The Proteome of Community Living Candida albicans Is Differentially Modulated by the Morphologic and Structural Features of the Bacterial Cohabitants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biofilm Formation
2.2. Bacterial Culture Conditions
2.3. Treatment of Candida albicans Biofilm with Heat-Killed Bacteria
2.4. Protein Extraction and Sample Preparation for Mass Spectrometry Analysis
2.5. LC-MS Analysis
2.6. Protein Identification
2.7. Protein Quantification from Spectral Counts
2.8. Differential Protein Expression Analysis and Cluster Analysis
2.9. Pathway Analysis
3. Results
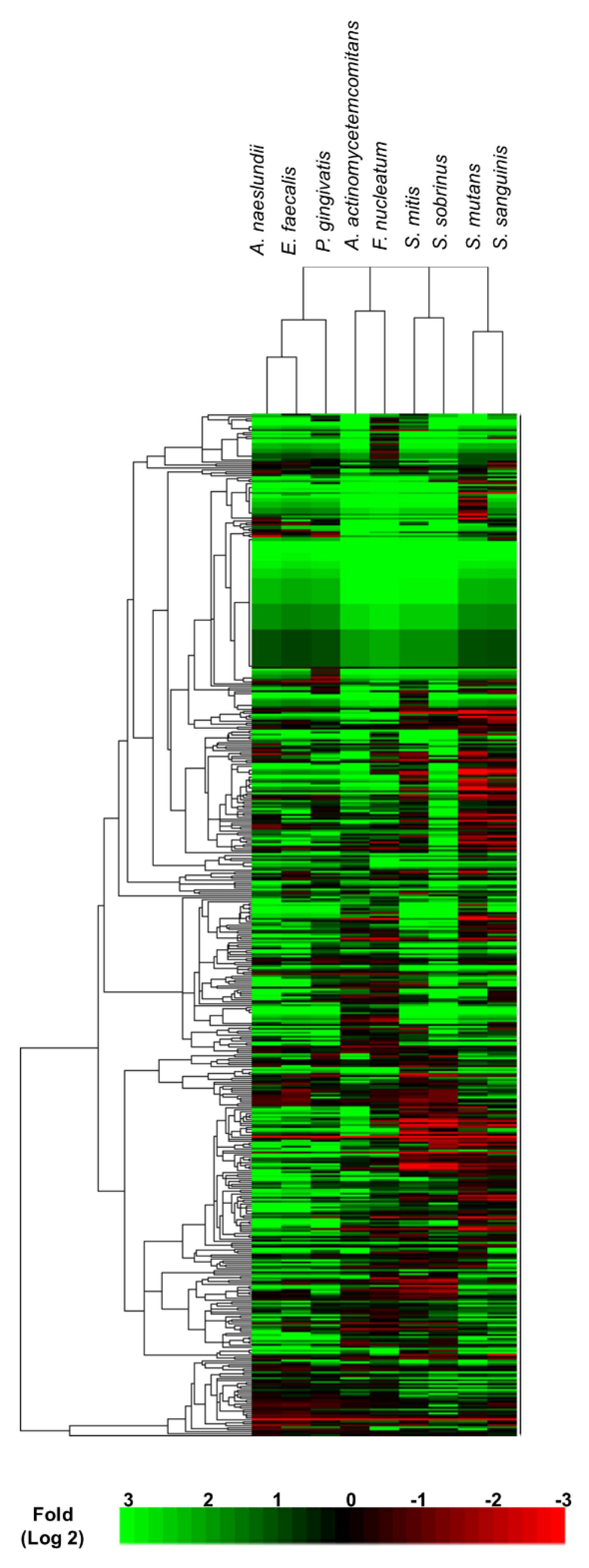
3.1. Proteomic Analysis of Candida albicans Biofilms Identified Proteins Expressed on Exposure to Heat-Killed Bacteria
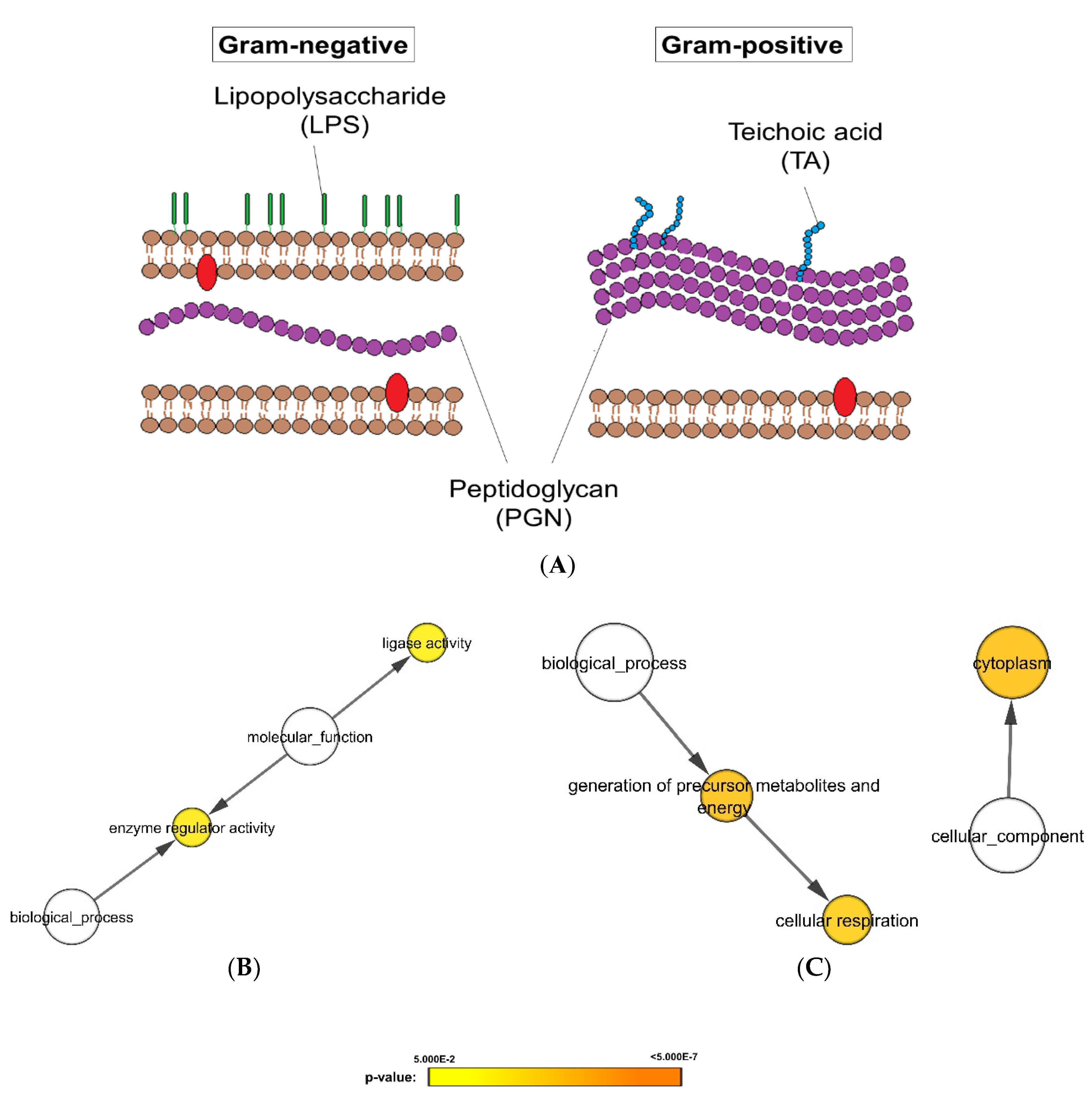
3.2. Differential Expression Analysis Uncovered Proteomic Features of Candida albicans Biofilms Responding to Gram-Positive and -Negative Bacteria
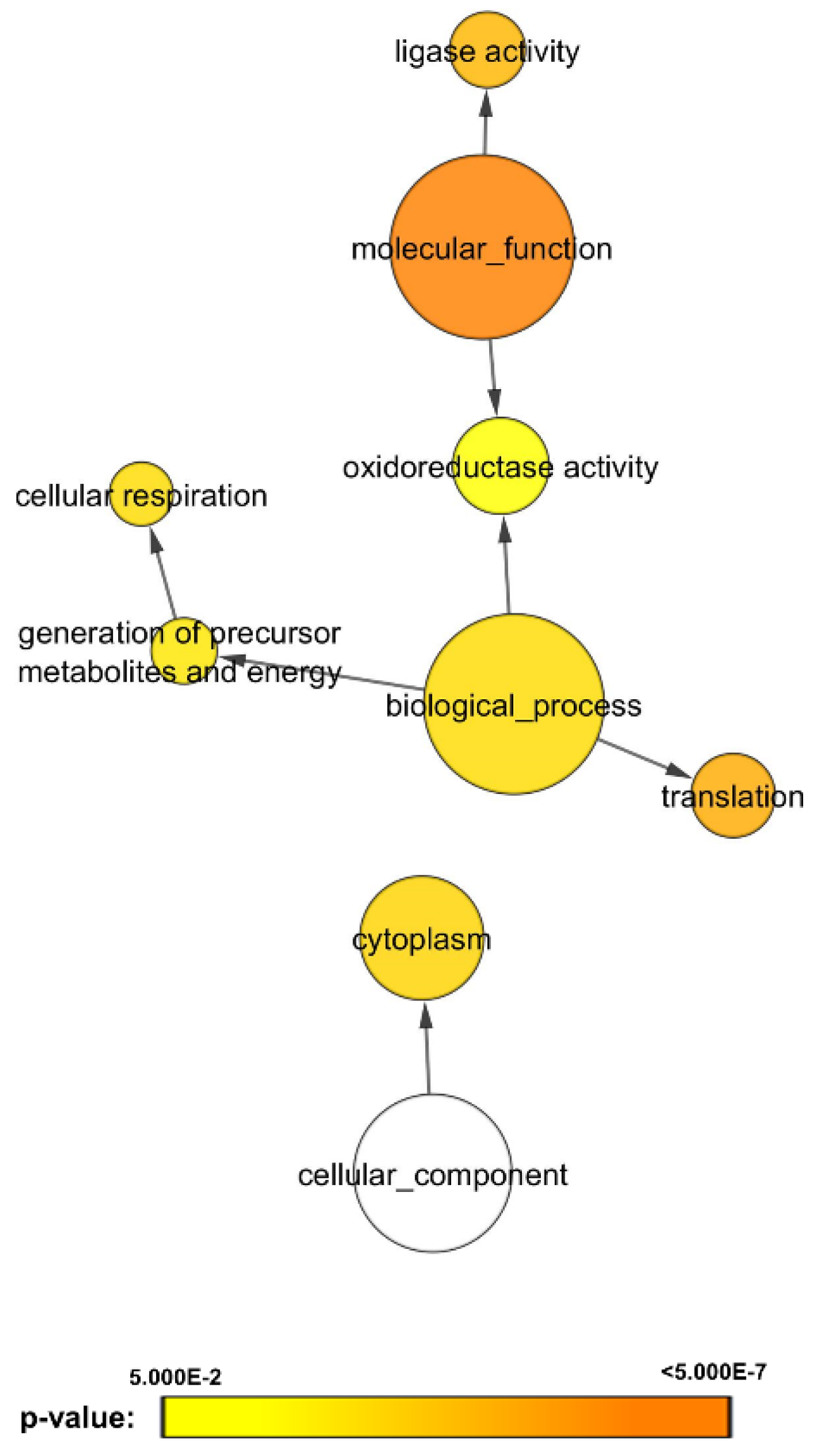
3.3. Cluster Analysis Revealed C. albicans Biofilm Proteins with Similar Expression Profiles on Bacterial Exposure
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mavor, A.L.; Thewes, S.; Hube, B. Systemic fungal infections caused by Candida species: Epidemiology, infection process and virulence attributes. Curr. Drug Targets 2005, 6, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, S.; Majumdar, B.; Sarode, S.C.; Sarode, G.S.; Awan, K.H. Oropharyngeal Candidosis in HIV-Infected Patients—An Update. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kullberg, B.J.; Arendrup, M.C. Invasive Candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramage, G.; Saville, S.P.; Thomas, D.P.; Lopez-Ribot, J.L. Candida biofilms: An update. Eukaryot. Cell 2005, 4, 633–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seneviratne, C.J.; Jin, L.; Samaranayake, L.P. Biofilm lifestyle of Candida: A mini review. Oral Dis. 2008, 14, 582–590. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Hogan, D.A.; Mylonakis, E. Medically important bacterial–fungal interactions. Nat. Rev. Microbiol. 2010, 8, 340–349. [Google Scholar] [CrossRef]
- Montelongo-Jauregui, D.; Srinivasan, A.; Ramasubramanian, A.K.; Lopez-Ribot, J.L. An In Vitro Model for Oral Mixed Biofilms of Candida albicans and Streptococcus gordonii in Synthetic Saliva. Front. Microbiol. 2016, 7, 686. [Google Scholar] [CrossRef]
- Seneviratne, C.J.; Wang, Y.; Jin, L.; Wong, S.S.; Herath, T.D.; Samaranayake, L.P. Unraveling the resistance of microbial biofilms: Has proteomics been helpful? Proteomics 2012, 12, 651–665. [Google Scholar] [CrossRef]
- Pandey, A.; Mann, M. Proteomics to study genes and genomes. Nature 2000, 405, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seneviratne, C.J.; Zeng, G.; Truong, T.; Sze, S.; Wong, W.; Samaranayake, L.; Chan, F.Y.; Wang, Y.M.; Wang, H.; Gao, J.; et al. New “haploid biofilm model” unravels IRA2 as a novel regulator of Candida albicans biofilm formation. Sci. Rep. 2015, 5, 12433. [Google Scholar] [CrossRef] [Green Version]
- Truong, T.; Zeng, G.; Qingsong, L.; Kwang, L.T.; Tong, C.; Chan, F.Y.; Wang, Y.; Seneviratne, C.J. Comparative Ploidy Proteomics of Candida albicans Biofilms Unraveled the Role of the AHP1 Gene in the Biofilm Persistence Against Amphotericin B. Mol. Cell. Proteom. 2016, 15, 3488–3500. [Google Scholar] [CrossRef] [Green Version]
- Vaudel, M.; Barsnes, H.; Berven, F.S.; Sickmann, A.; Martens, L. SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics 2011, 11, 996–999. [Google Scholar] [CrossRef]
