Insights into the Potential of Sourdough-Related Lactic Acid Bacteria to Degrade Proteins in Wheat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Isolation of Proteins
2.3. Tested Isolates
2.4. Growth Screening on a Gluten-Based Agar
2.5. Gliadin Degradation Capacity
2.6. Growth Determination on ATI-Based Medium
2.7. Protein Analysis and Determination of Free Amino Acids
2.8. Statistics
3. Results and Discussion
3.1. Protein Extraction and Characterization
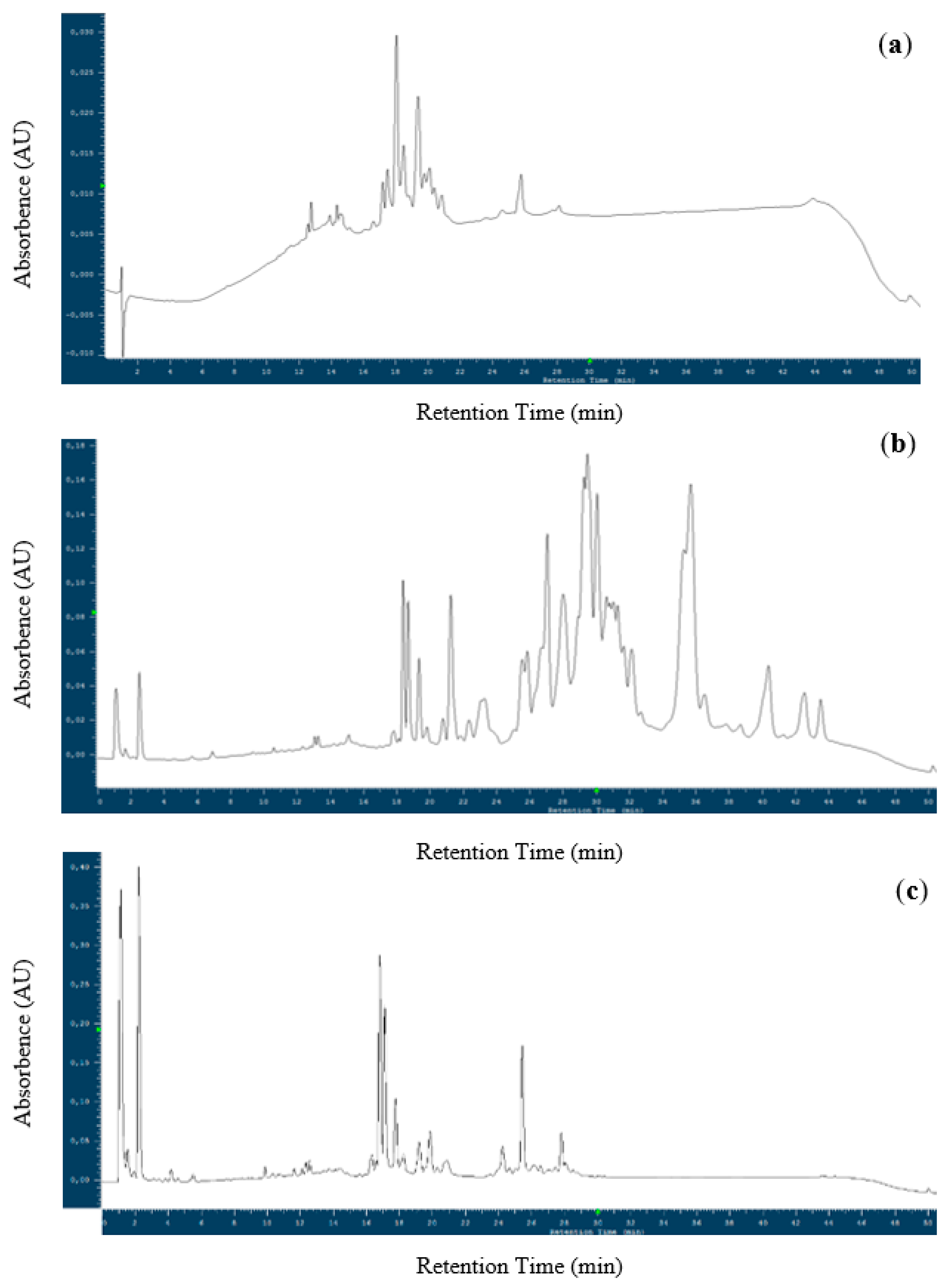
3.2. Gluten Degradation Capacity of LAB
