Inhibiting Methanogenesis in Rumen Batch Cultures Did Not Increase the Recovery of Metabolic Hydrogen in Microbial Amino Acids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Treatments and Incubations
2.2. Sampling and Analytical Procedures
2.3. Denaturing Gradient Gel Electrophoresis Analysis
2.4. Calculations
2.5. Statistical Analyses
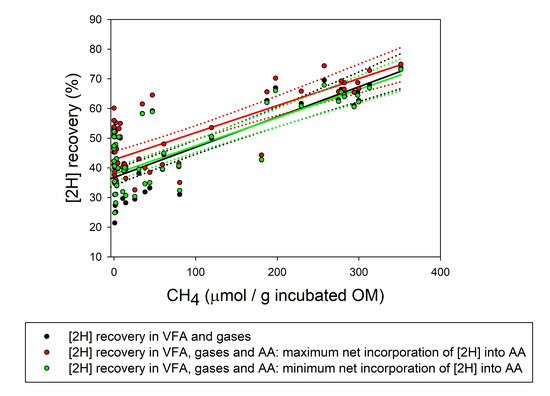
3. Results
3.1. Fermentation
3.2. Digestion and Microbial Biomass Production and Composition
3.3. Reducing Equivalents Balance
3.4. Denaturing Gradient Gel Electrophoresis Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wallace, R.J.; Onodera, R.; Cotta, M.A. Metabolism of nitrogen-containing compounds. In The Rumen Microbial Ecosystem, 2nd ed.; Hobson, P.N., Stewart, C.S., Eds.; Blackie Academic & Professional: London, UK, 1997; pp. 283–328. [Google Scholar]
- Wolin, M.J.; Miller, T.L.; Stewart, C.S. Microbe-microbe interactions. In The Rumen Microbial Ecosystem, 2nd ed.; Hobson, P.N., Stewart, C.S., Eds.; Blackie Academic & Professional: London, UK, 1997; pp. 467–491. [Google Scholar]
- Greening, C.; Geier, R.; Wang, C.; Woods, L.C.; Morales, S.E.; McDonald, M.J.; Rushton-Green, R.; Morgan, X.C.; Koike, S.; Leahy, S.C.; et al. Alternative hydrogen uptake pathways suppress methane production in ruminants. bioRxiv 2018, 486894. [Google Scholar] [CrossRef]
- Moss, A.R.; Jouany, J.-P.; Newbold, J. Methane production by ruminants: Its contribution to global warming. Ann. Zootech. 2000, 49, 231–253. [Google Scholar] [CrossRef]
- Johnson, K.A.; Johnson, D.E. Methane emissions from cattle. J. Anim. Sci. 1995, 73, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Eckard, R.J.; Grainger, C.; de Klein, C.A.M. Options for the abatement of methane and nitrous oxide from ruminant production: A review. Livestock Sci. 2010, 130, 47–56. [Google Scholar] [CrossRef]
- Guyader, J.; Eugene, M.; Meunier, B.; Doreau, M.; Morgavi, D.P.; Silberberg, M.; Rochette, Y.; Gerard, C.; Loncke, C.; Martin, C. Additive methane-mitigating effect between linseed oil and nitrate fed to cattle. J. Anim. Sci. 2015, 93, 3564–3577. [Google Scholar] [CrossRef] [PubMed]
- Ungerfeld, E.M. Inhibition of rumen methanogenesis and ruminant productivity: A meta-analysis. Front. Vet. Sci. 2018, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ungerfeld, E.M. Shifts in metabolic hydrogen sinks in the methanogenesis-inhibited ruminal fermentation: A meta-analysis. Front. Microbiol. 2015, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hino, T.; Russell, J.B. Effect of reducing-equivalent disposal and NADH/NAD on deamination of amino acids by intact rumen microorganisms and their cell extracts. Appl. Environ. Microbiol. 1985, 50, 1368–1374. [Google Scholar] [PubMed]
- Voet, D.; Voet, J.G. Biochemistry; John Wiley & Sons, Inc.: New York, NY, USA, 1995; p. 1392. [Google Scholar]
- Goering, H.K.; van Soest, P.J. Forage Fiber Analyses (Apparatus, Reagents, Procedures, and Some Applications); United States Department of Agriculture: Washington, DC, USA, 1975; p. 24.