- Vaudel, M.; Burkhart, J.M.; Zahedi, R.P.; Oveland, E.; Berven, F.S.; Sickmann, A.; Martens, L.; Barsnes, H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015, 33, 22–24. [Google Scholar] [CrossRef]
- Wong, J.W.; Cagney, G. An overview of label-free quantitation methods in proteomics by mass spectrometry. Methods Mol. Biol. 2010, 604, 273–283. [Google Scholar] [CrossRef]
- Old, W.M.; Meyer-Arendt, K.; Aveline-Wolf, L.; Pierce, K.G.; Mendoza, A.; Sevinsky, J.R.; Resing, K.A.; Ahn, N.G. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteom. 2005, 4, 1487–1502. [Google Scholar] [CrossRef]
- Beissbarth, T.; Hyde, L.; Smyth, G.K.; Job, C.; Boon, W.M.; Tan, S.S.; Scott, H.S.; Speed, T.P. Statistical modeling of sequencing errors in SAGE libraries. Bioinformatics 2004, 20 (Suppl. 1), i31–i39. [Google Scholar] [CrossRef]
- Truong, T.; Suriyanarayanan, T.; Zeng, G.; Le, T.D.; Liu, L.; Li, J.; Tong, C.; Wang, Y.; Seneviratne, C.J. Use of Haploid Model of Candida albicans to Uncover Mechanism of Action of a Novel Antifungal Agent. Front. Cell. Infect. Microbiol. 2018, 8, 164. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truong, T.; Zeng, G.; Lim, T.K.; Cao, T.; Pang, L.M.; Lee, Y.M.; Lin, Q.; Wang, Y.; Seneviratne, C.J. Proteomics Analysis of Candida albicans dnm1 Haploid Mutant Unraveled the Association between Mitochondrial Fission and Antifungal Susceptibility. Proteomics 2020, 20, e1900240. [Google Scholar] [CrossRef]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–214. [Google Scholar] [CrossRef]
- Morales, D.K.; Grahl, N.; Okegbe, C.; Dietrich, L.E.; Jacobs, N.J.; Hogan, D.A. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. MBio 2013, 4, e00526-12. [Google Scholar] [CrossRef] [Green Version]
- Calderone, R.; Li, D.; Traven, A. System-level impact of mitochondria on fungal virulence: To metabolism and beyond. FEMS Yeast Res. 2015, 15, fov027. [Google Scholar] [CrossRef] [Green Version]