3.2.1. Determination of Gluten Degradation by MALDI-TOF MS
3.2.2. Determination of Acidification Parameters
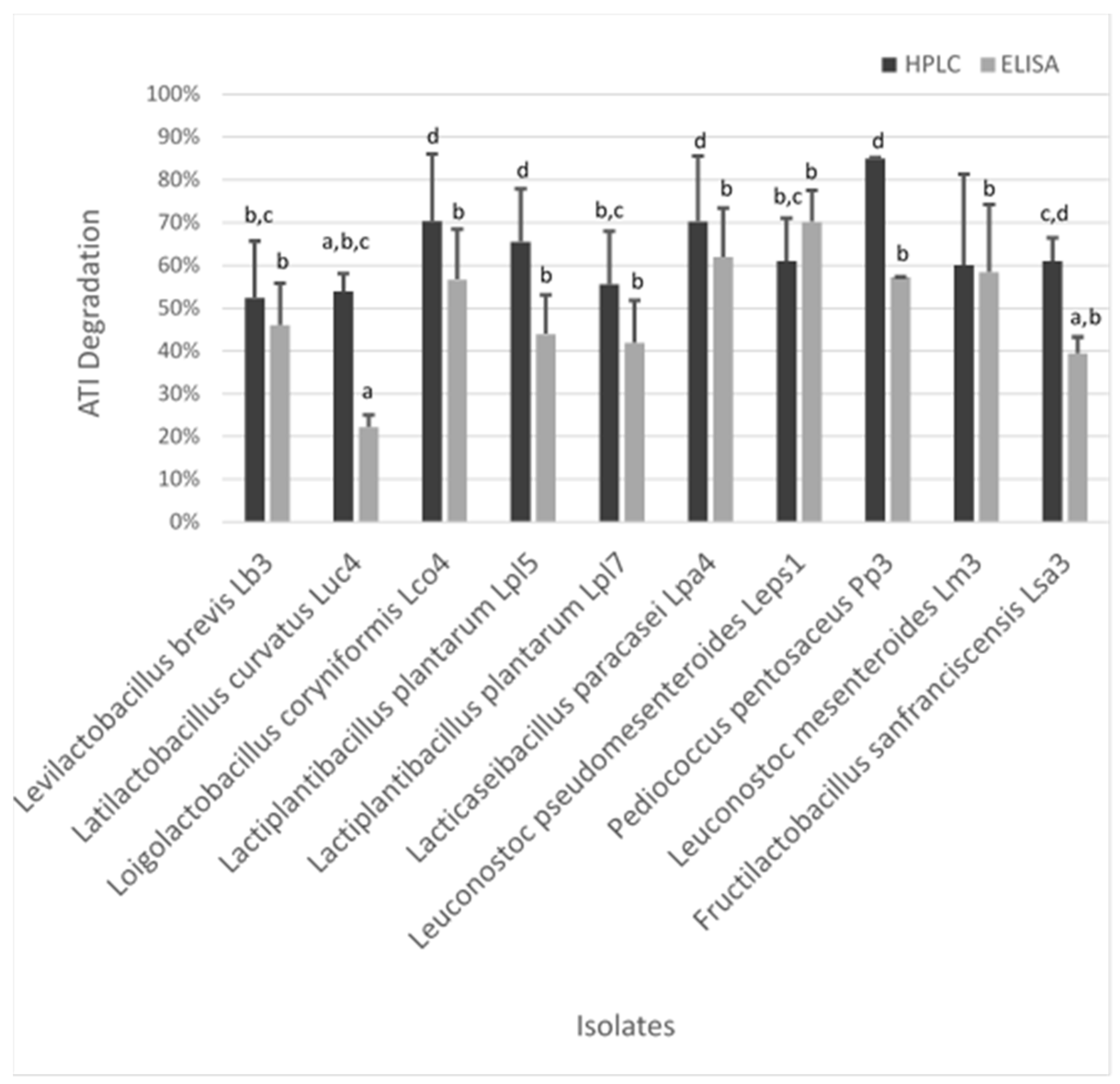
3.3. Degradation of ATIs by LAB
3.3.1. Growth Curve Determination
3.3.2. Determination of Acidification Parameters
3.3.3. Determination of the Degradation Capacity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. Cereal Supplies to Remain Ample in 2020/21 Despite This Month’s Cut in Global Production Forecast. Available online: http://www.fao.org/worldfoodsituation/csdb/en/ (accessed on 5 October 2020).
