Study on the Isolation of Two Atrazine-Degrading Bacteria and the Development of a Microbial Agent
Abstract
:1. Introduction
2. Materials and Methods
2.1. Media
2.2. Determination of Atrazine and Its Metabolites
2.3. Isolation and Identification of Atrazine-Degrading Bacteria
2.4. Preparation of Bacterial Inocula
2.5. Degradation of Atrazine by Strains ATLJ-5 and ATLJ-11
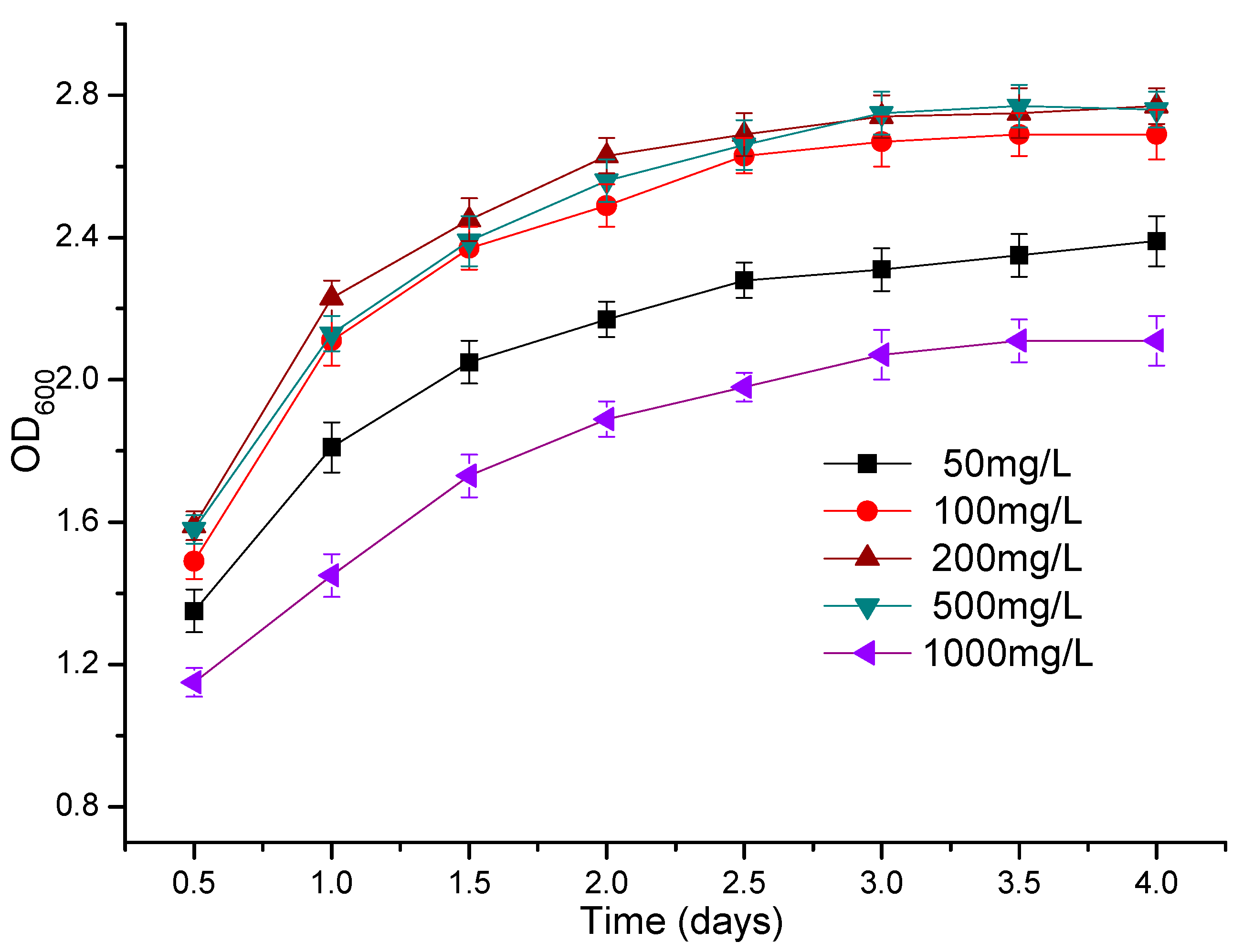
2.6. The Effect of Atrazine Concentration on the Growth of Bacteria
2.7. Construction of a Simple Microbial Consortium and Development of a Microbial Agent
3. Results and Discussion
3.1. Characterization of Atrazine-Degrading Bacteria
3.2. Degradation of Atrazine by Strains ATLJ-5 and ATLJ-11
3.3. The Effect of Atrazine Concentration on the Growth of Strains ATLJ-5 and ATLJ-11
3.4. Degradation of Atrazine by a Simple Microbial Consortium or Microbial Agent
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Singh, P.; Suri, C.R.; Cameotra, S.S. Isolation of a member of acinetobacter species involved in atrazine degradation. Biochem. Biophys. Res. Commun. 2004, 317, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Dong, C.; Kong, D.; Lu, J. New insights into atrazine degradation by cobalt catalyzed peroxymonosulfate oxidation: kinetics, reaction products and transformation mechanisms. J. Hazard Mater. 2015, 285, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Siripattanakul, S.; Wirojanagud, W.; Mcevoy, J.; Limpiyakorn, T.; Khan, E. Atrazine degradation by stable mixed cultures enriched from agricultural soil and their characterization. J. Appl. Microbiol. 2010, 106, 986–992. [Google Scholar] [CrossRef]
- Elsheekh, M.M.; Kotkat, H.M.; Hammouda, O.H. Effect of atrazine herbicide on growth, photosynthesis, protein synthesis, and fatty acid composition in the unicellular green alga Chlorella kessleri. Ecotoxicol. Environ. Saf. 1994, 29, 349–358. [Google Scholar] [CrossRef]
- Wüst, S.; Hock, B. A sensitive enzyme immunoassay for the detection of atrazine based upon sheep antibodies. Anal. Lett. 1992, 25, 1025–1037. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, V.; Chauhan, A.; Datta, S.; Wani, A.B.; Singh, N. Toxicity, degradation and analysis of the herbicide atrazine. Environ. Chem. Lett. 2018, 16, 211–237. [Google Scholar] [CrossRef]
- Yang, C.Y.; Yang, L.; Zhang, K.; Xia, W.; Ma, C.Q.; Tang, H.Z. Atrazine degradation by a simple consortium of Klebsiella sp. A1 and Comamonas sp. A2 in nitrogen enriched medium. Biodegradation 2010, 21, 97–105. [Google Scholar] [CrossRef]
- Roustan, A.; Aye, M.; Meo, M.D.; Giorgio, C.D. Genotoxicity of mixtures of glyphosate and atrazine and their environmental transformation products before and after photoactivation. Chemosphere 2014, 108, 93–100. [Google Scholar] [CrossRef]
- Yang, Y.; Cao, H.; Peng, P.; Bo, H. Degradation and transformation of atrazine under catalyzed ozonation process with TiO2 as catalyst. J. Hazard Mater. 2014, 279, 444–451. [Google Scholar] [CrossRef]