- Coico, R. Gram staining. Curr. Protoc. Microbiol. 2005, Appendix 3, Appendix 3C. [Google Scholar] [CrossRef]
- Henry-Stanley, M.J.; Hess, D.J.; Erickson, E.A.; Garni, R.M.; Wells, C.L. Effect of lipopolysaccharide on virulence of intestinal candida albicans. J. Surg. Res. 2003, 113, 42–49. [Google Scholar] [CrossRef]
- Bandara, H.M.; Yau, J.Y.; Watt, R.M.; Jin, L.J.; Samaranayake, L.P. Escherichia coli and its lipopolysaccharide modulate in vitro Candida biofilm formation. J. Med. Microbiol. 2009, 58 Pt 12, 1623–1631. [Google Scholar] [CrossRef] [Green Version]
- Livins’ka, O.P.; Garmasheva, I.L.; Kovalenko, N.K. The influence of teichoic acids from probiotic lactobacilli on microbial adhesion to epithelial cells. Mikrobiol. Zhurnal. 2012, 74, 16–22. [Google Scholar]
- Xu, X.-L.; Lee, R.T.H.; Fang, H.-M.; Wang, Y.-M.; Li, R.; Zou, H.; Zhu, Y.; Wang, Y. Bacterial Peptidoglycan Triggers Candida albicans Hyphal Growth by Directly Activating the Adenylyl Cyclase Cyr1p. Cell Host Microbe 2008, 4, 28–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grahl, N.; Demers, E.G.; Lindsay, A.K.; Harty, C.E.; Willger, S.D.; Piispanen, A.E.; Hogan, D.A. Mitochondrial Activity and Cyr1 Are Key Regulators of Ras1 Activation of C. albicans Virulence Pathways. PLoS Pathog. 2015, 11, e1005133. [Google Scholar] [CrossRef] [Green Version]
- Zou, H.; Fang, H.-M.; Zhu, Y.; Wang, Y. Candida albicans Cyr1, Cap1 and G-actin form a sensor/effector apparatus for activating cAMP synthesis in hyphal growth. Mol. Microbiol. 2010, 75, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.R.; Graham, C.E.; Gagliano, B.C.; Lorenz, M.C.; Garsin, D.A. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect. Immun. 2013, 81, 189–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Wei, C.; Liu, C.; Li, D.; Sun, J.; Huang, H.; Zhou, H. Inhibitory effects of oral Actinomyces on the proliferation, virulence and biofilm formation of Candida albicans. Arch. Oral Biol. 2015, 60, 1368–1374. [Google Scholar] [CrossRef]