- Tchewonpi Sagu, S.; Huschek, G.; Bonick, J.; Homann, T.; Rawel, H.M. A new approach of extraction of alpha-amylase/trypsin inhibitors from wheat (Triticum aestivum L.), based on optimization using Plackett-Burman and Box-Behnken designs. Molecules 2019, 24, 3589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wieser, H. Chemistry of gluten proteins. Food Microbiol. 2007, 24, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Ozuna, C.V.; Barro, F. Characterization of gluten proteins and celiac disease-related immunogenic epitopes in the Triticeae: Cereal domestication and breeding contributed to decrease the content of gliadins and gluten. Mol. Breed. 2018, 38, 22. [Google Scholar] [CrossRef]
- Barbaro, M.R.; Cremon, C.; Stanghellini, V.; Barbara, G. Recent advances in understanding non-celiac gluten sensitivity. F1000Research 2018, 7. [Google Scholar] [CrossRef] [Green Version]
- Halmos, E.P.; Power, V.A.; Shepherd, S.J.; Gibson, P.R.; Muir, J.G. A diet low in FODMAPs reduces symptoms of irritable bowel syndrome. Gastroenterology 2014, 146, 67–75.e65. [Google Scholar] [CrossRef]
- Junker, Y.; Zeissig, S.; Kim, S.J.; Barisani, D.; Wieser, H.; Leffler, D.A.; Zevallos, V.; Libermann, T.A.; Dillon, S.; Freitag, T.L.; et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J. Exp. Med. 2012, 209, 2395–2408. [Google Scholar] [CrossRef]
- Scherf, K.A. Immunoreactive cereal proteins in wheat allergy, non-celiac gluten/wheat sensitivity (NCGS) and celiac disease. Curr. Opin. Food Sci. 2019, 25, 35–41. [Google Scholar] [CrossRef]
- Caminero, A.; McCarville, J.L.; Zevallos, V.F.; Pigrau, M.; Yu, X.B.; Jury, J.; Galipeau, H.J.; Clarizio, A.V.; Casqueiro, J.; Murray, J.A.; et al. Lactobacilli degrade wheat amylase trypsin inhibitors to reduce intestinal dysfunction induced by immunogenic wheat proteins. Gastroenterology 2019, 156, 2266–2280. [Google Scholar] [CrossRef] [Green Version]
- Catassi, C.; Bai, J.C.; Bonaz, B.; Bouma, G.; Calabro, A.; Carroccio, A.; Castillejo, G.; Ciacci, C.; Cristofori, F.; Dolinsek, J.; et al. Non-Celiac Gluten sensitivity: The new frontier of gluten related disorders. Nutrients 2013, 5, 3839–3853. [Google Scholar] [CrossRef]
