Vegetation-Dependent Response to Drought in Salt Marsh Ammonia-Oxidizer Communities
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Sediment Conditions
3.2. Gene Abundance
3.3. Community Composition
3.4. MiSeq Analysis of 16S rRNA Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bernhard, A.E.; Bollmann, A. Estuarine nitrifiers: New players, patterns and processes. Estuar. Coast. Shelf Sci. 2010, 88, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Mosier, A.C.; Francis, C.A. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 2008, 10, 3002–3016. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, A.E.; Tucker, J.; Giblin, A.E.; Stahl, D.A. Functionally distinct communities of ammonia-oxidizing bacteria along an estuarine salinity gradient. Environ. Microbiol. 2007, 9, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- Santoro, A.E.; Francis, C.A.; De Sieyes, N.R.; Boehm, A.B. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ. Microbiol. 2008, 10, 1068–1079. [Google Scholar] [CrossRef]
- Moin, N.S.; Nelson, K.A.; Bush, A.; Bernhard, A.E. Distribution and diversity of archaeal and bacterial ammonia oxidizers in salt marsh sediments. Appl. Environ. Microbiol. 2009, 75. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Yando, E.; Hildebrand, E.; Dwyer, C.; Kearney, A.; Waciega, A.; Valiela, I.; Bernhard, A.E. Differential responses of ammonia-oxidizing archaea and bacteria to long-term fertilization in a New England salt marsh. Front. Microbiol. 2013, 3, 445. [Google Scholar] [CrossRef] [Green Version]