- Ximenez-Fyvie, L.A.; Haffajee, A.D.; Socransky, S.S. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 2000, 27, 648–657. [Google Scholar] [CrossRef]
- Bor, B.; Cen, L.; Agnello, M.; Shi, W.; He, X. Morphological and physiological changes induced by contact-dependent interaction between Candida albicans and Fusobacterium nucleatum. Sci. Rep. 2016, 6, 27956. [Google Scholar] [CrossRef]
- Bachtiar, E.W.; Bachtiar, B.M.; Jarosz, L.M.; Amir, L.R.; Sunarto, H.; Ganin, H.; Meijler, M.M.; Krom, B.P. AI-2 of Aggregatibacter actinomycetemcomitans inhibits Candida albicans biofilm formation. Front. Cell. Infect. Microbiol. 2014, 4, 94. [Google Scholar] [CrossRef]
- Fragkou, S.; Balasouli, C.; Tsuzukibashi, O.; Argyropoulou, A.; Menexes, G.; Kotsanos, N.; Kalfas, S. Streptococcus mutans, Streptococcus sobrinus and Candida albicans in oral samples from caries-free and caries-active children. Eur. Arch. Paediatr. Dent. 2016, 17, 367–375. [Google Scholar] [CrossRef]
- Diaz, P.I.; Xie, Z.; Sobue, T.; Thompson, A.; Biyikoglu, B.; Ricker, A.; Ikonomou, L.; Dongari-Bagtzoglou, A. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 2012, 80, 620–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Sobue, T.; Thompson, A.; Xie, Z.; Poon, K.; Ricker, A.; Cervantes, J.; Diaz, P.I.; Dongari-Bagtzoglou, A. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell. Microbiol. 2014, 16, 214–231. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.; Liu, Y.; Kim, D.; Li, Y.; Krysan, D.J.; Koo, H. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 2017, 13, e1006407. [Google Scholar] [CrossRef] [PubMed]



