Small Sample Stress: Probing Oxygen-Deprived Ammonia-Oxidizing Bacteria with Raman Spectroscopy In Vivo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Cultures
2.2. Culture Conditions
2.3. Resonance Raman Measurements
2.4. Data Preparation and Spectral Analysis
3. Results and Discussion
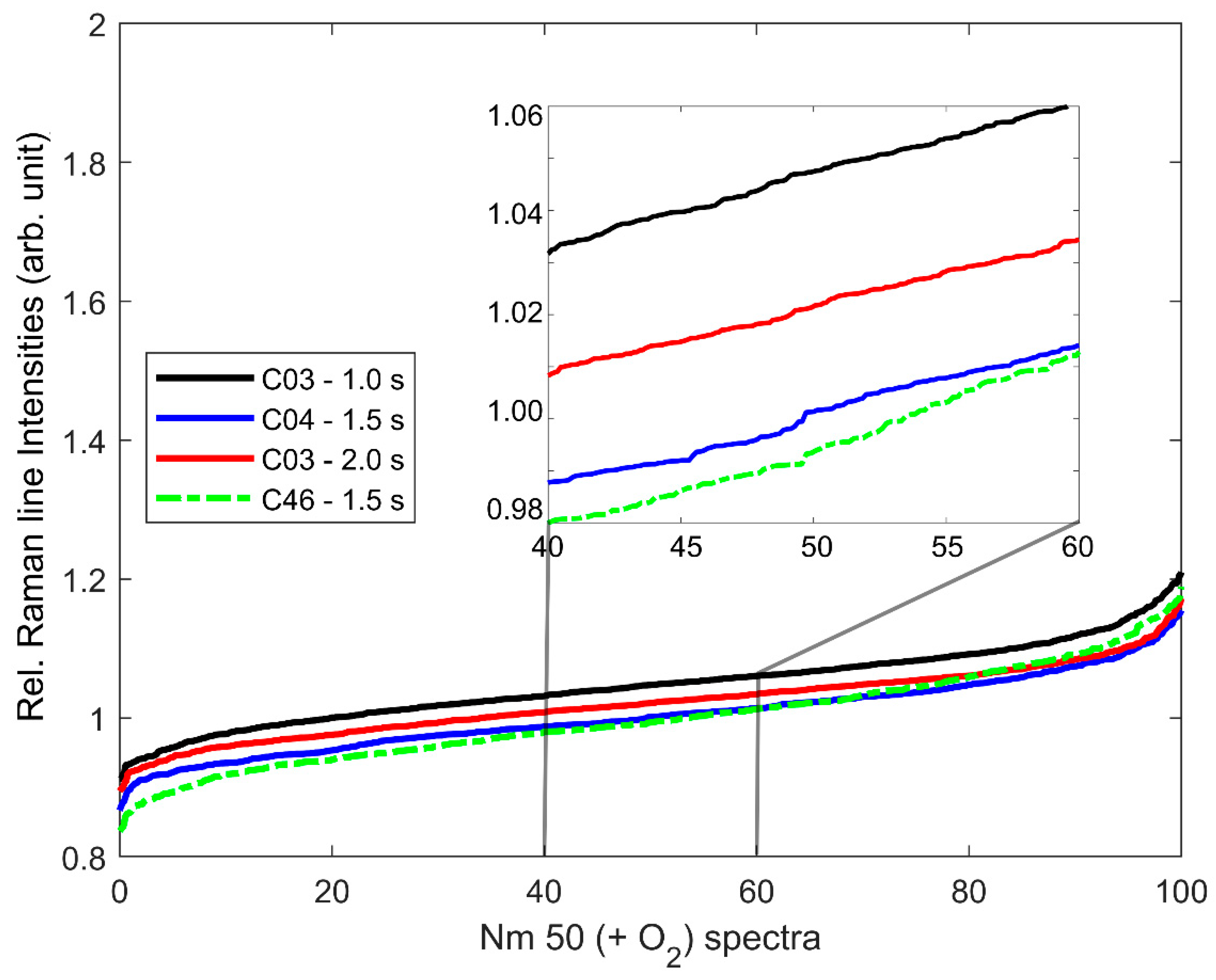
3.1. Cytochrome-C-Resonant Raman Spectra and Relative Raman Line Intensity (relRI)
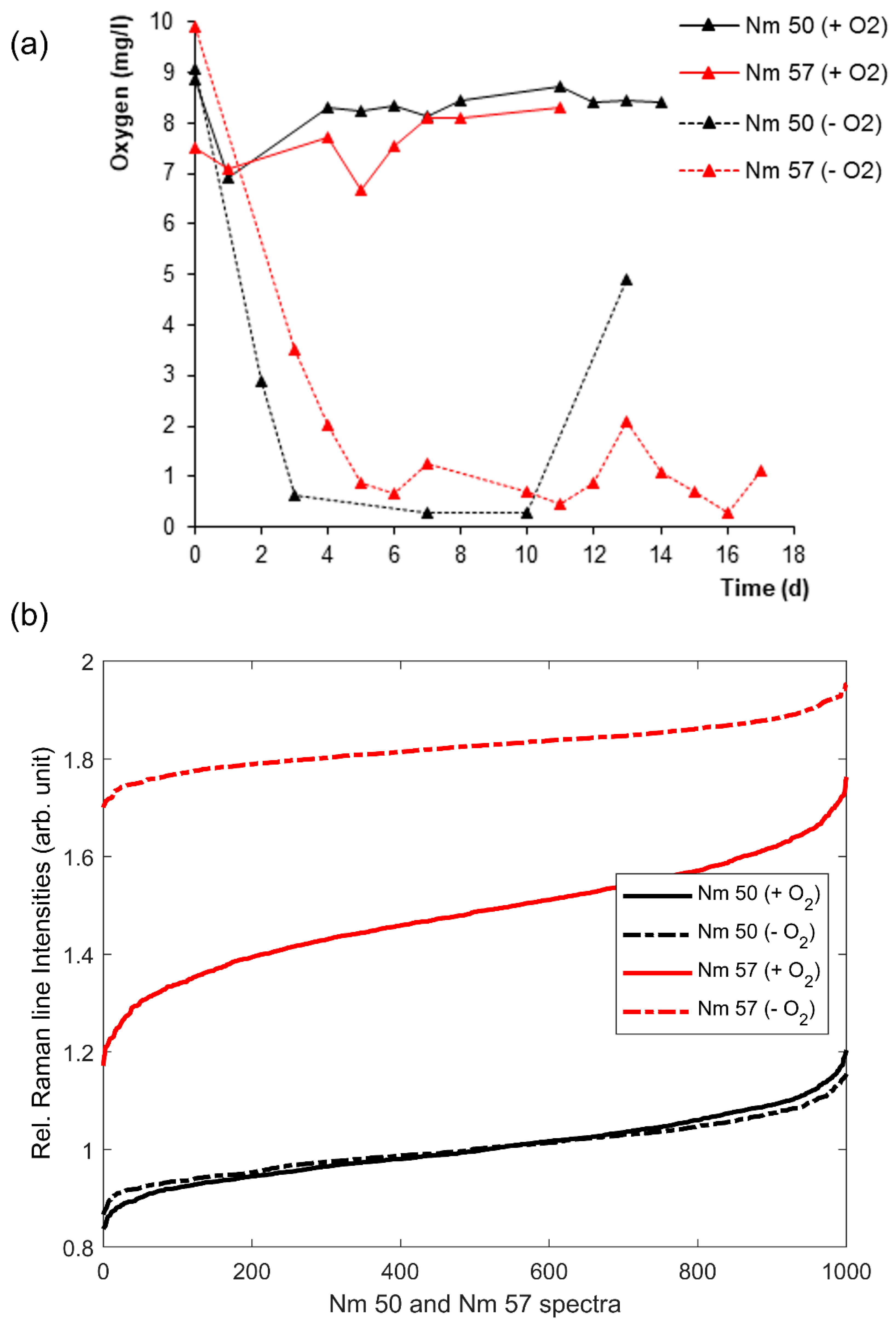
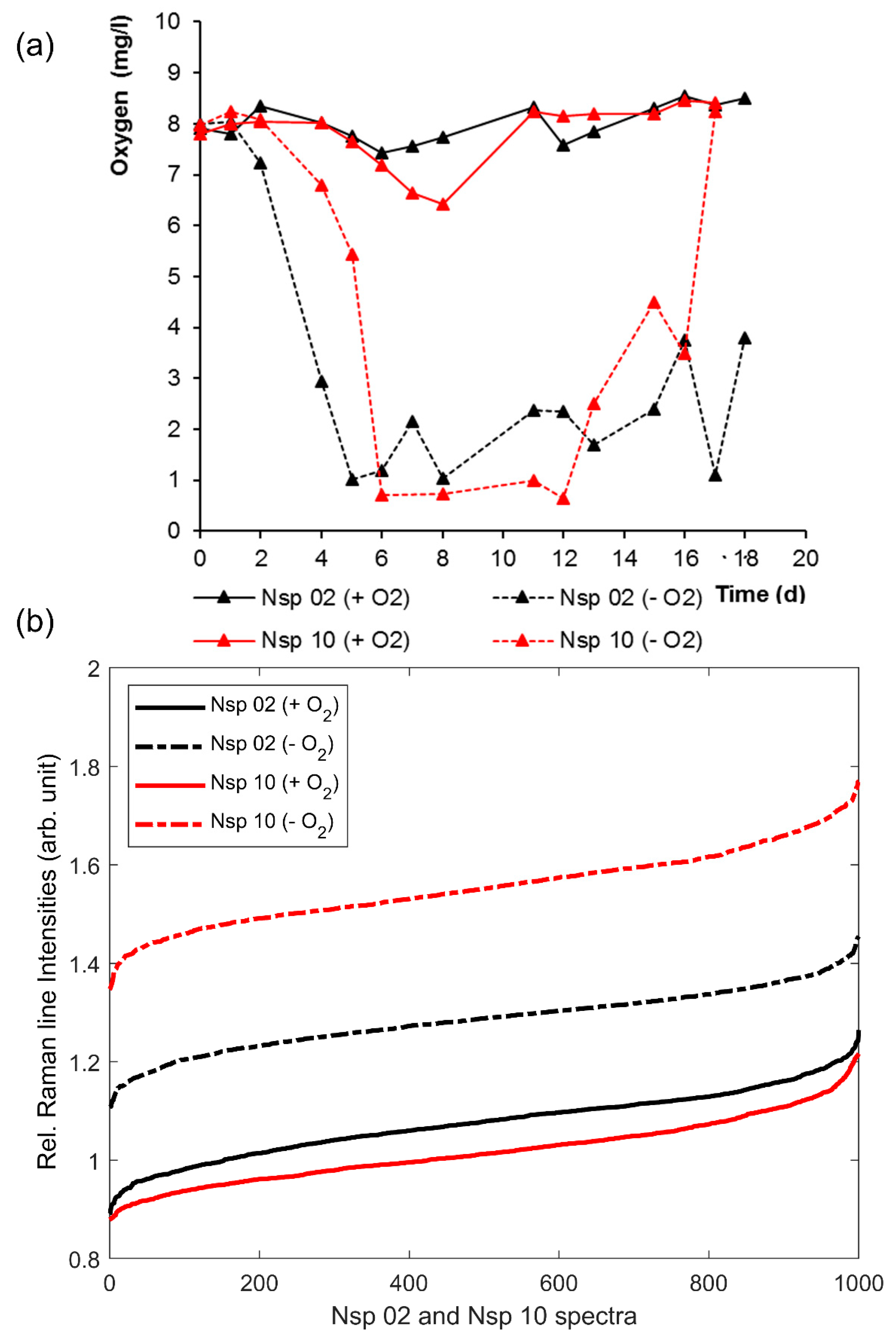
3.2. Effects of Oxygen Stress on the Cytochrome-C Content of Nitrosomonas and Nitrosospira
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Monteiro, M.; Seneca, J.; Magalhaes, C. The history of aerobic ammonia oxidizers: From the first discoveries to today. J. Microbiol. 2014, 52, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Lehtovirta-Morley, L.E. Ammonia oxidation: Ecology, physiology, biochemistry and why they must all come together. FEMS Microbiol. Lett. 2018, 365, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koops, H.P.; Purkhold, U.; Pommerening-Röser, A.; Timmermann, G.; Wagner, M. The lithoautotrophic ammonia-oxidizing bacteria. In The Prokaryotes: An Evoluting Electronic Resource for the Microbiological Community; Dworking, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2003; ISBN 978-0-387-25495-1. [Google Scholar] [CrossRef]
- Purkhold, U.; Pommerening-Röser, A.; Juretschko, S.; Schmid, M.C.; Koops, H.P.; Wagner, M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: Implications for molecular diversity surveys. Appl. Environ. Microbiol. 2000, 66, 5368–5382. [Google Scholar] [CrossRef] [Green Version]
- Koops, H.-P.; Böttcher, B.; Möller, U.C.; Pommerening-Röser, A.; Stehr, G. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotroph asp. Nov. and Nitrosomonas halophila sp. nov. Microbiology 1991, 137, 1689–1699. [Google Scholar] [CrossRef] [Green Version]