- Fraberger, V.; Call, L.M.; Domig, K.J.; D’Amico, S. Applicability of yeast fermentation to reduce fructans and other FODMAPs. Nutrients 2018, 10, 1247. [Google Scholar] [CrossRef] [Green Version]
- Gobbetti, M.; Rizzello, C.G.; Di Cagno, R.; De Angelis, M. How the sourdough may affect the functional features of leavened baked goods. Food Microbiol. 2014, 37, 30–40. [Google Scholar] [CrossRef]
- Poutanen, K.; Flander, L.; Katina, K. Sourdough and cereal fermentation in a nutritional perspective. Food Microbiol. 2009, 26, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Rollan, G.; De Angelis, M.; Gobbetti, M.; de Valdez, G.F. Proteolytic activity and reduction of gliadin-like fractions by sourdough lactobacilli. J. Appl. Microbiol. 2005, 99, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Zotta, T.; Piraino, P.; Ricciardi, A.; McSweeney, P.L.; Parente, E. Proteolysis in model sourdough fermentations. J. Agric. Food Chem. 2006, 54, 2567–2574. [Google Scholar] [CrossRef] [PubMed]
- Gerez, C.L.; Rollan, G.C.; de Valdez, G.F. Gluten breakdown by lactobacilli and pediococci strains isolated from sourdough. Lett. Appl. Microbiol. 2006, 42, 459–464. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, J.; Yang, S.; Feng, J.; Jia, F.; Zhang, C. Protein degradation in wheat sourdough fermentation with Lactobacillus plantarum M616. Interdiscip. Sci. 2015, 7, 205–210. [Google Scholar] [CrossRef]
- Di Cagno, R.; De Angelis, M.; Lavermicocca, P.; De Vincenzi, M.; Giovannini, C.; Faccia, M.; Gobbetti, M. Proteolysis by sourdough lactic acid bacteria: Effects on wheat flour protein fractions and gliadin peptides involved in human cereal intolerance. Appl. Environ. Microbiol. 2002, 68, 623–633. [Google Scholar] [CrossRef] [Green Version]
- Laatikainen, R.; Koskenpato, J.; Hongisto, S.M.; Loponen, J.; Poussa, T.; Huang, X.; Sontag-Strohm, T.; Salmenkari, H.; Korpela, R. Pilot Study: Comparison of sourdough wheat bread and yeast-fermented wheat bread in individuals with wheat sensitivity and irritable bowel syndrome. Nutrients 2017, 9, 1215. [Google Scholar] [CrossRef] [Green Version]