- Bernhard, A.E.; Dwyer, C.; Idrizi, A.; Bender, G.; Zwick, R. Long-term impacts of disturbance on nitrogen-cycling bacteria in a New England salt marsh. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Thion, C.; Prosser, J.I. Differential response of nonadapted ammonia-oxidising archaea and bacteria to drying-rewetting stress. FEMS Microbiol. Ecol. 2014, 90, 380–389. [Google Scholar] [CrossRef]
- Bello, M.O.; Thion, C.; Gubry-Rangin, C.; Prosser, J.I. Differential sensitivity of ammonia oxidising archaea and bacteria to matric and osmotic potential. Soil Biol. Biochem. 2019, 129, 184–190. [Google Scholar] [CrossRef] [Green Version]
- Ye, F.; Ma, M.-H.; Op den Camp, H.J.M.; Chatzinotas, A.; Li, L.; Lv, M.-Q.; Wu, S.-J.; Wang, Y. Different Recovery Processes of Soil Ammonia Oxidizers from Flooding Disturbance. Microb. Ecol. 2018, 76, 1041–1052. [Google Scholar] [CrossRef]
- Urakawa, H.; Garcia, J.C.; Barreto, P.D.; Molina, G.A.; Barreto, J.C. A sensitive crude oil bioassay indicates that oil spills potentially induce a change of major nitrifying prokaryotes from the Archaea to the Bacteria. Environ. Pollut. 2012, 164, 42–45. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, P.R.; Kitchens, W.M. Vegetation change from chronic stress events: Detection of the effects of tide gate removal and long-term drought on a tidal marsh. J. Veg. Sci. 2007, 18, 431–442. [Google Scholar] [CrossRef]
- Hughes, A.L.H.; Wilson, A.M.; Morris, J.T. Hydrologic variability in a salt marsh: Assessing the links between drought and acute marsh dieback. Estuar. Coast. Shelf Sci. 2012, 111, 95–106. [Google Scholar] [CrossRef]
- Davis, D.A.; Malone, S.L.; Lovell, C.R. Responses of salt marsh plant rhizosphere diazotroph assemblages to drought. Microorganisms 2018, 6, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naylor, D.; Degraaf, S.; Purdom, E.; Coleman-Derr, D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017, 11, 2691–2704. [Google Scholar] [CrossRef] [Green Version]
