Nitrifying Microbes in the Rhizosphere of Perennial Grasses Are Modified by Biological Nitrification Inhibition
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
4.1. BNI Capacity of Australian Perennial Grasses
4.2. The Effect of BNI Capacity on Suppressing Potential Nitrifiers in the Rhizosphere Reflect
4.3. Rhizospheric Selection for Microbiome Composition between Different Grass Species
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tilman, D.; Fargione, J.; Wolff, B.; D’Antonio, C.; Dobson, A.; Howarth, R.; Schindler, D.; Schlesinger, W.H.; Simberloff, D.; Swackhamer, D. Forecasting agriculturally driven global environmental change. Science 2001, 292, 281–284. [Google Scholar] [CrossRef]
- Ladha, J.K.; Pathak, H.; Krupnik, T.J.; Six, J.; van Kessel, C. Efficiency of fertilizer nitrogen in cereal production: Retrospects and prospects. Adv. Agron. 2005, 87, 85–156. [Google Scholar]
- Coskun, D.; Britto, D.T.; Shi, W.M.; Kronzucker, H.J. Nitrogen transformations in modern agriculture and the role of biological nitrification inhibition. Nat. Plants 2017, 3, 17074. [Google Scholar] [CrossRef]
- Sylvester-Bradley, R.; Mosquera, D.; Mendez, J. Inhibition of nitrate accumulation in tropical grassland soils: Effect of nitrogen fertilization and soil disturbance. J. Soil Sci. 1988, 39, 407–416. [Google Scholar] [CrossRef]
- Subbarao, G.V.; Rondon, M.; Ito, O.; Ishikawa, T.; Rao, I.M.; Nakahara, K.; Lascano, C.; Berry, W.L. Biological nitrification inhibition (bni)—Is it a widespread phenomenon? Plant Soil 2007, 294, 5–18. [Google Scholar] [CrossRef]
- Boudsocq, S.; Lata, J.-C.; Mathieu, J.; Abbadie, L.; Barot, S. Modelling approach to analyse the effects of nitrification inhibition on primary production. Funct. Ecol. 2009, 23, 220–230. [Google Scholar] [CrossRef]
- Zakir, H.; Subbarao, G.V.; Pearse, S.J.; Gopalakrishnan, S.; Ito, O.; Ishikawa, T.; Kawano, N.; Nakahara, K.; Yoshihashi, T.; Ono, H.; et al. Detection, isolation and characterization of a root-exuded compound, methyl 3-(4-hydroxyphenyl) propionate, responsible for biological nitrification inhibition by sorghum (sorghum bicolor). New Phytol. 2008, 180, 442–451. [Google Scholar] [CrossRef]