- Call, L.; Reiter, E.V.; Wenger-Oehn, G.; Strnad, I.; Grausgruber, H.; Schoenlechner, R.; D’Amico, S. Development of an enzymatic assay for the quantitative determination of trypsin inhibitory activity in wheat. Food Chem. 2019, 299, 125038. [Google Scholar] [CrossRef]
- Fraberger, V.; Unger, C.; Kummer, C.; Domig, K.J. Insights into microbial diversity of traditional Austrian sourdough. LWT-Food Sci. Technol. 2020, 127, 109358. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Call, L.; Kapeller, M.; Grausgruber, H.; Reiter, E.; Schoenlechner, R.; D’Amico, S. Effects of species and breeding on wheat protein composition. J. Cereal Sci. 2020, 93, 102974. [Google Scholar] [CrossRef]
- Bender, D.; Fraberger, V.; Szepasvari, P.; D’Amico, S.; Tomoskozi, S.; Cavazzi, G.; Jager, H.; Domig, K.J.; Schoenlechner, R. Effects of selected lactobacilli on the functional properties and stability of gluten-free sourdough bread. Eur. Food Res. Technol. 2018, 244, 1037–1046. [Google Scholar] [CrossRef] [Green Version]
- Gänzle, M.G.; Zheng, J. Lifestyles of sourdough lactobacilli—Do they matter for microbial ecology and bread quality? Int. J. Food Microbiol. 2019, 302, 15–23. [Google Scholar] [CrossRef]
- Belina-Aldemita, M.D.; Opper, C.; Schreiner, M.; D’Amico, S. Nutritional composition of pot-pollen produced by stingless bees (Tetragonula biroi Friese) from the Philippines. J. Food Compos. Anal. 2019, 82, 103215. [Google Scholar] [CrossRef]
- Geisslitz, S.; Ludwig, C.; Scherf, K.A.; Koehler, P. Targeted LC-MS/MS reveals similar contents of alpha-amylase/trypsin-inhibitors as putative triggers of nonceliac gluten sensitivity in all wheat species except Einkorn. J. Agric. Food Chem. 2018, 66, 12395–12403. [Google Scholar] [CrossRef]
- Altenbach, S.B.; Vensel, W.H.; Dupont, F.M. The spectrum of low molecular weight alpha-amylase/protease inhibitor genes expressed in the US bread wheat cultivar Butte 86. BMC Res. Notes 2011, 4, 242. [Google Scholar] [CrossRef] [Green Version]