| Bacterial Species | Group | Gram | Size | Morphology |
|---|---|---|---|---|
| E. faecalis | 1 | Positive | 0.6–2.5 µm | Cocci (spherical or ovoid) |
| P. gingivalis | 1 | Negative | 0.5–5.0 µm in length | Bacillus |
| A. naeslundii | 1 | Positive | 5–10 µm in length | Bacillus |
| F. nucleatum | 2 | Negative | 5–50 µm in length, varied | Bacillus |
| A. actinomycetemcomitans | 2 | Negative | 0.2–1 µm in diameter, varied | Coccobacillus |
| S. mutans | 3 | Positive | 0.5–0.75 µm | Cocci (spherical) |
| S. sanguinis | 3 | Positive | 0.5–0.75 µm | Cocci (spherical) |
| S. mitis | 4 | Positive | 0.5–0.75 µm | Cocci (spherical) |
| S. sobrinus | 4 | Positive | 0.5–0.75 µm | Cocci (spherical) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Truong, T.; Pang, L.M.; Rajan, S.; Wong, S.S.W.; Fung, Y.M.E.; Samaranayake, L.; Seneviratne, C.J. The Proteome of Community Living Candida albicans Is Differentially Modulated by the Morphologic and Structural Features of the Bacterial Cohabitants. Microorganisms 2020, 8, 1541. https://doi.org/10.3390/microorganisms8101541
Truong T, Pang LM, Rajan S, Wong SSW, Fung YME, Samaranayake L, Seneviratne CJ. The Proteome of Community Living Candida albicans Is Differentially Modulated by the Morphologic and Structural Features of the Bacterial Cohabitants. Microorganisms. 2020; 8(10):1541. https://doi.org/10.3390/microorganisms8101541
Chicago/Turabian StyleTruong, Thuyen, Li Mei Pang, Suhasini Rajan, Sarah Sze Wah Wong, Yi Man Eva Fung, Lakshman Samaranayake, and Chaminda Jayampath Seneviratne. 2020. "The Proteome of Community Living Candida albicans Is Differentially Modulated by the Morphologic and Structural Features of the Bacterial Cohabitants" Microorganisms 8, no. 10: 1541. https://doi.org/10.3390/microorganisms8101541