- Sun, L.; Lu, Y.F.; Yu, F.W.; Kronzucker, H.J.; Shi, W.M. Biological nitrification inhibition by rice root exudates and its relationship with nitrogen-use efficiency. New Phytol. 2016, 212, 646–656. [Google Scholar] [CrossRef]
- O’Sullivan, C.A.; Whisson, K.; Treble, K.; Roper, M.M.; Micin, S.F.; Ward, P.R. Biological nitrification inhibition by weeds: Wild radish, brome grass, wild oats and annual ryegrass decrease nitrification rates in their rhizospheres. Crop Pasture Sci. 2017, 68, 798–804. [Google Scholar] [CrossRef]
- O’Sullivan, C.A.; Fillery, I.R.P.; Roper, M.M.; Richards, R.A. Identification of several wheat landraces with biological nitrification inhibition capacity. Plant Soil 2016, 404, 61–74. [Google Scholar] [CrossRef]
- Nunez, J.; Arevalo, A.; Karwat, H.; Egenolf, K.; Miles, J.; Chirinda, N.; Cadisch, G.; Rasche, F.; Rao, I.; Subbarao, G.; et al. Biological nitrification inhibition activity in a soil-grown biparental population of the forage grass, Brachiaria humidicola. Plant Soil 2018, 426, 401–411. [Google Scholar] [CrossRef]
- Lata, J.C.; Degrange, V.; Raynaud, X.; Maron, P.A.; Lensi, R.; Abbadie, L. Grass populations control nitrification in savanna soils. Funct. Ecol. 2004, 18, 605–611. [Google Scholar] [CrossRef]
- McKenzie, N.; Jacquier, D.; Isbell, R.; Brown, K. Australian Soils and Landscapes: An Illustrated Compendium; CSIRO Publishing: Melbourne, Australia, 2004. [Google Scholar]
- Janke, C.K.; Wendling, L.A.; Fujinuma, R. Biological nitrification inhibition by root exudates of native species, Hibiscus splendens and Solanum echinatum. PeerJ 2018, 6, e4960. [Google Scholar] [CrossRef]
- Still, C.J.; Berry, J.A.; Collatz, G.J.; DeFries, R.S. Global distribution of C3 and C4 vegetation: Carbon cycle implications. Glob. Biogeochem. Cycles 2003, 17, 6-1–6-14. [Google Scholar] [CrossRef]
- Jones, D.L.; Nguyen, C.; Finlay, R.D. Carbon flow in the rhizosphere: Carbon trading at the soil–root interface. Plant Soil 2009, 321, 5–33. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470. [Google Scholar] [CrossRef]