- Schaefer, D.M.; Davis, C.L.; Bryant, M.P. Ammonia saturation constants for predominant species of rumen bacteria. J. Dairy Sci. 1980, 63, 1248–1263. [Google Scholar] [CrossRef]
- Caldwell, D.R.; Bryant, M.P. Medium without rumen fluid for nonselective enumeration and isolation of rumen bacteria. Appl. Microbiol. 1966, 14, 794–801. [Google Scholar] [PubMed]
- O’Brien, M.; Navarro-Villa, A.; Purcell, P.J.; Boland, T.M.; O’Kiely, P. Reducing in vitro rumen methanogenesis for two contrasting diets using a series of inclusion rates of different additives. Anim. Prod. Sci. 2013, 54, 141–157. [Google Scholar] [CrossRef]
- Sauer, F.D.; Teather, R.M. Changes in oxidation reduction potentials and volatile fatty acids production by rumen bacteria when methane synthesis is inhibited. J. Dairy Sci. 1987, 70, 1835–1840. [Google Scholar] [CrossRef]
- García-López, P.M.; Kung, L., Jr.; Odom, J.M. In vitro inhibition of microbial methane production by 9,10-anthraquinone. J. Anim. Sci. 1996, 74, 2276–2284. [Google Scholar] [CrossRef] [PubMed]
- Bauchop, T. Inhibition of rumen methanogenesis by methane analogues. J. Bacteriol. 1967, 94, 171–175. [Google Scholar] [PubMed]
- Ungerfeld, E.M.; Rust, S.R.; Burnett, R. Effects of butyrate precursors on electron relocation when methanogenesis is inhibited in ruminal mixed cultures. Let. Appl. Microbiol. 2006, 42, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Guyader, J.; Tavendale, M.; Martin, C.; Muetzel, S. Dose-response effect of nitrate on hydrogen distribution between rumen fermentation end products: an in vitro approach. Anim. Prod. Sci. 2016, 56, 224–230. [Google Scholar] [CrossRef]
- Guyader, J.; Ungerfeld, E.M.; Beauchemin, K.A. Redirection of metabolic hydrogen by inhibiting methanogenesis in the rumen simulation technique (RUSITEC). Front. Microbiol. 2017, 8, 393. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis, 16th ed.; Association of Official Analytical Chemists (AOAC) International: Airlington, VA, USA, 1995; p. 771. [Google Scholar]
- Kaplan, A. The determination of urea, ammonia, and urease. Methods Biochem. Anal. 1969, 17, 311–324. [Google Scholar] [PubMed]
- Molina-Alcaide, E.; Moumen, A.; Martin-Garcia, I.; Carro, M.D. Comparison of bacterial pellets and microbial markers for the estimation of the microbial nitrogen and amino acids flows from single flow continuous culture fermenters fed diets containing two-stage olive cake. J. Anim. Physiol. Anim. Nutr. (Berl.) 2009, 93, 527–537. [Google Scholar] [CrossRef]
- Yu, Z.; Forster, R.J. Nucleic acid extraction, oligonucleotide probes and PCR methods. In Methods in Gut Microbial Ecology for Ruminants; Makkar, H.P.S., McSweeney, C.S., Eds.; International Atomic Energy Agency, FAO, Springer: Dordrecht, The Netherlands, 2005; pp. 81–104. [Google Scholar]
- Martínez, E.D.; Turnbull, K.E.; Quigley, S.P.; Streeter, S.J.; Swain, A.; Klieve, A.V.; Ouwerkerk, D.; Poppi, D.P. Liquid phase DGGE profiles of rumen bacteria from Brahman cross steers selected into two groups on the basis of post-weaning live weight gain on low crude protein pasture. Anim. Prod. Sci. 2012, 52, 647–652. [Google Scholar] [CrossRef]
- Kocherginskaya, S.A.; Aminov, R.I.; White, B.A. Analysis of the rumen bacterial diversity under two different diet conditions using denaturing gradient gel electrophoresis, random sequencing, and statistical ecology approaches. Anaerobe 2001, 7, 119–134. [Google Scholar] [CrossRef]
- Singh, R.; White, D.; Demirel, Y.; Kelly, R.; Noll, K.; Blum, P. Uncoupling Fermentative Synthesis of Molecular Hydrogen from Biomass Formation in Thermotoga maritima. Appl. Environ. Microbiol. 2018, 84, e00998-18. [Google Scholar] [CrossRef] [PubMed]
- Czerkawski, J.W. An Introduction to Rumen Studies; Pergamon Press: Oxford, UK, 1986; p. 246. [Google Scholar]
- Silver Chloride Electrode. Available online: https://en.wikipedia.org/wiki/Silver_chloride_electrode#cite_note-5 (accessed on 24 March 2019).