- Arce, M.I.; Von Schiller, D.; Bengtsson, M.M.; Hinze, C.; Jung, H.; Alves, R.J.E.; Urich, T.; Singer, G. Drying and rainfall shape the structure and functioning of nitrifying microbial communities in riverbed sediments. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Fuchslueger, L.; Kastl, E.-M.; Bauer, F.; Kienzl, S.; Hasibeder, R.; Ladreiter-Knauss, T.; Schmitt, M.; Bahn, M.; Schloter, M.; Richter, A.; et al. Effects of drought on nitrogen turnover and abundances of ammonia-oxidizers in mountain grassland. Biogeosciences 2014, 11, 6003–6015. [Google Scholar] [CrossRef] [Green Version]
- Warren, R.S.; Neiring, W.A. Vegetation changes on a northeast tidal marsh: Interaction of sea-level rise and accretion. Ecology 1993, 74, 96–103. [Google Scholar] [CrossRef]
- Bernhard, A.E.; Marshall, D.; Yiannos, L. Increased variability of microbial communities in restored salt marshes nearly 30 years after tidal flow restoration. Estuar. Coasts 2012, 35, 1049–1059. [Google Scholar] [CrossRef] [Green Version]
- Strickland, J.D.H.; Parsons, T.R. A Practical Handbook of Seawater Analysis; Fisheries Research Board: Ottawa, ON, Canada, 1972. [Google Scholar]
- Rotthauwe, J.H.; Witzel, K.P.; Liesack, W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 1997, 63, 4704–4712. [Google Scholar]
- Nicolaisen, M.H.; Ramsing, N.B. Denaturing gradient gel electrophoresis (DGGE) approached to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 2002, 50, 189–203. [Google Scholar] [CrossRef]