- D’Amico, S.; Call, L.; Edlmayr, M.; Marzak, S.; Jäger, H.; Domig, K.; Reiter, E.; Grausgruber, H. Characterization of non-gluten proteins in Austrian wheat varieties from 19th century until today. Cereal Technol. 2018, 2, 4–12. [Google Scholar] [CrossRef]
- Juhasz, A.; Belova, T.; Florides, C.G.; Maulis, C.; Fischer, I.; Gell, G.; Birinyi, Z.; Ong, J.; Keeble-Gagnere, G.; Maharajan, A.; et al. Genome mapping of seed-borne allergens and immunoresponsive proteins in wheat. Sci. Adv. 2018, 4, eaar8602. [Google Scholar] [CrossRef] [Green Version]
- Vermeulen, N.; Pavlovic, M.; Ehrmann, M.A.; Ganzle, M.G.; Vogel, R.F. Functional characterization of the proteolytic system of Lactobacillus sanfranciscensis DSM 20451T during growth in sourdough. Appl. Environ. Microbiol. 2005, 71, 6260–6266. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Sieiro, P.; Redruello, B.; Ladero, V.; Martin, M.C.; Fernandez, M.; Alvarez, M.A. Screening sourdough samples for gliadin-degrading activity revealed Lactobacillus casei strains able to individually metabolize the coeliac-disease-related 33-mer peptide. Can. J. Microbiol. 2016, 62, 422–430. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Coda, R.; Silano, M.; Minervini, F.; Rizzello, C.G.; Di Cagno, R.; Vicentini, O.; De Vincenzi, M.; Gobbetti, M. Fermentation by selected sourdough lactic acid bacteria to decrease coeliac intolerance to rye flour. J. Cereal Sci. 2006, 43, 301–314. [Google Scholar] [CrossRef]
- Gerez, C.L.; Font de Valdez, G.; Rollan, G.C. Functionality of lactic acid bacteria peptidase activities in the hydrolysis of gliadin-like fragments. Lett. Appl. Microbiol. 2008, 47, 427–432. [Google Scholar] [CrossRef]
- Dallagnol, A.M.; Pescuma, M.; De Valdez, G.F.; Rollan, G. Fermentation of quinoa and wheat slurries by Lactobacillus plantarum CRL 778: Proteolytic activity. Appl. Microbiol. Biotechnol. 2013, 97, 3129–3140. [Google Scholar] [CrossRef] [PubMed]