- Sauer, F.D.; Erfle, J.D.; Mahadevan, S. Amino acid biosynthesis in mixed rumen cultures. Annu. Rev. Biochem. 1975, 150, 357–372. [Google Scholar] [CrossRef] [Green Version]
- Umbarger, H.E. Amino acid biosynthesis and its regulation. Annu. Rev. Biochem. 1978, 47, 532–606. [Google Scholar] [CrossRef]
- Allison, M.J. Biosynthesis of amino acids by ruminal microorganisms. J. Anim. Sci. 1969, 29, 797–807. [Google Scholar] [CrossRef] [PubMed]
- JMP®, version 13.2.1; SAS Institute: Cary, NC, USA, 2016.
- Ungerfeld, E.M.; Instituto de Investigaciones Agropecuarias INIA, Temuco, Chile. Preliminary experiment. 2016; unpublished. [Google Scholar]
- Janssen, P.H. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Technol. 2010, 160, 1–22. [Google Scholar] [CrossRef]
- Prasad, K. Flaxseed and cardiovascular health. J. Cardiovasc. Pharmacol. 2009, 54, 369–377. [Google Scholar] [CrossRef]
- Zhang, C.M.; Guo, Y.Q.; Yuan, Z.P.; Wu, Y.M.; Wang, J.K.; Liu, J.X.; Zhu, W.Y. Effect of octadeca carbon fatty acids on microbial fermentation, methanogenesis and microbial flora in vitro. Anim. Feed Sci. Technol. 2008, 146, 259–269. [Google Scholar] [CrossRef]
- Yang, C.; Rooke, J.A.; Cabeza, I.; Wallace, R.J. Nitrate and inhibition of ruminal methanogenesis: Microbial ecology, obstacles, and opportunities for lowering methane emissions from ruminant livestock. Front. Microbiol. 2016, 7, 132. [Google Scholar] [CrossRef] [PubMed]
- Petersen, S.O.; Hellwing, A.L.; Brask, M.; Hojberg, O.; Poulsen, M.; Zhu, Z.; Baral, K.R.; Lund, P. Dietary nitrate for methane mitigation leads to nitrous oxide emissions from dairy cows. J. Environ. Qual. 2015, 44, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Ungerfeld, E.M.; Rust, S.R.; Burnett, R. Use of some novel alternative electron sinks to inhibit ruminal methanogenesis. Reprod. Nutr. Dev. 2003, 43, 189–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Meng, Q.; Yu, Z. Effects of methanogenic inhibitors on methane production and abundances of methanogens and cellulolytic bacteria in in vitro ruminal cultures. Appl. Environ. Microbiol. 2011, 77, 2634–2639. [Google Scholar] [CrossRef] [PubMed]



| Response | Treatment | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | BES 1 | AQ | CL | BTCM | PA | E2B | LO | SN | SEM | p = | |
| Gas production (mmol) | 1.80 a3 | 1.63 a | 0.91 cd | 1.67 a | 1.59 ab | 0.79 d | 1.24 bc | 1.71 a | 0.52 d | 0.17 | <0.001 |
| CH4 2 (mol/100 mol total gas) | 10.9 a | 0.55 bc | 2.34 bc | 1.36 bc | 2.70 bc | 0.076 bc | 3.13 b | 8.05a | ND 4 | 1.05 | <0.001 |