- Kudjordjie, E.N.; Sapkota, R.; Steffensen, S.K.; Fomsgaard, I.S.; Nicolaisen, M. Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome 2019, 7, 59. [Google Scholar] [CrossRef]
- Corral-Lugo, A.; Daddaoua, A.; Ortega, A.; Espinosa-Urgel, M.; Krell, T. Rosmarinic acid is a homoserine lactone mimic produced by plants that activates a bacterial quorum-sensing regulator. Sci. Signal. 2016, 9, ra1. [Google Scholar] [CrossRef]
- Turner, T.R.; Ramakrishnan, K.; Walshaw, J.; Heavens, D.; Alston, M.; Swarbreck, D.; Osbourn, A.; Grant, A.; Poole, P.S. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 2013, 7, 2248–2258. [Google Scholar] [CrossRef]
- Subbarao, G.V.; Nakahara, K.; Ishikawa, T.; Ono, H.; Yoshida, M.; Yoshihashi, T.; Zhu, Y.Y.; Zakir, H.; Deshpande, S.P.; Hash, C.T.; et al. Biological nitrification inhibition (BNI) activity in sorghum and its characterization. Plant Soil 2013, 366, 243–259. [Google Scholar] [CrossRef]
- Subbarao, G.V.; Nakahara, K.; Hurtado, M.P.; Ono, H.; Moreta, D.E.; Salcedo, A.F.; Yoshihashi, A.T.; Ishikawa, T.; Ishitani, M.; Ohnishi-Kameyama, M.; et al. Evidence for biological nitrification inhibition in Brachiaria pastures. Proc. Natl. Acad. Sci. USA 2009, 106, 17302–17307. [Google Scholar] [CrossRef] [PubMed]
- Sarr, P.S.; Ando, Y.; Nakamura, S.; Deshpande, S.; Subbarao, G.V. Sorgoleone release from sorghum roots shapes the composition of nitrifying populations, total bacteria, and archaea and determines the level of nitrification. Biol. Fertil. Soils 2020, 56, 145–166. [Google Scholar] [CrossRef]
- Shi, S.; Nuccio, E.E.; Shi, Z.J.; He, Z.; Zhou, J.; Firestone, M.K. The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecol. Lett. 2016, 19, 926–936. [Google Scholar] [CrossRef]
- Zhou, Y.; Coventry, D.R.; Denton, M.D. Soil surface pressure reduces post-emergent shoot growth in wheat. Plant Soil 2017, 413, 127–144. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. Dada2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M. The vegan package. Community Ecol. Package 2007, 10, 631–637. [Google Scholar]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. Stamp: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Zhou, Y.; Lambrides, C.J.; Kearns, R.; Ye, C.; Fukai, S. Water use, water use efficiency and drought resistance among warm-season turfgrasses in shallow soil profiles. Funct. Plant Biol. 2012, 39, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Loch, D.S.; Roche, M.B.; Sun-Yue, J.; Arief, V.; Delacy, I.H.; Lambrides, C.J. Characterization of commercial cultivars and naturalized genotypes of Stenotaphrum secundatum (walter) kuntze in Australia. Int. Turfgrass Soc. Res. J. 2009, 11, 549–561. [Google Scholar]
- Zhou, Y.; Lambrides, C.J.; Fukai, S. Drought resistance of C4 grasses under field conditions: Genetic variation among a large number of bermudagrass (Cynodon spp.) ecotypes collected from different climatic zones. J. Agron. Crop Sci. 2013, 199, 253–263. [Google Scholar] [CrossRef]
- Jewell, M.C.; Zhou, Y.; Loch, D.S.; Godwin, I.D.; Lambrides, C.J. Maximizing genetic, morphological, and geographic diversity in a core collection of Australian bermudagrass. Crop Sci. 2012, 52, 879–889. [Google Scholar] [CrossRef]
- Jewell, M.; Frère, C.H.; Harris-Shultz, K.; Anderson, W.F.; Godwin, I.D.; Lambrides, C.J. Phylogenetic analysis reveals multiple introductions of Cynodon species in Australia. Mol. Phylogenet. Evol. 2012, 65, 390–396. [Google Scholar] [CrossRef]
- Tanaka, J.P.; Nardi, P.; Wissuwa, M. Nitrification inhibition activity, a novel trait in root exudates of rice. AoB Plants 2010, 2010, plq014. [Google Scholar]
- Daims, H. The Family Nitrospiraceae. In The Prokaryotes: Other Major Lineages of Bacteria and the Archaea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Pester, M.; Maixner, F.; Berry, D.; Rattei, T.; Koch, H.; Lücker, S.; Nowka, B.; Richter, A.; Spieck, E.; Lebedeva, E. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol. 2014, 16, 3055–3071. [Google Scholar] [CrossRef]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A. Complete nitrification by Nitrospira bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef]