- Loponen, J.; Mikola, M.; Katina, K.; Sontag-Strohm, T.; Salovaara, H. Degradation of HMW glutenins during wheat sourdough fermentations. Cereal Chem. J. 2004, 81, 87–93. [Google Scholar] [CrossRef]
- Gänzle, M.G. Lactic metabolism revisited: Metabolism of lactic acid bacteria in food fermentations and food spoilage. Curr. Opin. Food Sci. 2015, 2, 106–117. [Google Scholar] [CrossRef]
- Montemurro, M.; Coda, R.; Rizzello, C.G. Recent advances in the use of sourdough biotechnology in pasta making. Foods 2019, 8, 129. [Google Scholar] [CrossRef] [Green Version]
- Filannino, P.; Cardinali, G.; Rizzello, C.G.; Buchin, S.; De Angelis, M.; Gobbetti, M.; Di Cagno, R. Metabolic responses of Lactobacillus plantarum strains during fermentation and storage of vegetable and fruit juices. Appl. Environ. Microbiol. 2014, 80, 2206–2215. [Google Scholar] [CrossRef] [Green Version]
- Tavaria, F.K.; Dahl, S.; Carballo, F.J.; Malcata, F.X. Amino acid catabolism and generation of volatiles by lactic acid bacteria. J. Dairy Sci. 2002, 85, 2462–2470. [Google Scholar] [CrossRef]
- Loponen, J.; Sontag-Strohm, T.; Venalainen, J.; Salovaara, H. Prolamin hydrolysis in wheat sourdoughs with differing proteolytic activities. J. Agric. Food Chem. 2007, 55, 978–984. [Google Scholar] [CrossRef]
- Sanchez, D.; Stepanova Honzova, S.; Hospodkova, M.; Hoffmanova, I.; Habova, V.; Halada, P.; Tlaskalova-Hogenova, H.; Tuckova, L. Occurrence of serum antibodies against wheat alpha-amylase inhibitor 0.19 in celiac disease. Physiol. Res. 2018, 67, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Huebener, S.; Tanaka, C.K.; Uhde, M.; Zone, J.J.; Vensel, W.H.; Kasarda, D.D.; Beams, L.; Briani, C.; Green, P.H.; Altenbach, S.B.; et al. Specific nongluten proteins of wheat are novel target antigens in celiac disease humoral response. J. Proteome Res. 2015, 14, 503–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lexhaller, B.; Colgrave, M.L.; Scherf, K.A. Characterization and relative quantitation of wheat, rye, and barley gluten protein types by liquid chromatography-tandem mass spectrometry. Front. Plant Sci. 2019, 10, 1530. [Google Scholar] [CrossRef] [Green Version]