- Koops, H.-P.; Pommerening-Röser, A. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 2001, 37, 1–9. [Google Scholar] [CrossRef]
- Bollmann, A.; Bär-Gilissen, M.-J.; Laanbroek, H.J. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 2002, 68, 4751–4757. [Google Scholar] [CrossRef] [Green Version]
- Wilhelm, R.; Abeliovich, A.; Nejidat, A. Effect of long-term ammonia starvation on the oxidation of ammonia and hydroxylamine by Nitrosomonas europaea. J. Biochem. 1998, 124, 811–815. [Google Scholar] [CrossRef]
- Sedlacek, C.J.; Giguere, A.T.; Dobie, M.D.; Mellbye, B.L.; Ferrell, R.V.; Woebken, D.; Sayavedra-Soto, L.A.; Bottomley, P.J.; Daims, H.; Wagner, M.; et al. Transcriptomic Response of Nitrosomonas europaea Transitioned from Ammonia- to Oxygen-Limited Steady-State Growth. mSystems 2020, 5. [Google Scholar] [CrossRef] [Green Version]
- Kozlowski, J.A.; Kits, K.D.; Stein, L.Y. Comparison of Nitrogen Oxide Metabolism among Diverse Ammonia-Oxidizing Bacteria. Front. Microbiol. 2016, 7, 1090. [Google Scholar] [CrossRef] [Green Version]
- Lehnert, N.; Dong, H.T.; Harland, J.B.; Hunt, A.P.; White, C.J. Reversing nitrogen fixation. Nat. Rev. Chem. 2018, 2, 278–289. [Google Scholar] [CrossRef]
- Chu, H.; Fujii, T.; Morimoto, S.; Lin, X.; Yagi, K. Population size and specific nitrification potential of soil ammonia-oxidizing bacteria under long-term fertilizer management. Soil Biol. Biochem. 2008, 40, 1960–1963. [Google Scholar] [CrossRef]
- Schepers, J.S. Nitrogen in Agricultural Systems; American Soc. of Agronomy: Madison, WI, USA, 2008; ISBN 9780891181644. [Google Scholar]