| CH4 production (µmol/g incubated OM) | 242 a | 14.3 b | 29.3 b | 46.7 b | 51.1 b | 1.79 b | 62.1 b | 205 a | ND | 28.3 | <0.001 |
| CH4 production (µmol/g truly digested OM) | 523 a | 30.4 bc | 121 bc | 68.3 bc | 110 bc | 9.26 c | 257 b | 575 a | ND | 57.5 | <0.001 |
| H2 accumulation (mol/100 mol total gas) | 0.91 cd | 2.91 b | 5.60 a | 2.34 bc | 2.42 bc | 1.68 bcd | 0.36 d | 0.84 cd | 0.39 d | 0.64 | <0.001 |
| H2 accumulation (µmol/g incubated OM) | 14.7 c | 60.2 a | 76.4 a | 51.2 ab | 52.3 a | 21.7 bc | 6.79 c | 19.5 c | 2.97 c | 9.95 | <0.001 |
| H2 accumulation (µmol/g truly digested OM) | 37.4 bc | 155 abc | 307 a | 213 abc | 257 ab | 84.6 abc | 26.8 c | 68.3 bc | 8.52 c | 65.3 | <0.001 |
| Final pH | 6.72 c | 6.70 c | 6.83 a | 6.72 c | 6.72 c | 6.79 ab | 6.76 bc | 6.70 c | 6.84 a | 0.042 | <0.001 |
| Final Eh | −163 ab | −167 ab | −220 c | −171 b | −170 b | −167 ab | −165 ab | −157 ab | −140 a | 6.06 | <0.001 |
| Total VFA (mM) | 27.2 a | 26.1 ab | 20.7 bc | 26.0 ab | 25.5 ab | 17.3 c | 23.7 ab | 27.2 a | 17.7 c | 7.95 | <0.001 |
| Acetate (mol/100 mol) | 60.2 cd | 55.3 d | 62.3 c | 56.7 cd | 57.0 cd | 70.0 a | 63.2 bc | 57.8 cd | 69.7 ab | 2.18 | <0.001 |
| Propionate (mol/100 mol) | 26.8 a | 30.0 a | 19.3 b | 29.5 a | 29.5 a | 15.8 b | 19.6 b | 29.4 a | 16.8 b | 2.06 | <0.001 |
| Butyrate (mol/100 mol) | 8.04 c | 9.65 bc | 11.6 ab | 8.63 c | 8.55 c | 7.56 c | 12.7 a | 7.92 c | 7.13 c | 1.06 | <0.001 |
| Isobutyrate (mol/100 mol) | 1.37 | 1.48 | 1.73 | 1.67 | 1.49 | 1.70 | 1.66 | 1.47 | 1.68 | 0.32 | 0.11 |
| 2- and 3-methylbutyrate (mol/100 mol) | 1.86 | 1.75 | 2.31 | 1.77 | 1.63 | 1.88 | 2.53 | 1.88 | 1.92 | 1.02 | 0.081 |
| Valerate (mol/100 mol) | 1.30 c | 1.24 c | 1.80 a | 1.22 c | 1.32 c | 1.43 bc | 1.69 ab | 1.19 c | 1.78 a | 0.22 | <0.001 |
| Caproate (mol/100 mol) | 0.44 b | 0.56 b | 0.88 ab | 0.50 b | 0.50 b | 1.75 a | 0.42 b | 0.43 b | 1.00 ab | 0.44 | 0.008 |
| Acetate/propionate (mol/mol) | 2.36 bcd | 1.96 d | 3.27 abc | 2.02 d | 2.04 cd | 4.49 a | 3.57 ab | 2.07 cd | 4.30 a | 0.30 | <0.001 |
| NH4+ (mM) | 8.26 | 8.77 | 9.29 | 8.64 | 8.95 | 9.09 | 9.09 | 8.87 | 9.24 | 1.00 | 0.096 |
| Response | Treatment | SEM | p = | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | BES 1 | AQ | CL | BTCM | PA | E2B | LO | SN | |||
| True OM digestibility (%) 2 | 42.7 a 3 | 41.4 ab | 31.2 ab | 46.4 a | 34.2 ab | 27.2 ab | 22.8 b | 31.4 ab | 31.2 ab | 6.13 | 0.008 |
| Microbial OM (mg) | 170 | 215 | 223 | 164 | 155 | 209 | 136 | 120 | 188 | 28.7 | 0.14 |
| Microbial N (mg) | 3.13 ab | 2.12 bc | 1.75 bc | 3.20 ab | 3.55 ab | 1.19 c | 1.97 bc | 3.97 a | 1.37 c | 0.47 | <0.001 |