- Francis, C.A.; Roberts, K.J.; Beman, J.M.; Santoro, A.E.; Oakley, B.B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 2005, 102, 14683–14688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernhard, A.E.; Sheffer, R.; Giblin, A.E.; Marton, J.M.; Roberts, B.J. Population dynamics and community composition of ammonia oxidizers in salt marshes after the Deepwater Horizon oil spill. Front. Microbiol. 2016, 7, 854. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruskal, J.B. Nonmetric multidimensional scaling: A numerical method. Psychometrika 1964, 29, 115–129. [Google Scholar] [CrossRef]
- McCune, B.; Mefford, M.J. PC-ORD, Multivariate Analysis of Ecological Data; Version 4; MjM Software: Gleneden Beach, OR, USA, 1999. [Google Scholar]
- McCune, B.; Grace, J. Analysis of Ecological Communities; MjM Software Design: Gleneden Beach, OR, USA, 2002. [Google Scholar]
- Trenberth, K.E.; Dai, A.; van der Schrier, G.; Jones, P.D.; Barichivich, J.; Briffa, K.R.; Sheffield, J. Global warming and changes in drought. Nat. Clim. Chang. 2014, 4, 17–22. [Google Scholar] [CrossRef]
- Placella, S.A.; Firestone, M.K. Transcriptional response of nitrifying communities to wetting of dry soil. Appl. Environ. Microbiol. 2013, 79, 3294–3302. [Google Scholar] [CrossRef] [Green Version]
- Gleeson, D.B.; Müller, C.; Banerjee, S.; Ma, W.; Siciliano, S.D.; Murphy, D.V. Response of ammonia oxidizing archaea and bacteria to changing water filled pore space. Soil Biol. Biochem. 2010, 42, 1888–1891. [Google Scholar] [CrossRef]
- Hink, L.; Nicol, G.W.; Prosser, J.I. Archaea produce lower yields of N2O than bacteria during aerobic ammonia oxidation in soil. Environ. Microbiol. 2017, 19, 4829–4837. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Veldsink, J.H.; Dini-Andreote, F.; Salles, J.F. Compositional and abundance changes of nitrogen-cycling genes in plant-root microbiomes along a salt marsh chronosequence. Antonie Leeuwenhoek 2018, 111, 2061–2078. [Google Scholar] [CrossRef] [PubMed]