- Geets, J.; Boon, N.; Verstraete, W. Strategies of aerobic ammonia-oxidizing bacteria for coping with nutrient and oxygen fluctuations. FEMS Microbiol. Ecol. 2006, 58, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Yu, R.; Chandran, K. Strategies of Nitrosomonas europaea 19718 to counter low dissolved oxygen and high nitrite concentrations. BMC Microbiol. 2010, 10, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellitteri-Hahn, M.C.; Halligan, B.D.; Scalf, M.; Smith, L.; Hickey, W.J. Quantitative proteomic analysis of the chemolithoautotrophic bacterium Nitrosomonas europaea: Comparison of growing- and energy-starved cells. J. Proteomics 2011, 74, 411–419. [Google Scholar] [CrossRef]
- Arp, D.J.; Sayavedra-Soto, L.A.; Hommes, N.G. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol. 2002, 178, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Caranto, J.D.; Vilbert, A.C.; Lancaster, K.M. Nitrosomonas europaea cytochrome P460 is a direct link between nitrification and nitrous oxide emission. Proc. Natl. Acad. Sci. USA 2016, 113, 14704–14709. [Google Scholar] [CrossRef] [Green Version]
- Iverson, T.M.; Arciero, D.M.; Hooper, A.B.; Rees, D.C. High-resolution structures of the oxidized and reduced states of cytochrome c554 from Nitrosomonas europaea. J. Biol. Inorg. Chem. 2001, 6, 390–397. [Google Scholar] [CrossRef]
- Upadhyay, A.K.; Hooper, A.B.; Hendrich, M.P. NO reductase activity of the tetraheme cytochrome C554 of Nitrosomonas europaea. J. Am. Chem. Soc. 2006, 128, 4330–4337. [Google Scholar] [CrossRef] [Green Version]
- Bewley, K.D.; Ellis, K.E.; Firer-Sherwood, M.A.; Elliott, S.J. Multi-heme proteins: nature’s electronic multi-purpose tool. Biochim. Biophys. Acta 2013, 1827, 938–948. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Yan, T.; Hommes, N.G.; Liu, X.; Wu, L.; McAlvin, C.; Klotz, M.G.; Sayavedra-Soto, L.A.; Zhou, J.; Arp, D.J. Transcript profiles of Nitrosomonas europaea during growth and upon deprivation of ammonia and carbonate. FEMS Microbiol. Lett. 2006, 257, 76–83. [Google Scholar] [CrossRef] [Green Version]