- Hu, H.-W.; He, J.-Z. Comammox—A newly discovered nitrification process in the terrestrial nitrogen cycle. J. Soils Sediments 2017, 17, 2709–2717. [Google Scholar] [CrossRef]
- Li, C.; Hu, H.-W.; Chen, Q.-L.; Chen, D.; He, J.-Z. Growth of comammox Nitrospira is inhibited by nitrification inhibitors in agricultural soils. J. Soils Sediments 2020, 20, 621–628. [Google Scholar] [CrossRef]
- O’Sullivan, C.A.; Duncan, E.G.; Whisson, K.; Treble, K.; Ward, P.R.; Roper, M.M. A colourimetric microplate assay for simple, high throughput assessment of synthetic and biological nitrification inhibitors. Plant Soil 2017, 413, 275–287. [Google Scholar] [CrossRef]
- Leininger, S.; Urich, T.; Schloter, M.; Schwark, L.; Qi, J.; Nicol, G.W.; Prosser, J.I.; Schuster, S.; Schleper, C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 2006, 442, 806–809. [Google Scholar] [CrossRef]
- Jia, Z.; Conrad, R. Bacteria rather than archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 2009, 11, 1658–1671. [Google Scholar] [CrossRef] [PubMed]
- Briones, A.M.; Okabe, S.; Umemiya, Y.; Ramsing, N.-B.; Reichardt, W.; Okuyama, H. Influence of different cultivars on populations of ammonia-oxidizing bacteria in the root environment of rice. Appl. Environ. Microbiol. 2002, 68, 3067–3075. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, K.S.; Cassman, N.A.; Pijl, A.S.; van Veen, J.A.; Cantarella, H.; Kuramae, E.E. Nitrosospira sp. govern nitrous oxide emissions in a tropical soil amended with residues of bioenergy crop. Front. Microbiol. 2018, 9, 674. [Google Scholar] [CrossRef] [PubMed]
- Cassman, N.A.; Soares, J.R.; Pijl, A.; Lourenço, K.S.; van Veen, J.A.; Cantarella, H.; Kuramae, E.E. Nitrification inhibitors effectively target N2O-producing Nitrosospira spp. in tropical soil. Environ. Microbiol. 2019, 21, 1241–1254. [Google Scholar] [CrossRef]
- Pommerening-Röser, A.; Koops, H.-P. Environmental pH as an important factor for the distribution of urease positive ammonia-oxidizing bacteria. Microbiol. Res. 2005, 160, 27–35. [Google Scholar] [CrossRef]