- Amadori, M.F.; Rodrigues, M.B.; Rebouças, C.C.; Peralta-Zamora, P.G.; Grassi, M.T.; Abate, G. Behavior of atrazine and its degradation products deethylatrazine and deisopropylatrazine in oxisol samples. Water Air Soil Pollut. 2016, 227, 380. [Google Scholar] [CrossRef]
- Satsuma, K. Mineralization of s-triazine herbicides by a newly isolated Nocardioides species strain DN36. Appl. Microbiol. Biotechnol. 2010, 86, 1585–1592. [Google Scholar] [CrossRef] [PubMed]
- Accinelli, C.; Dinelli, G.; Vicari, A.; Catizone, P. Atrazine and metolachlor degradation in subsoils. Biol. Fertil. Soils 2001, 33, 495–500. [Google Scholar] [CrossRef]
- Vaishampayan, P.A.; Kanekar, P.P.; Dhakephalkar, P.K. Isolation and characterization of Arthrobacter sp. strain MCM B-436, an atrazine-degrading bacterium, from rhizospheric soil. Int. Biodeterior. Biodegrad. 2007, 60, 273–278. [Google Scholar] [CrossRef]
- Vibber, L.L.; Pressler, M.J.; Colores, G.M. Isolation and characterization of novel atrazine-degrading microorganisms from an agricultural soil. Appl. Microbiol. Biotechnol. 2007, 75, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Wenk, M.; Baumgartner, T.; Dobovsek, J.; Fuchs, T.; Kucsera, J.; Zopfi, J. Rapid atrazine mineralisation in soil slurry and moist soil by inoculation of an atrazine-degrading Pseudomonas sp. strain. Appl. Microbiol. Biotechnol. 1998, 49, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Hu, E.; Cheng, H. Rapid extraction and determination of atrazine and its degradation;products from microporous mineral sorbents using microwave-assisted; solvent extraction followed by ultra-HPLC-MS/MS. Microchim. Acta 2013, 180, 703–710. [Google Scholar] [CrossRef]
- Barchanska, H.; Babilas, B.; Gluzicka, K.; Zralek, D.; Baranowska, I. Rapid determination of mesotrione, atrazine and its main degradation products in selected plants by MSPD-HPLC and indirect estimation of herbicides phytotoxicity by chlorophyll quantification. Int. J. Environ. Anal. Chem. 2014, 94, 99–114. [Google Scholar] [CrossRef]
- Dong, X.Z. Identification Manual of Systematic Bacteriology; Science China Press Ltd.: Beijing, China, 2001. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1998. [Google Scholar]
- Shi, X.Z.; Guo, R.J.; Takagi, K.; Miao, Z.Q.; Li, S.D. Chlorothalonil degradation by Ochrobactrum lupinistrain TP-D1 and identification of its metabolites. World J. Microbiol. Biotechnol. 2011, 27, 1755–1764. [Google Scholar] [CrossRef]
- Cho, K.M.; Math, R.K.; Islam, S.M.A.; Lim, W.J.; Hong, S.Y.; Kim, J.M. Biodegradation of chlorpyrifos by lactic acid bacteria during kimchi fermentation. J. Agric. Food Chem. 2009, 57, 1882–1889. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, C.; Yan, Y. Biodegradation of methyl parathion andp-nitrophenol by a newly isolated Agrobacterium sp. strain Yw12. Biodegradation 2012, 23, 107–116. [Google Scholar] [CrossRef]