- Bollmann, A.; Schmidt, I.; Saunders, A.M.; Nicolaisen, M.H. Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl. Environ. Microbiol. 2005, 71, 1276–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Kniggendorf, A.-K.; Schmidt, D.; Roth, B.; Plettenburg, O.; Zeilinger, C. pH-Dependent Conformational Changes of KcsA Tetramer and Monomer Probed by Raman Spectroscopy. Int. J. Mol. Sci. 2019, 20, 2736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kniggendorf, A.-K.; Meinhardt-Wollweber, M.; Yuan, X.; Roth, B.; Seifert, A.; Fertig, N.; Zeilinger, C. Temperature-sensitive gating of hCx26: High-resolution Raman spectroscopy sheds light on conformational changes. Biomed. Opt. Express 2014, 5, 2054–2065. [Google Scholar] [CrossRef] [Green Version]
- Kniggendorf, A.-K.; Gaul, T.W.; Meinhardt-Wollweber, M. Effects of ethanol, formaldehyde, and gentle heat fixation in confocal resonance Raman microscopy of purple nonsulfur bacteria. Microsc. Res. Tech. 2011, 74, 177–183. [Google Scholar] [CrossRef]
- Kniggendorf, A.-K.; Meinhardt-Wollweber, M. Of microparticles and bacteria identification--(resonance) Raman micro-spectroscopy as a tool for biofilm analysis. Water Res. 2011, 45, 4571–4582. [Google Scholar] [CrossRef]
- Dina, N.E.; Leş, A.; Baricz, A.; Szöke-Nagy, T.; Leopold, N.; Sârbu, C.; Banciu, H.L. Discrimination of haloarchaeal genera using Raman spectroscopy and robust methods for multivariate data analysis. J. Raman Spectrosc. 2017, 48, 1122–1126. [Google Scholar] [CrossRef]
- Maquelin, K.; Dijkshoorn, L.; van der Reijden, T.J.K.; Puppels, G.J. Rapid epidemiological analysis of Acinetobacter strains by Raman spectroscopy. J. Microbiol. Methods 2006, 64, 126–131. [Google Scholar] [CrossRef]