- Finkel, O.M.; Delmont, T.O.; Post, A.F.; Belkin, S. Metagenomic signatures of bacterial adaptation to life in the phyllosphere of a salt-secreting desert tree. Appl. Environ. Microbiol. 2016, 82, 2854–2861. [Google Scholar] [CrossRef]
- Zhang, Q.; Acuna, J.J.; Inostroza, N.G.; Mora, M.L.; Radic, S.; Sadowsky, M.J.; Jorquera, M.A. Endophytic bacterial communities associated with roots and leaves of plants growing in Chilean extreme environments. Sci. Rep. 2019, 9, 4950. [Google Scholar] [CrossRef]
- Furtado, B.U.; Golebiewski, M.; Skorupa, M.; Hulisz, P.; Hrynkiewicz, K. Bacterial and fungal endophytic microbiomes of Salicornia europaea. Appl. Environ. Microbiol. 2019, 85, e00305-19. [Google Scholar] [CrossRef]
- Eida, A.A.; Ziegler, M.; Lafi, F.F.; Michell, C.T.; Voolstra, C.R.; Hirt, H.; Saad, M.M. Desert plant bacteria reveal host influence and beneficial plant growth properties. PLoS ONE 2018, 13, e0208223. [Google Scholar] [CrossRef]
- Yamamoto, K.; Shiwa, Y.; Ishige, T.; Sakamoto, H.; Tanaka, K.; Uchino, M.; Tanaka, N.; Oguri, S.; Saitoh, H.; Tsushima, S. Bacterial diversity associated with the rhizosphere and endosphere of two halophytes: Glaux maritima and Salicornia europaea. Front. Microbiol. 2018, 9, 2878. [Google Scholar] [CrossRef]
- Zhou, Y.; Coventry, D.R.; Gupta, V.V.; Fuentes, D.; Merchant, A.; Kaiser, B.N.; Li, J.; Wei, Y.; Liu, H.; Wang, Y. The preceding root system drives the composition and function of the rhizosphere microbiome. Genome Biol. 2020, 21, 1–19. [Google Scholar] [CrossRef] [PubMed]
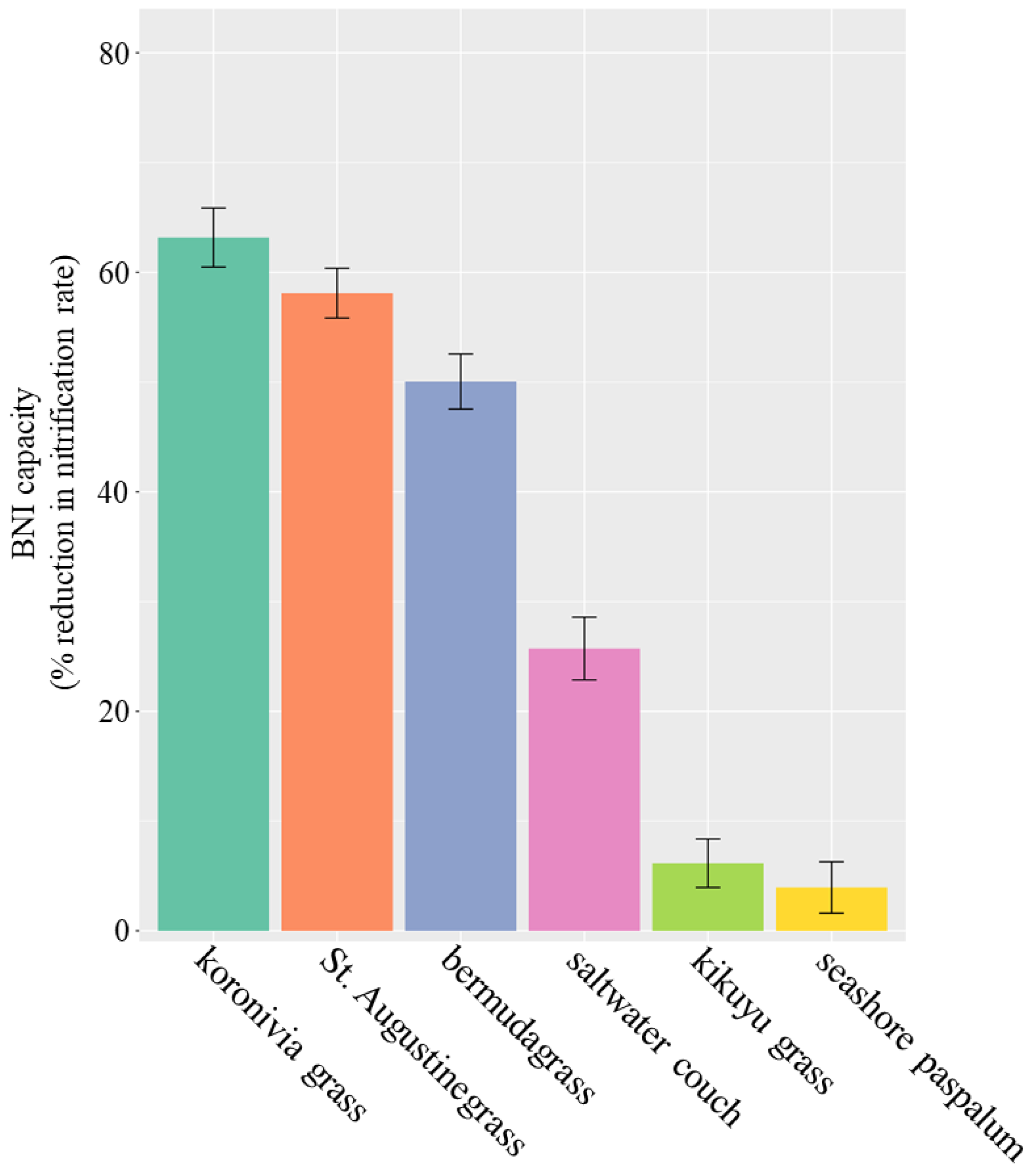
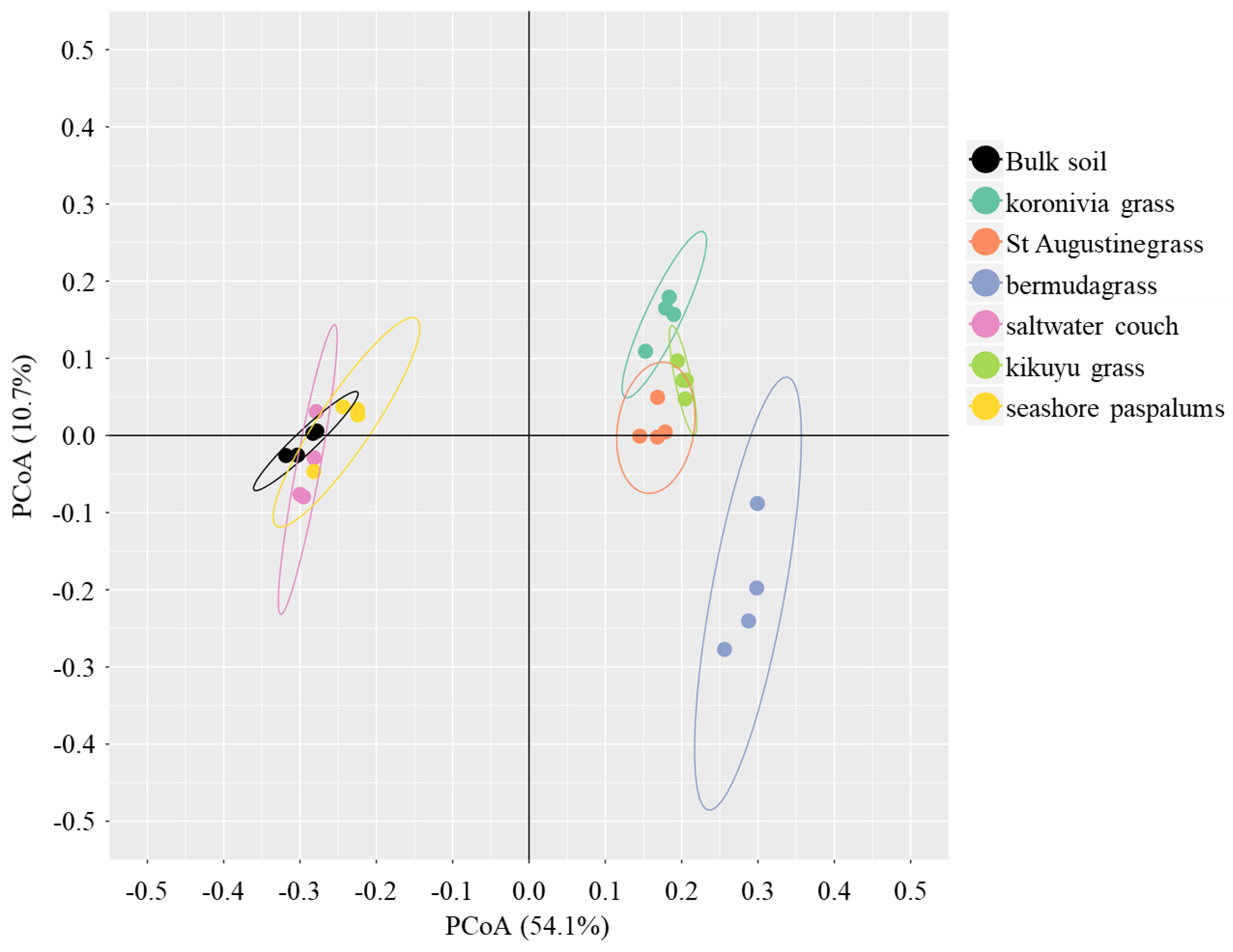


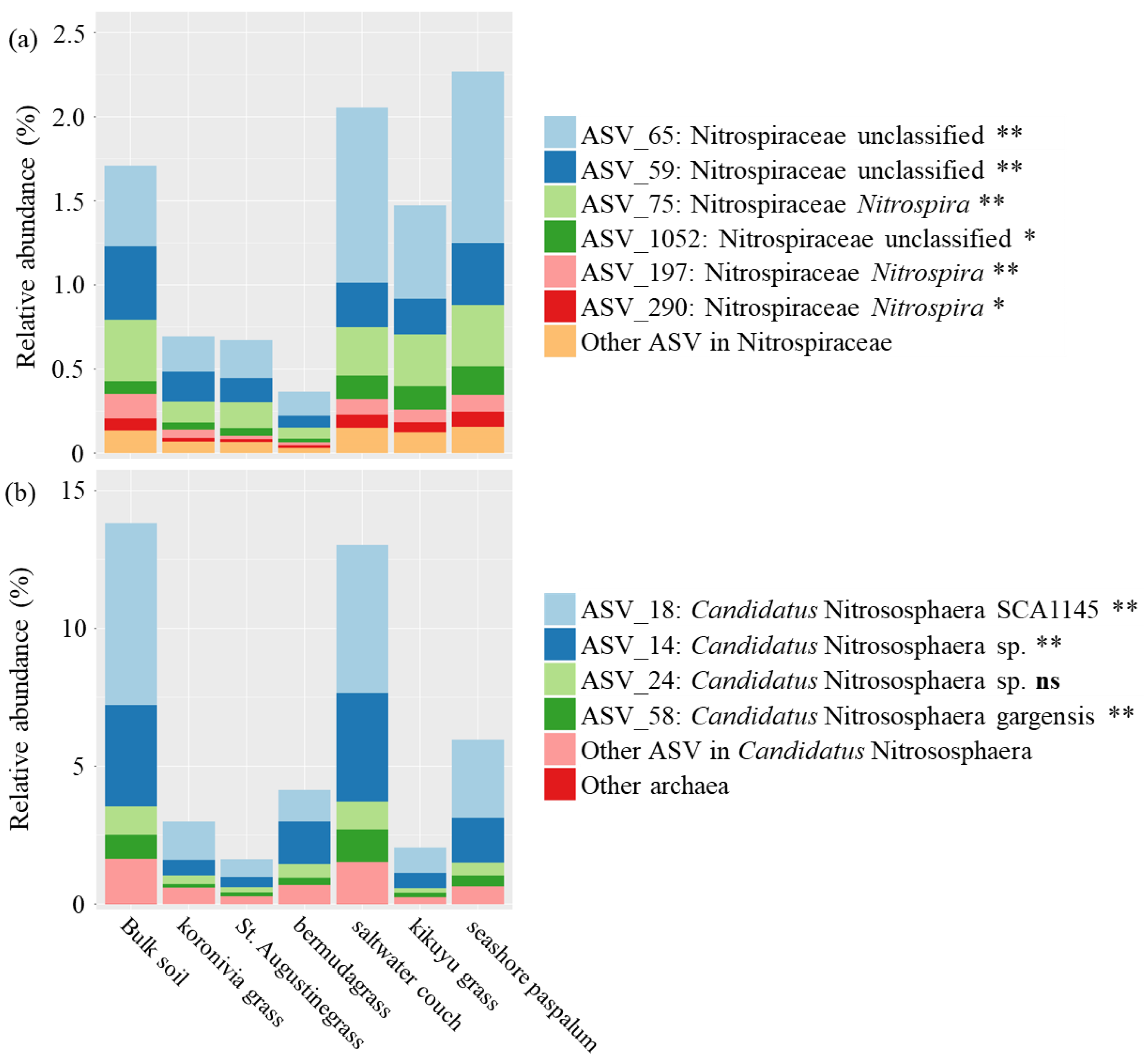
| R2 | p-Value | |
|---|---|---|
| Block effect | 0.32 | <0.01 |
| Grass species | 0.44 | <0.01 |
| Taxonomic Genus | Nitrification Enzymes | Nitrifier Group | Results in Present Study |
|---|---|---|---|
| Bacteria; Nitrospirae; Nitrospira; Nitrospirales; Nitrospiraceae; Nitrospira | NXR | NOB | Figure 5a |
| Bacteria; Nitrospirae; Nitrospira; Nitrospirales; Nitrospiraceae; Nitrospira; certain species | AOM, HAO, and NXR | Comammox bacteria | Figure 5a |