| Total microbial AA-N (mg) | 1.48 | 1.20 | 0.87 | 1.51 | 1.56 | 0.94 | 0.89 | 1.59 | 1.48 | 0.16 | 0.001 |
| Amino acid (g/100 g total AA) | |||||||||||
| Asp | 11.7 bc | 12.8 ab | 5.73 c | 11.9 abc | 12.0 abc | 9.43 bc | 13.8 ab | 13.3 ab | 16.9 a | 1.23 | <0.001 |
| Glu | 18.1 | 10.9 | 15.0 | 19.8 | 19.9 | 18.0 | 11.8 | 18.0 | 28.5 | 3.95 | 0.067 |
| Ser | 5.28 a | 5.55 a | 4.85 abc | 4.81 abc | 5.14 ab | 3.34 c | 6.12 a | 5.23 a | 3.53 bc | 0.42 | <0.001 |
| Gly | 6.27 ab | 6.67 ab | 6.33 ab | 5.32 b | 6.14 ab | 7.17 ab | 8.11 a | 6.32 ab | 6.14 ab | 0.70 | 0.008 |
| His | 1.18 ab | 2.25 ab | 2.21 ab | 1.71 ab | 2.38 a | 0.62 b | 0.52 b | 1.47 ab | 0.73 ab | 0.47 | 0.005 |
| Arg | 4.26 ab | 4.98 a | 4.41 ab | 3.38 bcd | 4.26 abc | 2.82 cd | 3.23 bcd | 3.65 abc | 2.03 d | 0.48 | <0.001 |
| Thr | 5.50 a | 5.77 a | 5.33 a | 4.97 a | 5.05 a | 2.86 b | 5.46 a | 4.94 a | 2.37 b | 0.43 | <0.001 |
| Ala | 8.58 b | 10.2 ab | 13.8 a | 8.34 b | 8.42 b | 10.8 ab | 14.2 a | 10.1 ab | 8.93 b | 1.67 | <0.001 |
| Pro | 3.26 bc | 3.83 abc | 4.46 ab | 3.93 abc | 3.61 abc | 4.74 a | 3.57 abc | 3.39 abc | 2.78 c | 0.36 | 0.002 |
| Tyr | 4.42 a | 4.57 a | 3.93 ab | 4.57 a | 3.75 ab | 1.83 b | 3.13 ab | 3.91 ab | 2.01 b | 0.60 | <0.001 |
| Val | 6.03 a | 7.42 a | 8.13 a | 5.65 a | 6.09 a | 6.50 a | 7.33 a | 5.58 a | 5.82 a | 0.63 | 0.032 |
| Ile | 5.50 | 6.45 | 6.75 | 5.01 | 5.35 | 5.95 | 5.94 | 6.10 | 4.89 | 0.41 | 0.058 |
| Leu | 7.02 a | 7.95 a | 7.55 a | 6.14 ab | 6.54 a | 3.28 c | 7.75 a | 6.77 a | 3.95 bc | 0.66 | <0.001 |
| Lys | 7.61 | 8.81 | 9.35 | 7.00 | 7.32 | 7.40 | 7.77 | 8.54 | 7.78 | 0.61 | 0.17 |
| Phe | 4.49 a | 4.71 a | 4.35 ab | 3.80 ab | 3.92 ab | 3.55 ab | 2.98 b | 3.98 ab | 3.57 ab | 0.37 | 0.009 |
| Response | Treatment | SEM | p = | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | BES 1 | AQ | CL | BTCM | PA | E2B | LO | SN | |||
| VFA and Gases | |||||||||||
| [2H] Produced | |||||||||||
| Acetate (mmol) | 4.17 a3 | 3.79 ab | 3.33 bc | 3.81 ab | 3.76 ab | 2.99 c | 4.38 a | 4.14 a | 3.00 c | 1.27 | <0.001 |
| Propionate (mmol) | 0.91 a | 0.97 a | 0.53 b | 0.95 a | 0.93 a | 0.38 b | 0.56 b | 1.02 a | 0.41 b | 0.21 | <0.001 |
| Butyrate (mmol) | 1.28 b | 1.39 b | 1.26 bc | 1.27 b | 1.25 b | 0.81 c | 2.01 a | 1.35 b | 0.83 c | 0.53 | <0.001 |
| Valerate (mmol) | 0.16 ab | 0.15 abc | 0.15 abc | 0.14 abc | 0.14 abc | 0.11 c | 0.18 a | 0.14 abc | 0.13 bc | 0.073 | <0.001 |
| Total [2H] produced (mmol) 2 | 6.52 ab | 6.30 ab | 5.27 bc | 6.17 ab | 6.08 ab | 4.28 c | 7.13 a | 6.66 ab | 4.38 c | 2.06 | <0.001 |
| [2H] incorporated | |||||||||||