- Kniggendorf, A.-K.; Nogueira, R.; Kelb, C.; Schadzek, P.; Meinhardt-Wollweber, M.; Ngezahayo, A.; Roth, B. Confocal Raman microscopy and fluorescent in situ hybridization—A complementary approach for biofilm analysis. Chemosphere 2016, 161, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Kniggendorf, A.-K.; Gaul, T.W.; Meinhardt-Wollweber, M. Hierarchical Cluster Analysis (HCA) of Microorganisms: An Assessment of Algorithms for Resonance Raman Spectra. Appl. Spectrosc. 2011, 65, 165–173. [Google Scholar] [CrossRef]
- Hu, S.; Morris, I.K.; Singh, J.P.; Smith, K.M.; Spiro, T.G. Complete assignment of cytochrome c resonance Raman spectra via enzymic reconstitution with isotopically labeled hemes. J. Am. Chem. Soc. 1993, 115, 12446–12458. [Google Scholar] [CrossRef]
- Li, X.Y.; Czernuszewicz, R.S.; Kincaid, J.R.; Stein, P.; Spiro, T.G. Consistent porphyrin force field. 2. Nickel octaethylporphyrin skeletal and substituent mode assignments from nitrogen-15, meso-d4, and methylene-d16 Raman and infrared isotope shifts. J. Phys. Chem. 1990, 94, 47–61. [Google Scholar] [CrossRef]
- Russo, V.; Candeloro, P.; Malara, N.; Perozziello, G.; Iannone, M.; Scicchitano, M.; Mollace, R.; Musolino, V.; Gliozzi, M.; Carresi, C.; et al. Key Role of Cytochrome C for Apoptosis Detection Using Raman Microimaging in an Animal Model of Brain Ischemia with Insulin Treatment. Appl. Spectrosc. 2019, 73, 1208–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakita, M.; Kaliaperumal, V.; Hamaguchi, H. Resonance Raman quantification of the redox state of cytochromes b and c in-vivo and in-vitro. J. Biophotonics 2012, 5, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.J.; Lee, J.H.; Kim, Y.D.; Shin, I.; Sim, T.; Lim, D.-K. Raman-Based in Situ Monitoring of Changes in Molecular Signatures during Mitochondrially Mediated Apoptosis. ACS Omega 2019, 4, 8188–8195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Ma, X.; Wang, Z. Surface-Enhanced Raman Scattering-Fluorescence Dual-Mode Nanosensors for Quantitative Detection of Cytochrome c in Living Cells. Anal. Chem. 2019, 91, 6600–6607. [Google Scholar] [CrossRef]