| Archaea; Crenarchaeota; Thaumarchaeota; Nitrososphaerales; Nitrososphaeraceae; Candidatus Nitrososphaera | AOM and HAO | AOA | Figure 5b |
| Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Nitrosomonadaceae; Nitrosomonas | AOM and HAO | AOB | genus not detected |
| Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Nitrosomonadaceae; Nitrosospira | AOM and HAO | AOB | genus not detected |
| Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Bradyrhizobiaceae; Nitrobacter | NXR | NOB | genus not detected |
| Microbial Community | |
|---|---|
| Bulk soil | 5.547 ± 0.062 b |
| Rhizosphere soil: | |
| koronivia grass | 5.446 ± 0.048 b |
| St. Augustinegrass | 5.463 ± 0.054 b |
| bermudagrass | 4.732 ± 0.123 c |
| saltwater couch | 5.769 ± 0.086 a |
| kikuyu grass | 5.374 ± 0.160 b |
| seashore paspalums | 5.925 ± 0.116 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Lambrides, C.J.; Li, J.; Xu, Q.; Toh, R.; Tian, S.; Yang, P.; Yang, H.; Ryder, M.; Denton, M.D. Nitrifying Microbes in the Rhizosphere of Perennial Grasses Are Modified by Biological Nitrification Inhibition. Microorganisms 2020, 8, 1687. https://doi.org/10.3390/microorganisms8111687
Zhou Y, Lambrides CJ, Li J, Xu Q, Toh R, Tian S, Yang P, Yang H, Ryder M, Denton MD. Nitrifying Microbes in the Rhizosphere of Perennial Grasses Are Modified by Biological Nitrification Inhibition. Microorganisms. 2020; 8(11):1687. https://doi.org/10.3390/microorganisms8111687
Chicago/Turabian StyleZhou, Yi, Christopher J. Lambrides, Jishun Li, Qili Xu, Ruey Toh, Shenzhong Tian, Peizhi Yang, Hetong Yang, Maarten Ryder, and Matthew D. Denton. 2020. "Nitrifying Microbes in the Rhizosphere of Perennial Grasses Are Modified by Biological Nitrification Inhibition" Microorganisms 8, no. 11: 1687. https://doi.org/10.3390/microorganisms8111687
APA StyleZhou, Y., Lambrides, C. J., Li, J., Xu, Q., Toh, R., Tian, S., Yang, P., Yang, H., Ryder, M., & Denton, M. D. (2020). Nitrifying Microbes in the Rhizosphere of Perennial Grasses Are Modified by Biological Nitrification Inhibition. Microorganisms, 8(11), 1687. https://doi.org/10.3390/microorganisms8111687





