridum sporogenes and Ruminococcus gnavus [87]. | Generate precursors of biogenic amines such as serotonin [87]. | Bioamines such as serotonin play a role in intestinal motility and secretory activity [88]. |
| Vitamin | Physiological Effects Through Circadian Controlled Mechanisms | Examples of Microorganisms That Synthesize the Vitamin |
|---|---|---|
| B1, thiamine | Core body temperature, rat study [93] | Streptococcus thermophilus ST5, Lactobacillus helveticus R0052, Bifidobacterium longum R0175 [94] |
| B2, riboflavin | Affects metabolism by influencingcryptochrome 2 stability [95] | Bacillus subtilis, Escherichia coli [94] |
| B3, niacin | Lowered blood B3 associated with decreased duration of deep sleep in Parkinson’s disease [96] | E. coli [91] |
| B9, folic acid | Important for brain function by regulating brain clock genes [97]; lowered serum B9 associated with sleep disturbance [90] | Bifidobacterium spp., Lactobacillus spp. [94] |
| B12, methylcobalamine or cyanocobalamine | Lowered serum B12 associated with sleep disturbance [90]; also reverses hydrogen sulfide effects on substrate oxidation [98] | Propionibacterium freudenreichii; Lactobacillus reuteri [94] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parkar, S.G.; Kalsbeek, A.; Cheeseman, J.F. Potential Role for the Gut Microbiota in Modulating Host Circadian Rhythms and Metabolic Health. Microorganisms 2019, 7, 41. https://doi.org/10.3390/microorganisms7020041
Parkar SG, Kalsbeek A, Cheeseman JF. Potential Role for the Gut Microbiota in Modulating Host Circadian Rhythms and Metabolic Health. Microorganisms. 2019; 7(2):41. https://doi.org/10.3390/microorganisms7020041
Chicago/Turabian StyleParkar, Shanthi G., Andries Kalsbeek, and James F. Cheeseman. 2019. "Potential Role for the Gut Microbiota in Modulating Host Circadian Rhythms and Metabolic Health" Microorganisms 7, no. 2: 41. https://doi.org/10.3390/microorganisms7020041
APA StyleParkar, S. G., Kalsbeek, A., & Cheeseman, J. F. (2019). Potential Role for the Gut Microbiota in Modulating Host Circadian Rhythms and Metabolic Health. Microorganisms, 7(2), 41. https://doi.org/10.3390/microorganisms7020041