- Zhu, J.; Jiang, M.; Ma, H.; Zhang, H.; Cheng, W.; Li, J.; Cai, L.; Han, X.X.; Zhao, B. Redox-State-Mediated Regulation of Cytochrome c Release in Apoptosis Revealed by Surface-Enhanced Raman Scattering on Nickel Substrates. Angew. Chem. Int. Ed. Engl. 2019, 58, 16499–16503. [Google Scholar] [CrossRef]
- Krümmel, A.; Harms, H. Effect of organic matter on growth and cell yield of ammonia-oxidizing bacteria. Arch. Microbiol. 1982, 133, 50–54. [Google Scholar] [CrossRef]
- Bremner, J.M.; Keeney, D.R. Steam distillation methods for determination of ammonium, nitrate and nitrite. Anal. Chim. Acta 1965, 32, 485–495. [Google Scholar] [CrossRef]
- Koch, M.; Suhr, C.; Roth, B.; Meinhardt-Wollweber, M. Iterative morphological and mollifier-based baseline correction for Raman spectra. J. Raman Spectrosc. 2017, 48, 336–342. [Google Scholar] [CrossRef] [Green Version]
- Koch, M.; Kniggendorf, A.-K.; Meinhardt-Wollweber, M.; Roth, B. In vivo determination of carotenoid resonance excitation profiles of Chlorella vulgaris, Haematococcus pluvialis, and Porphyridium purpureum. J. Raman Spectrosc. 2018, 49, 404–411. [Google Scholar] [CrossRef]
- Koch, M.; Zagermann, S.; Kniggendorf, A.-K.; Meinhardt-Wollweber, M.; Roth, B. Violaxanthin cycle kinetics analysed in vivo with resonance Raman spectroscopy. J. Raman Spectrosc. 2017, 48, 686–691. [Google Scholar] [CrossRef]
- The Mathworks Inc. MATLAB (Version 2019a Update 6). Windows; The Mathworks Inc.: Natick, MA, USA, 2019. [Google Scholar]