| Propionate (mmol) | 1.81 a | 1.94 a | 1.07 b | 1.89 a | 1.86 a | 0.76 b | 1.12 b | 2.04 a | 0.82 b | 0.43 | <0.001 |
| Butyrate (mmol) | 0.64 b | 0.70 b | 0.63 bc | 0.63 b | 0.62 b | 0.41 c | 1.01 a | 0.68 b | 0.42 c | 0.26 | <0.001 |
| Valerate (mmol) | 0.22 ab | 0.20 abc | 0.20 abc | 0.19 abc | 0.18 abc | 0.14 c | 0.24 a | 0.19 abc | 0.18 bc | 0.097 | <0.001 |
| CH4 (mmol) | 0.78 a | 0.037 b | 0.090 b | 0.15 b | 0.25 b | 0.0060 b | 0.26 b | 0.70 a | ND 3 | 0.11 | <0.001 |
| H2 (mmol) | 0.012 c | 0.046 a | 0.048 a | 0.041 a | 0.039 ab | 0.017 bc | 0.0082 c | 0.016 bc | 0.0024 c | 0.0082 | <0.001 |
| Total [2H] incorporated (mmol) | 3.45 a | 2.91 ab | 2.09 bc | 2.85 ab | 2.97 ab | 1.33 c | 2.62 ab | 3.62 a | 1.35 c | 0.76 | <0.001 |
| [2H] recovery (%) | 59.2 a | 48.9 ab | 40.1 bc | 47.1 ab | 49.5 ab | 29.2 c | 37.4 bc | 60.8 a | 27.1 c4 | 3.72 | <0.001 |
| Amino acids | |||||||||||
| Max [2H] net incorporation (mmol) | 0.10 | 0.098 | 0.072 | 0.13 | 0.13 | 0.084 | 0.073 | 0.12 | 0.13 | 0.022 | 0.34 |
| Min [2H] net incorporation (mmol) | −0.11 | −0.097 | −0.060 | −0.13 | −0.14 | −0.091 | −0.060 | −0.13 | −0.15 | 0.024 | 0.16 |
| Overall (VFA + gases + AA) | |||||||||||
| Max [2H] recovery (%) | 62.0a | 52.1 ab | 41.9 abc | 52.2 ab | 55.8 ab | 35.6 c | 38.1 bc | 63.3 a | 37.4 c4 | 4.90 | <0.001 |
| Min [2H] recovery (%) | 58.7a | 49.1 ab | 40.1 abc | 47.5 ab | 51.3 ab | 31.0 c | 36.9 bc | 59.1 a | 29.3 c4 | 4.13 | <0.001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ungerfeld, E.M.; Aedo, M.F.; Martínez, E.D.; Saldivia, M. Inhibiting Methanogenesis in Rumen Batch Cultures Did Not Increase the Recovery of Metabolic Hydrogen in Microbial Amino Acids. Microorganisms 2019, 7, 115. https://doi.org/10.3390/microorganisms7050115
Ungerfeld EM, Aedo MF, Martínez ED, Saldivia M. Inhibiting Methanogenesis in Rumen Batch Cultures Did Not Increase the Recovery of Metabolic Hydrogen in Microbial Amino Acids. Microorganisms. 2019; 7(5):115. https://doi.org/10.3390/microorganisms7050115
Chicago/Turabian StyleUngerfeld, Emilio M., M. Fernanda Aedo, Emilio D. Martínez, and Marcelo Saldivia. 2019. "Inhibiting Methanogenesis in Rumen Batch Cultures Did Not Increase the Recovery of Metabolic Hydrogen in Microbial Amino Acids" Microorganisms 7, no. 5: 115. https://doi.org/10.3390/microorganisms7050115
APA StyleUngerfeld, E. M., Aedo, M. F., Martínez, E. D., & Saldivia, M. (2019). Inhibiting Methanogenesis in Rumen Batch Cultures Did Not Increase the Recovery of Metabolic Hydrogen in Microbial Amino Acids. Microorganisms, 7(5), 115. https://doi.org/10.3390/microorganisms7050115