- Boot, C.M.; Schaeffer, S.M.; Schimel, J.P. Static osmolyte concentrations in microbial biomass during seasonal drought in a California grassland. Soil Biol. Biochem. 2013, 57, 356–361. [Google Scholar] [CrossRef]
- Palomo, L.; Meile, C.; Joye, S.B. Drought impacts on biogeochemistry and microbial processes in salt marsh sediments: A flow-through reactor approach. Biogeochemistry 2013, 112, 389–407. [Google Scholar] [CrossRef]
- Stark, J.M.; Firestone, M.K. Mechanisms for soil moisture effects on activity of nitrifying bacteria. Appl. Environ. Microbiol. 1995, 61, 218–221. [Google Scholar] [PubMed]
- Orson, R.A.; Warren, R.S.; Niering, W.A. Interpreting sea level rise and rates of vertical marsh accretion in a southern New England tidal salt marsh. Estuar. Coast. Shelf Sci. 1998, 47, 419–429. [Google Scholar] [CrossRef]
- Lisa, J.A.; Song, B.; Tobias, C.R.; Hines, D.E. Genetic and biogeochemical investigation of sedimentary nitrogen cycling communities responding to tidal and seasonal dynamics in Cape Fear River Estuary. Estuar. Coast. Shelf Sci. 2015, 167, A313–A323. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.M.; Mosier, A.C.; Francis, C.A. Spatiotemporal Relationships Between the Abundance, Distribution, and Potential Activities of Ammonia-Oxidizing and Denitrifying Microorganisms in Intertidal Sediments. Microb. Ecol. 2015, 69, 13–24. [Google Scholar] [CrossRef]
- Marton, J.M.; Roberts, B.J.; Bernhard, A.E.; Giblin, A.E. Spatial and Temporal Variability of Nitrification Potential and Ammonia-Oxidizer Abundances in Louisiana Salt Marshes. Estuar. Coasts 2015, 38, 1824–1837. [Google Scholar] [CrossRef]