- Song, X.; Zhang, Z.; Liu, Y.; Zhan, N.; Xiang, D. Isolation and identification of microorganisms causing mildew of fiberboard in Nanning, Guangxi. J. Eng. 2016, 1, 78–82. [Google Scholar]
- Zhu, J.; Meng, Z.; Ge, J.; Wu, Y.; Jin, C. Degradation characteristics of isocarbophos and isolation of an isocarbophos-degrading bacterium, Bacillus pumilus SALL-7. Fresenius Environ. Bull. 2018, 27, 7552–7558. [Google Scholar]
- Rehan, M.; Kluge, M.; Kellner, H.; Ullrich, R.; Hofrichter, M. Degradation of atrazine by Frankia alni ACN14a: Gene regulation, dealkylation, and dechlorination. Appl. Microbiol. Biotechnol. 2014, 98, 6125–6135. [Google Scholar] [CrossRef]
- Solomon, R.D.J.; Kumar, A.; Santhi, V.S. Atrazine biodegradation efficiency, metabolite detection, and trzD gene expression by enrichment bacterial cultures from agricultural soil. J. Zhejiang Univ. Sci. B 2013, 14, 1162–1172. [Google Scholar] [CrossRef] [Green Version]
- Jablonowski, N.D.; Koeppchen, S.; Hofmann, D.; Schaeffer, A.; Burauel, P. Spatial distribution and characterization of long-term aged 14C-labeled atrazine residues in soil. J. Agric. Food Chem. 2008, 56, 9548–9554. [Google Scholar] [CrossRef]
- Batra, M.; Pandey, J.; Suri, C.R.; Jain, R.K. Isolation and characterization of an atrazine-degrading rhodococcus sp. strain MB-P1 from contaminated soil. Lett. Appl. Microbiol. 2009, 49, 721–729. [Google Scholar]
- Wang, G.; Zhang, R.; Zhang, H.; Cao, F. Effect of addition of microbial inoculants on co-composting of ginkgo used leaves with pig manure and wheat straw. J. Eng. 2015, 29, 12–15. [Google Scholar]
- Nousiainen, A.O.; Björklöf, K.; Sagarkar, S.; Nielsen, J.L.; Kapley, A. Bioremediation strategies for removal of residual atrazine in the boreal groundwater zone. Appl. Microbiol. Biotechnol. 2015, 99, 10249–10259. [Google Scholar] [CrossRef]
- Sparling, G.; Dragten, R.; Aislabie, J.; Fraser, R. Atrazine mineralisation in new zealand topsoils and subsoils: influence of edaphic factors and numbers of atrazine-degrading microbes. Aust. J. Soil Res. 1998, 36, 557–570. [Google Scholar] [CrossRef]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Fu, L.; Jin, C.; Meng, Z.; Yang, N. Study on the Isolation of Two Atrazine-Degrading Bacteria and the Development of a Microbial Agent. Microorganisms 2019, 7, 80. https://doi.org/10.3390/microorganisms7030080
Zhu J, Fu L, Jin C, Meng Z, Yang N. Study on the Isolation of Two Atrazine-Degrading Bacteria and the Development of a Microbial Agent. Microorganisms. 2019; 7(3):80. https://doi.org/10.3390/microorganisms7030080
Chicago/Turabian StyleZhu, Jiangwei, Li Fu, Caihua Jin, Zili Meng, and Ning Yang. 2019. "Study on the Isolation of Two Atrazine-Degrading Bacteria and the Development of a Microbial Agent" Microorganisms 7, no. 3: 80. https://doi.org/10.3390/microorganisms7030080