- Murgida, D.H.; Hildebrandt, P. Electron-transfer processes of cytochrome C at interfaces. New insights by surface-enhanced resonance Raman spectroscopy. Acc. Chem. Res. 2004, 37, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Murgida, D.H.; Hildebrandt, P. Disentangling interfacial redox processes of proteins by SERR spectroscopy. Chem. Soc. Rev. 2008, 37, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Maier, U.; Büchs, J. Characterisation of the gas-liquid mass transfer in shaking bioreactors. Biochem. Eng. J. 2001, 7, 99–106. [Google Scholar] [CrossRef]
- Laanbroek, H.J.; Bodelier, P.L.E.; Gerards, S. Oxygen consumption kinetics of Nitrosomonas europaea and Nitrobacter hamburgensis grown in mixed continuous cultures at different oxygen concentrations. Arch. Microbiol. 1994, 161, 156–162. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kniggendorf, A.-K.; Nogueira, R.; Nasiri Bahmanabad, S.; Pommerening-Röser, A.; Roth, B.W. Small Sample Stress: Probing Oxygen-Deprived Ammonia-Oxidizing Bacteria with Raman Spectroscopy In Vivo. Microorganisms 2020, 8, 432. https://doi.org/10.3390/microorganisms8030432
Kniggendorf A-K, Nogueira R, Nasiri Bahmanabad S, Pommerening-Röser A, Roth BW. Small Sample Stress: Probing Oxygen-Deprived Ammonia-Oxidizing Bacteria with Raman Spectroscopy In Vivo. Microorganisms. 2020; 8(3):432. https://doi.org/10.3390/microorganisms8030432
Chicago/Turabian StyleKniggendorf, Ann-Kathrin, Regina Nogueira, Somayeh Nasiri Bahmanabad, Andreas Pommerening-Röser, and Bernhard Wilhelm Roth. 2020. "Small Sample Stress: Probing Oxygen-Deprived Ammonia-Oxidizing Bacteria with Raman Spectroscopy In Vivo" Microorganisms 8, no. 3: 432. https://doi.org/10.3390/microorganisms8030432
APA StyleKniggendorf, A.-K., Nogueira, R., Nasiri Bahmanabad, S., Pommerening-Röser, A., & Roth, B. W. (2020). Small Sample Stress: Probing Oxygen-Deprived Ammonia-Oxidizing Bacteria with Raman Spectroscopy In Vivo. Microorganisms, 8(3), 432. https://doi.org/10.3390/microorganisms8030432