- Lage, M.D.; Reed, H.E.; Weihe, C.; Crain, C.M.; Martiny, J.B.H. Nitrogen and phosphorus enrichment alter the composition of ammonia-oxidizing bacteria in salt marsh sediments. ISME J. 2010, 4, 933–944. [Google Scholar] [CrossRef]
- Koops, H.P.; Pommerening-Roser, A. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 2001, 37, 1–9. [Google Scholar] [CrossRef]
- Holling, C.S. Resilience and stability of ecological systems. In The Future of Nature: Documents of Global Change; Yale University Press: New Haven, CT, USA, 2013; pp. 245–256. [Google Scholar]
- Sheik, C.S.; Beasley, W.H.; Elshahed, M.S.; Zhou, X.; Luo, Y.; Krumholz, L.R. Effect of warming and drought on grassland microbial communities. ISME J. 2011, 5, 1692–1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meisner, A.; Jacquiod, S.; Snoek, B.L.; Ten Hooven, F.C.; van der Putten, W.H. Drought legacy effects on the composition of soil fungal and prokaryote communities. Front. Microbiol. 2018, 9, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maestre, F.T.; Delgado-Baquerizo, M.; Jeffries, T.C.; Eldridge, D.J.; Ochoa, V.; Gozalo, B.; Quero, J.L.; García-Gómez, M.; Gallardo, A.; Ulrich, W.; et al. Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc. Natl. Acad. Sci. USA 2015, 112, 15684–15689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013, 7, 2229–2241. [Google Scholar] [CrossRef] [PubMed]
- Berga, M.; Szekely, A.J.; Langenheder, S. Effects of disturbance intensity and frequency on bacterial community composition and function. PLoS ONE 2012, 7, e36959. [Google Scholar] [CrossRef] [Green Version]
- Ager, D.; Evans, S.; Li, H.; Lilley, A.K.; van der Gast, C.J. Anthropogenic disturbance affects the structure of bacterial communitiesemi. Environ. Microbiol. 2010, 12, 670–678. [Google Scholar] [CrossRef]
- Nielsen, D.L.; Brock, M.A.; Rees, G.N.; Baldwin, D.S. Effects of increasing salinity on freshwater ecosystems in Australia. Aust. J. Bot. 2003, 51, 655–665. [Google Scholar] [CrossRef]
- Nuccio, E.E.; Anderson-Furgeson, J.; Estera, K.Y.; Pett-Ridge, J.; de Valpine, P.; Brodie, E.L.; Firestone, M.K. Climate and edaphic controllers influence rhizosphere community assembly for a wild annual grass. Ecology 2016, 97, 1307–1318. [Google Scholar] [CrossRef] [Green Version]
- Blankinship, J.C.; Niklaus, P.A.; Hungate, B.A. A meta-analysis of responses of soil biota to global change. Oecologia 2011, 165, 553–565. [Google Scholar] [CrossRef]
- Nave, L.E.; Gough, C.M.; Maurer, K.D.; Bohrer, G.; Hardiman, B.S.; Le Moine, J.; Munoz, A.B.; Nadelhoffer, K.J.; Sparks, J.P.; Strahm, B.D.; et al. Disturbance and the resilience of coupled carbon and nitrogen cycling in a north temperate forest. J. Geophys. Res. Biogeosciences 2011, 116. [Google Scholar] [CrossRef] [Green Version]
- Alber, M.; Swenson, E.M.; Adamowicz, S.C.; Mendelssohn, I.A. Salt Marsh Dieback: An overview of recent events in the US. Estuar. Coast. Shelf Sci. 2008, 80, 1–11. [Google Scholar] [CrossRef]
- Sharp, S.J.; Angelini, C. Whether disturbances alter salt marsh soil structure dramatically affects Spartina alterniflora recolonization rate. Ecosphere 2016, 7, e01540. [Google Scholar] [CrossRef]
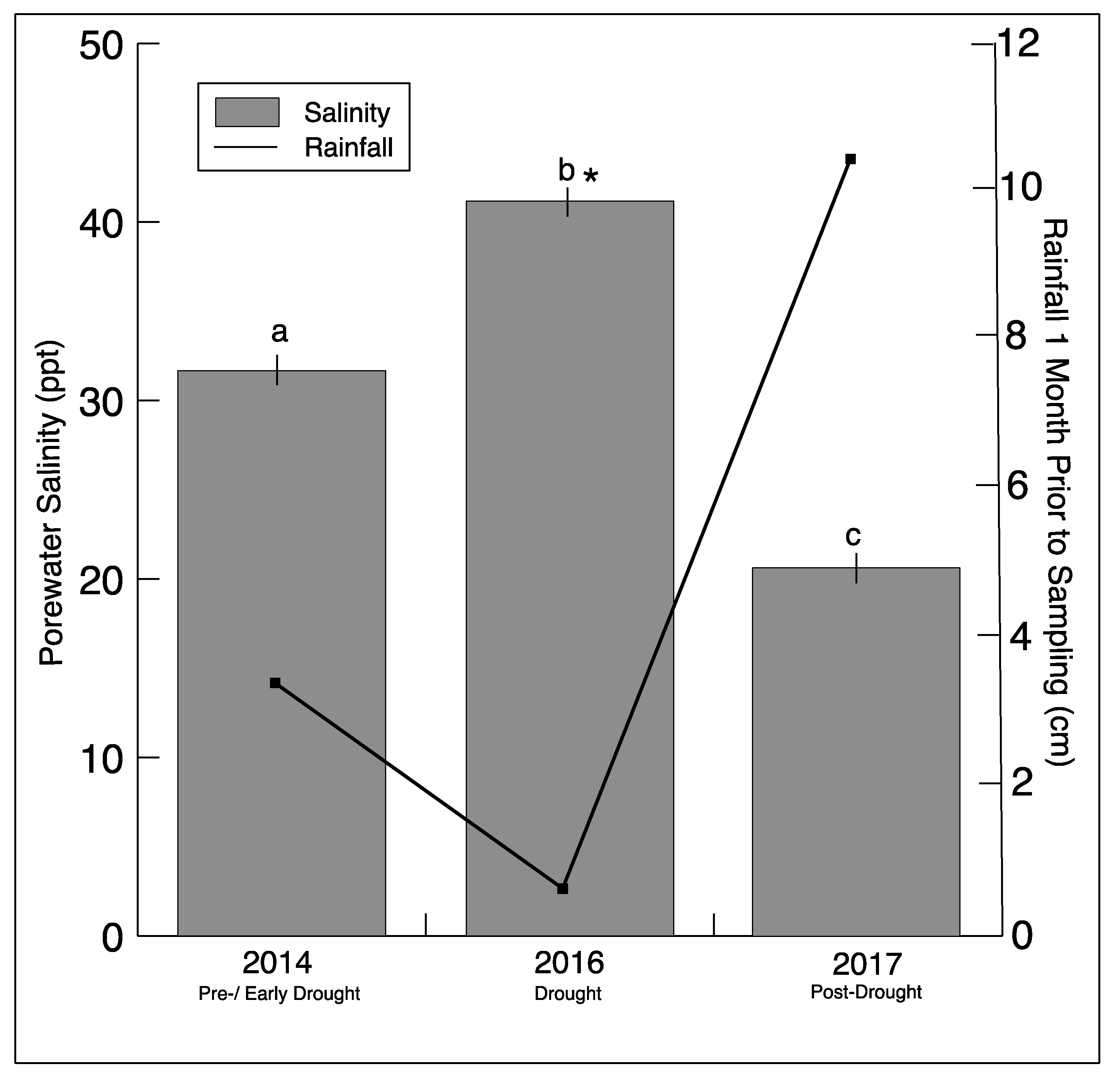
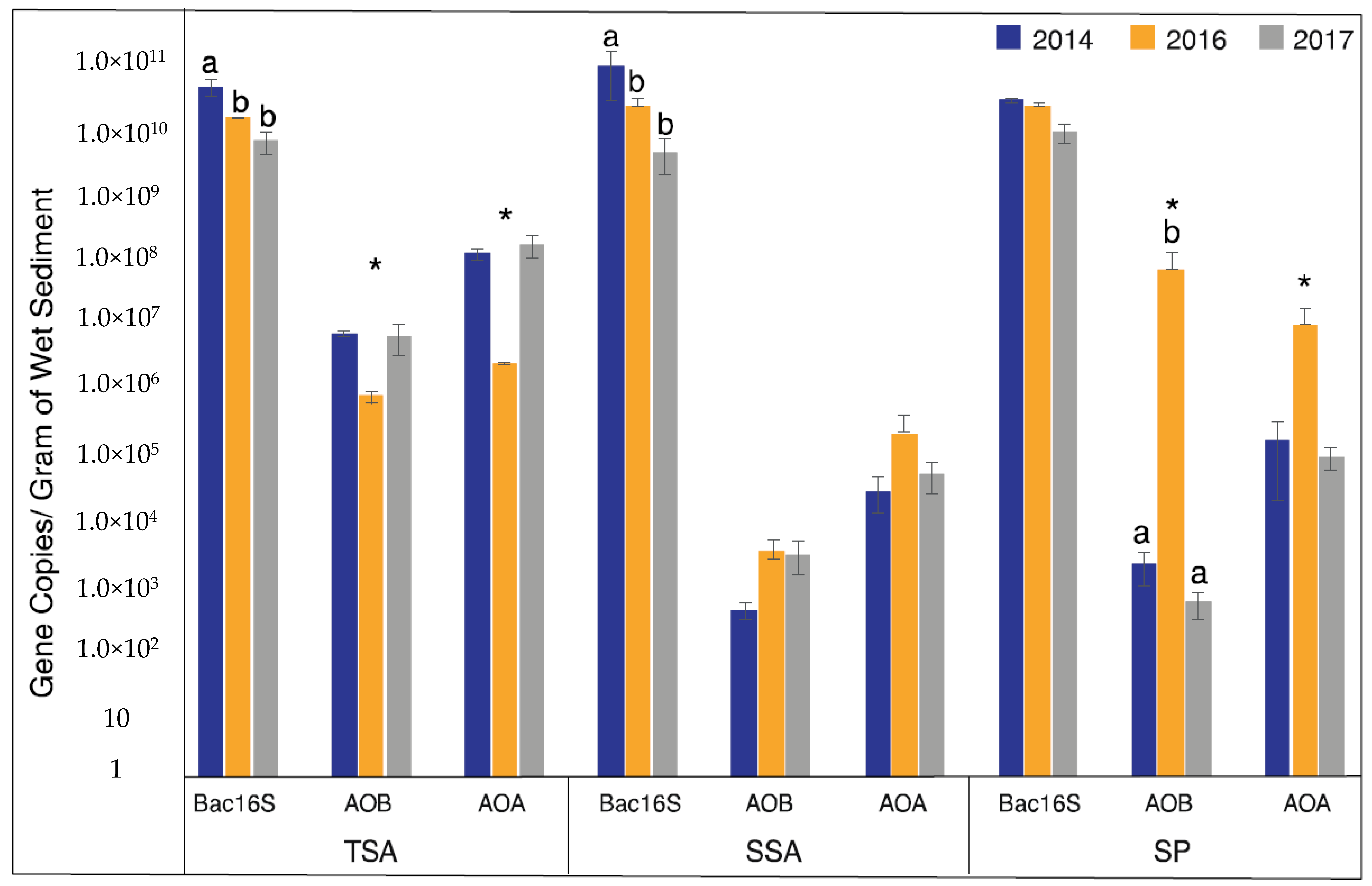
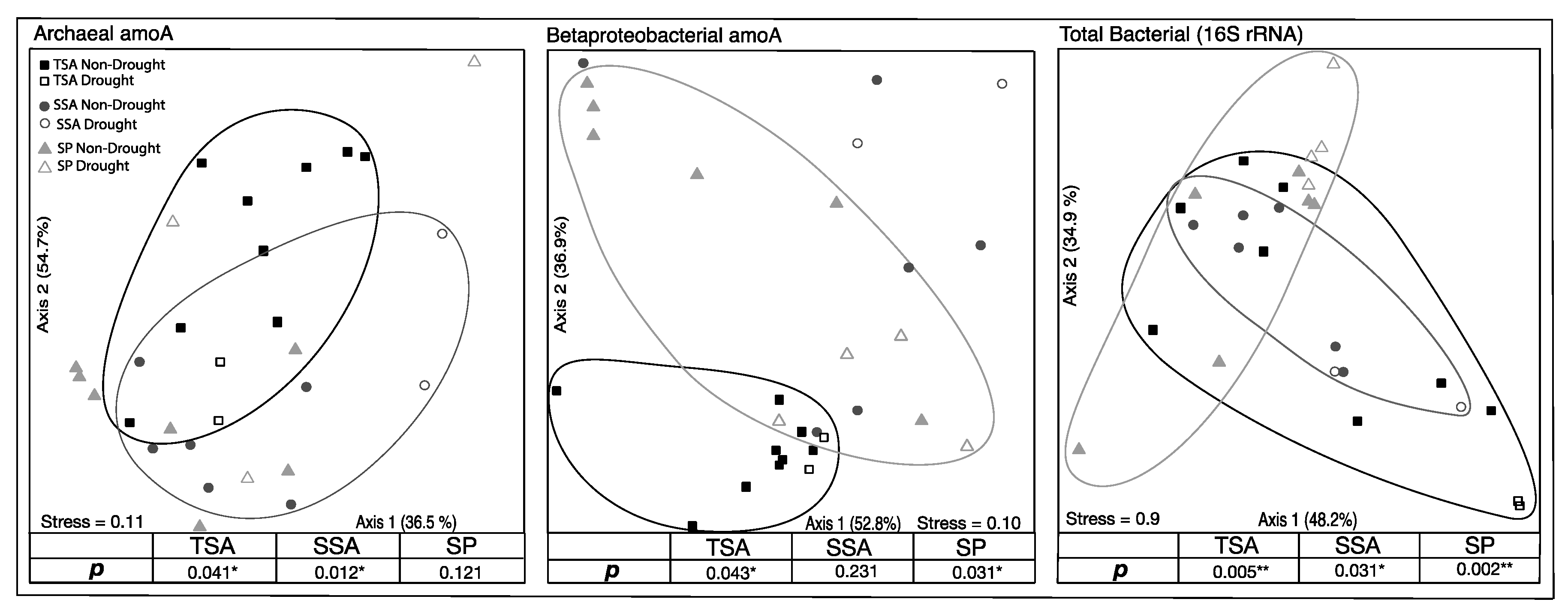

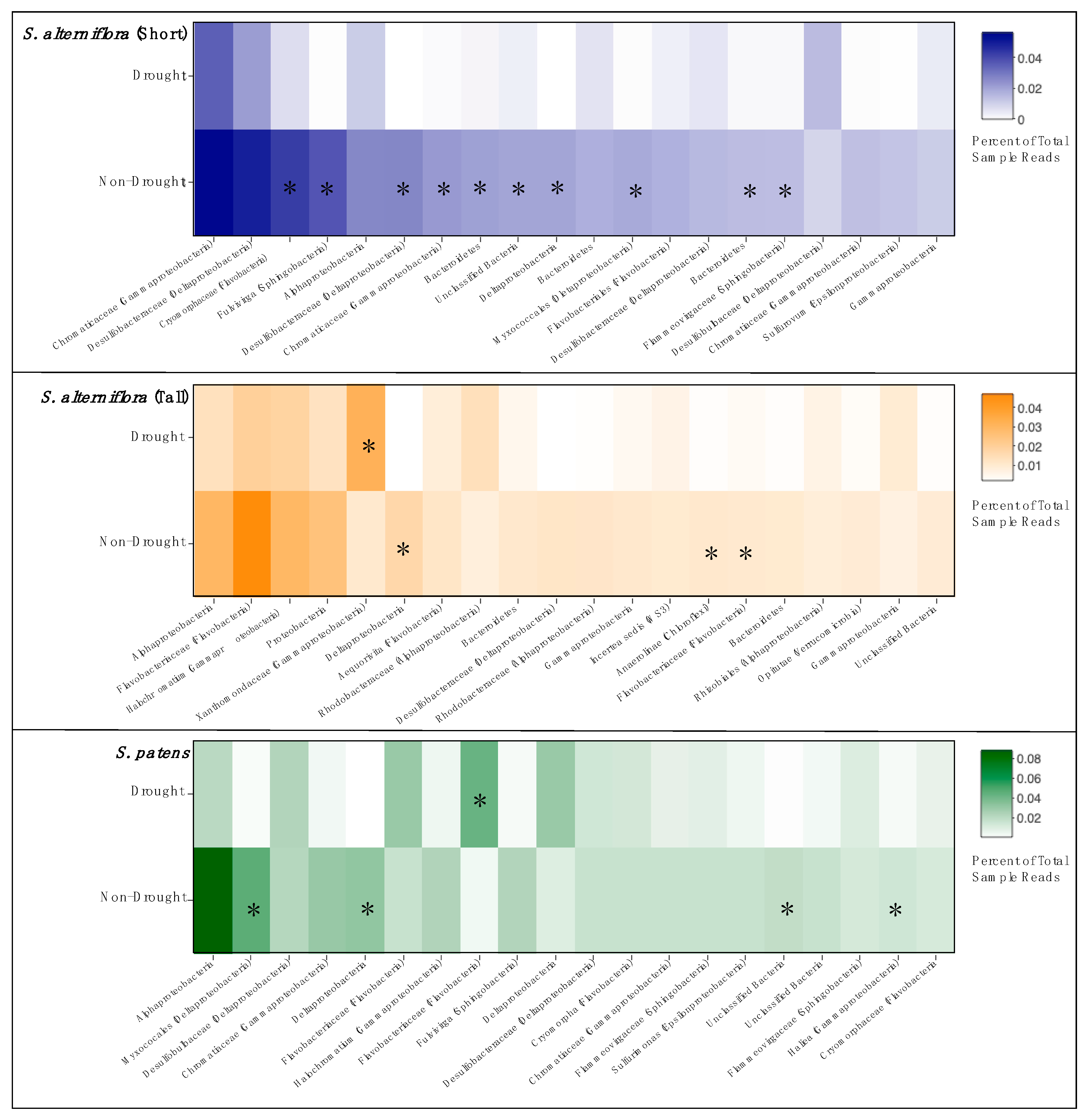
| Site | Year | Salinity | Soil Moisture (%) | Nitrate (µm NO3−-N) | Ammonium (µM NH4+-N) |
|---|---|---|---|---|---|
| SP | 2014 | 33.0 | 87.8 ± 0.8 | - | - |
| 2016 | 42.2 ± 13.6 a | 83.3 ± 0.04 | bd | bd | |
| 2017 | 24.3 ± 3.8 b | 85.4 ±2.1 | 2.8 ± 4.4 | bd | |
| SSA | 2014 | 34.0 | 87.8 ± 1.5 | - | - |
| 2016 | 39.0 ± 2.8 a | 87.8 ± 0.0 | bd | bd | |
| 2017 | 20.3 ± 1.2 b | 88.8 ± 2.2 | 9.9 ± 3.3 | 0.1 ± 0.1 | |
| TSA | 2014 | 28.0 | 62.3 ± 13.3 a | - | - |
| 2016 | na | 20.5 ± 0.1 b | na | bd | |
| 2017 | 23.3 ± 2.9 | 47.8 ± 6.1a | 23.7 ± 19.6 | 18.9 ± 10.9 |
| Inverse Simpsons | 2014 | 2016 | 2017 |
|---|---|---|---|
| ALL VEG | 253.5 ± 117.5 a | 332.3 ± 115.4 a | 236.2 ± 206.3 a |
| TSA | 370.2 ± 92.8 a | 356.1 ± 181.1 a | 479.9 ± 188.0 a |
| SSA | 158.3 ± 7.2 a | 233.5 ± 50.2 b | 129.7 ± 16.8 a |
| SP | 232.1 ± 108.9 a,b | 382.1 ± 90.6 b | 99.1 ± 18.3 a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beltz, J.K.; McMahon, H.; Torres Nunez, I.; Bernhard, A.E. Vegetation-Dependent Response to Drought in Salt Marsh Ammonia-Oxidizer Communities. Microorganisms 2020, 8, 9. https://doi.org/10.3390/microorganisms8010009
Beltz JK, McMahon H, Torres Nunez I, Bernhard AE. Vegetation-Dependent Response to Drought in Salt Marsh Ammonia-Oxidizer Communities. Microorganisms. 2020; 8(1):9. https://doi.org/10.3390/microorganisms8010009
Chicago/Turabian StyleBeltz, Jack K., Hayley McMahon, Isis Torres Nunez, and Anne E. Bernhard. 2020. "Vegetation-Dependent Response to Drought in Salt Marsh Ammonia-Oxidizer Communities" Microorganisms 8, no. 1: 9. https://doi.org/10.3390/microorganisms8010009
APA StyleBeltz, J. K., McMahon, H., Torres Nunez, I., & Bernhard, A. E. (2020). Vegetation-Dependent Response to Drought in Salt Marsh Ammonia-Oxidizer Communities. Microorganisms, 8(1), 9. https://doi.org/10.3390/microorganisms8010009