- Wieser, H.; Vermeulen, N.; Gaertner, F.; Vogel, R.F. Effects of different Lactobacillus and Enterococcus strains and chemical acidification regarding degradation of gluten proteins during sourdough fermentation. Eur. Food Res. Technol. 2007, 226, 1495–1502. [Google Scholar] [CrossRef]
- Huang, X.; Schuppan, D.; Rojas Tovar, L.E.; Zevallos, V.F.; Loponen, J.; Ganzle, M. Sourdough fermentation degrades wheat alpha-amylase/trypsin inhibitor (ATI) and reduces pro-inflammatory activity. Foods 2020, 9, 943. [Google Scholar] [CrossRef]
- Komen, G.; Baysal, A.H.; Harsa, H.S. Gliadin degradation ability of artisanal lactic acid bacteria, the potential probiotics from dairy products. J. Nutr. Ther. 2013, 2, 163–172. [Google Scholar]
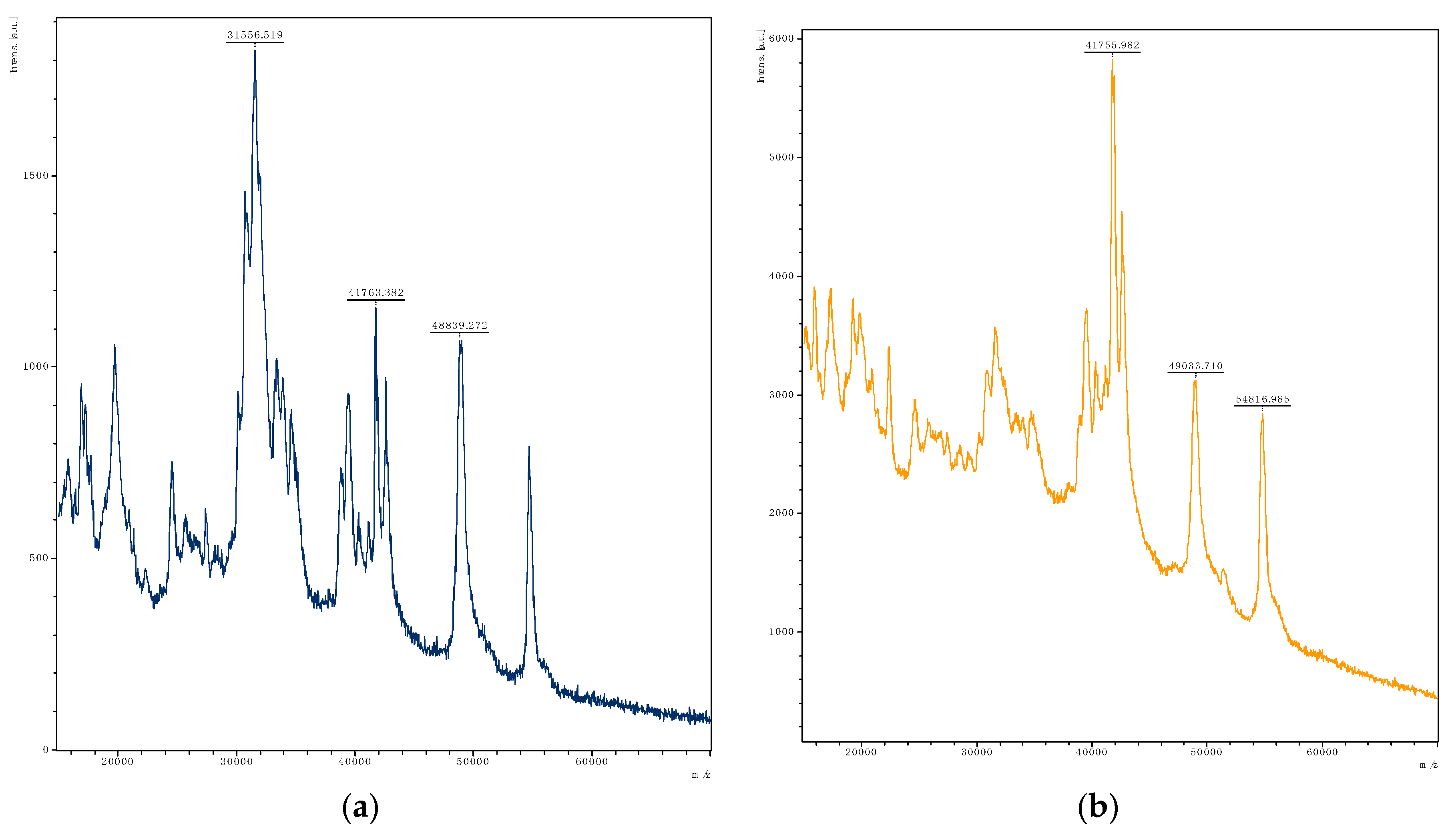
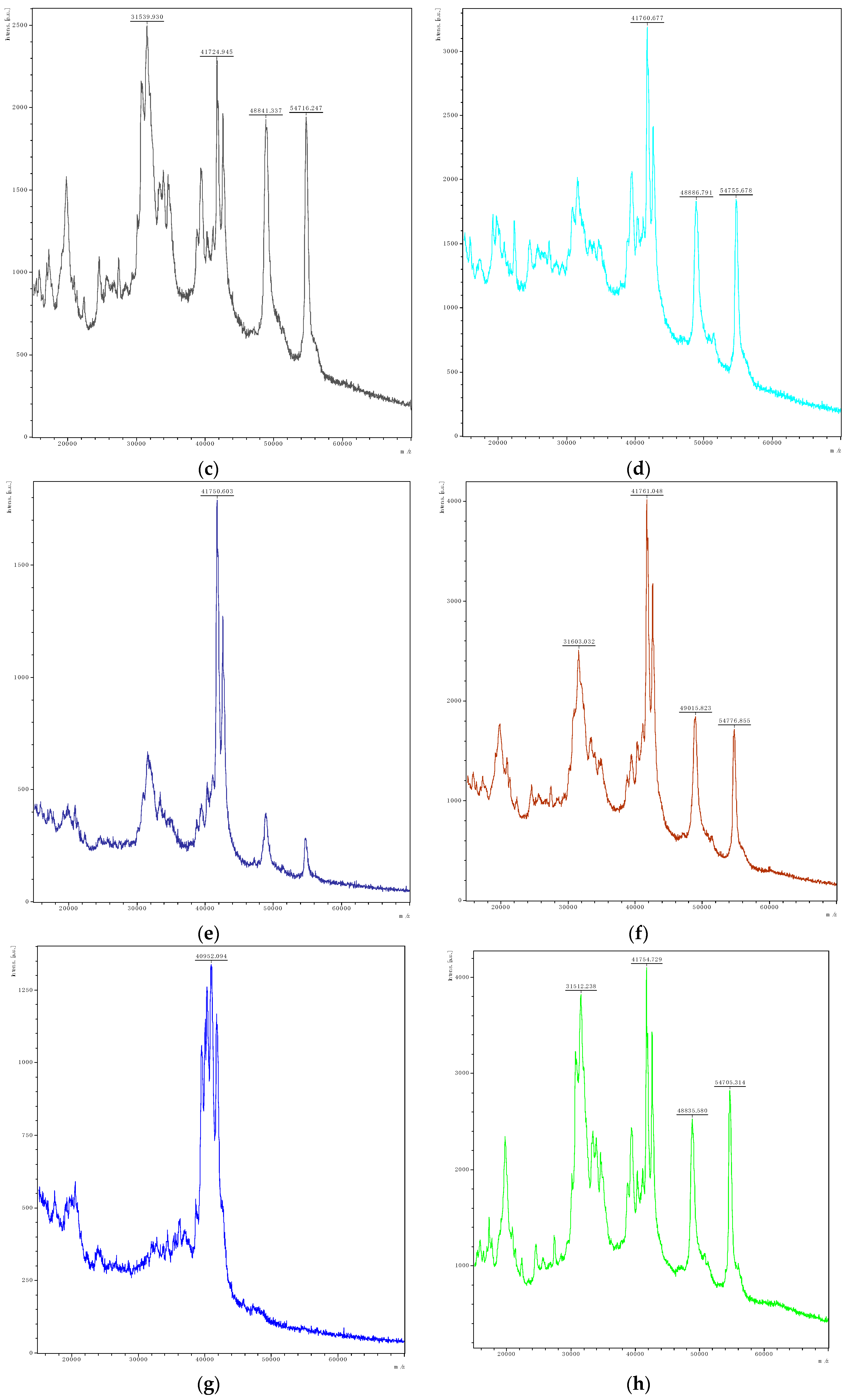
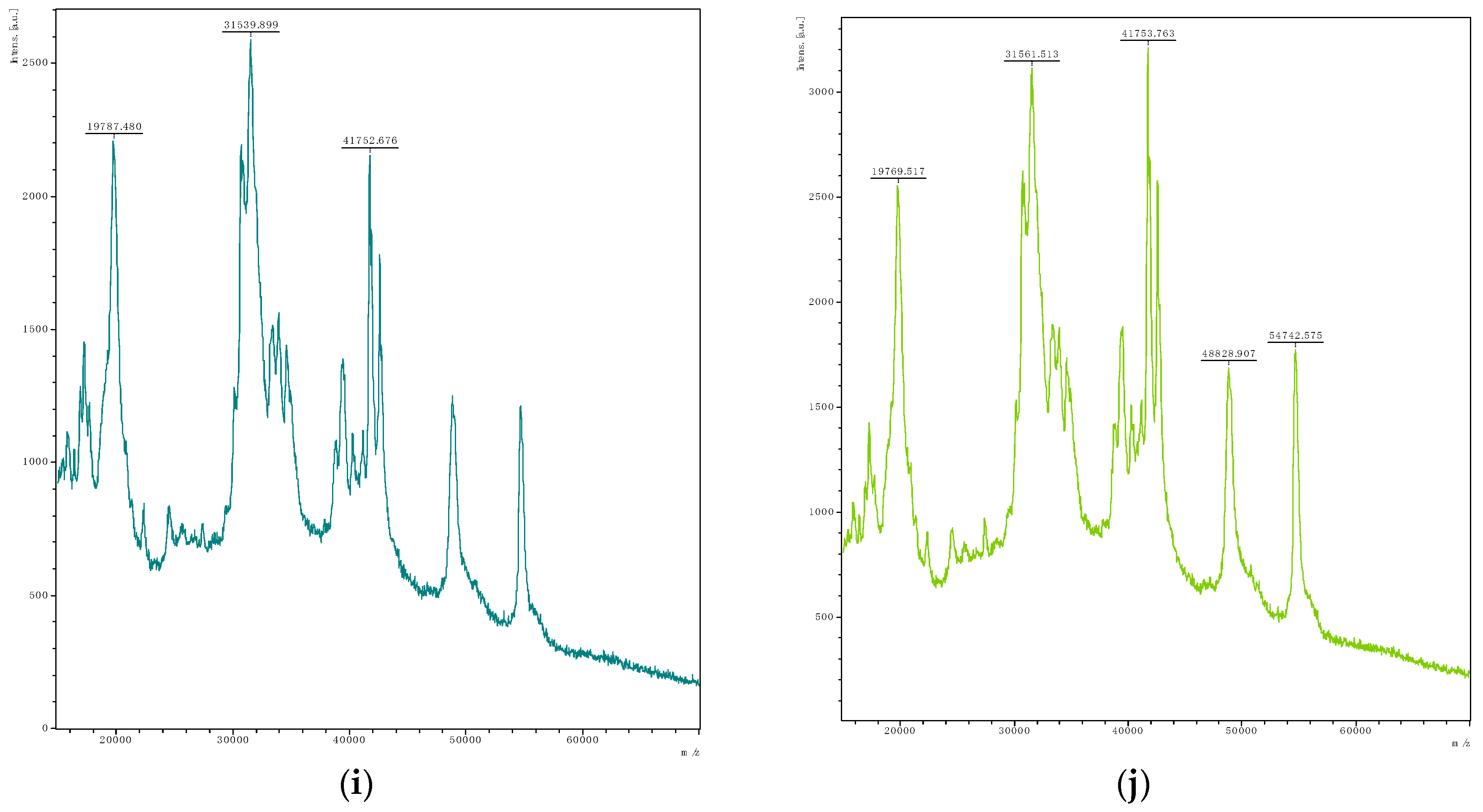
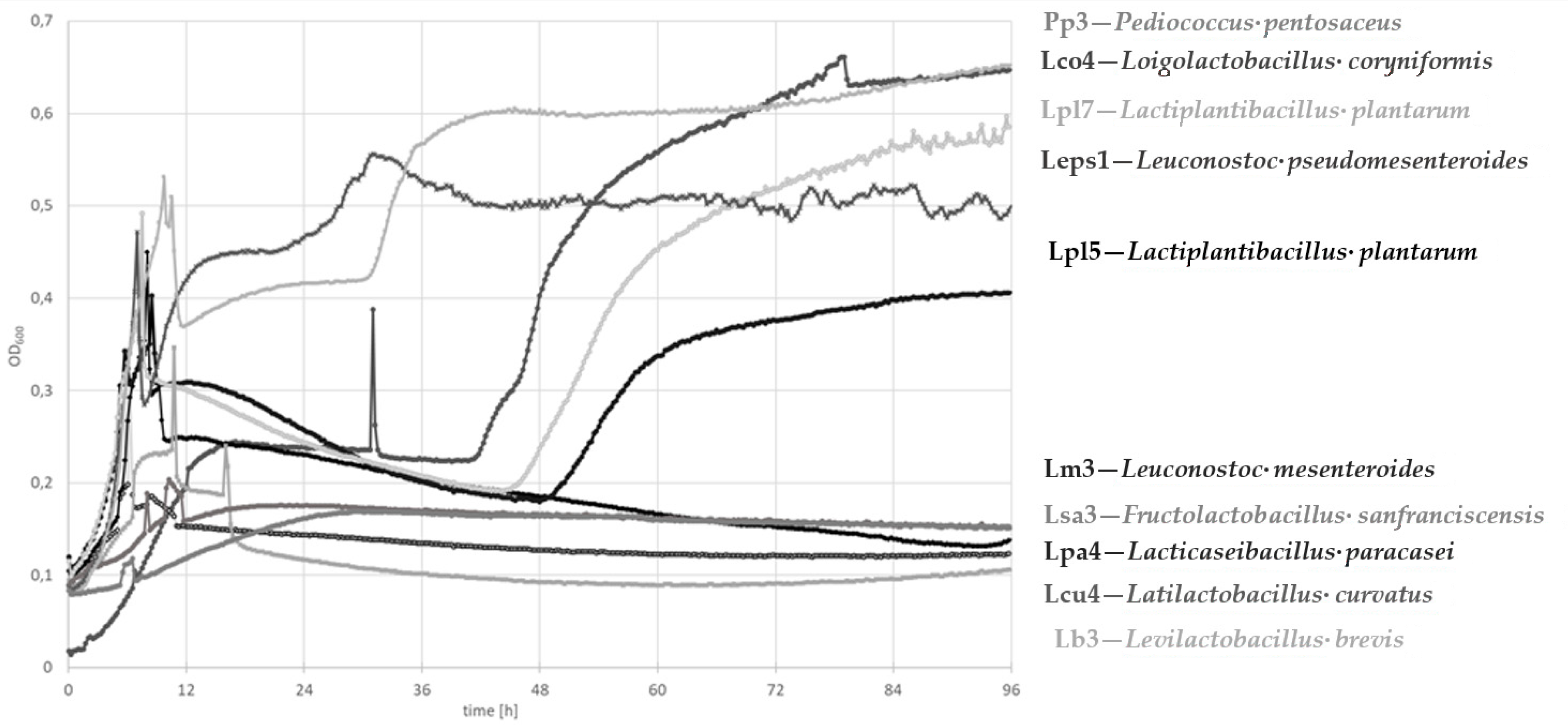
| Isolate | Associated Figure | pH | FAN (mg Glycine/L) | Lactic Acid (mg/L) | Acetic Acid (mg/L) | Gliadin Degradation |
|---|---|---|---|---|---|---|
| Levilactobacillus brevis Lb3 | Figure 2b | 6.67 | 6.77 ± 0.02 | 114 ± 3 | 88 ± 2 | ++ |
| Latilactobacillus curvatus Lcu4 | Figure 2c | 6.55 | 6.47 ± 0.14 | 392 ± 49 | <LOD | + |
| Loigolactobacillus coryniformis Lco4 | Figure 2d | 6.65 | 6.42 ± 0.27 | 108 ± 17 | 39 ± 3 | ++ |
| Lactiplantibacillus plantarum Lpl5 | Figure 2e | 4.49 | 5.37 ± 0.25 | 2786 ± 84 | 34 ± 1 | ++ |
| Lactiplantibacillus plantarum Lpl7 | Figure 2f | 4.84 | 5.58 ± 0.11 | 2392 ± 8 | 42 ± 2 | ++ |
| Lacticaseibacillus paracasei Lpa4 | Figure 2g | 4.77 | 55.67 ± 0.09 | 2442 ± 12 | 58 ± 2 | +++ |
| Leuconostoc pseudomesenteroides Leps1 | Figure 2h | 6.64 | 5.01 ± 0.23 | 223 ± 13 | 34 ± 0 | + |
| Pediococcus pentosaceus Pp3 | Figure 2i | 6.67 | 5.87 ± 0.21 | 190 ± 7 | <LOD | + |
| Leuconostoc mesenteroides Lm3 | Figure 2j | 6.44 | 3.85 ± 0.13 | 650 ± 55 | 184 ± 15 | + |
| Isolate | pH | Lactic Acid (mg/L) | Acetic Acid (mg/L) | FAN (mg Glycine/L) |
|---|---|---|---|---|
| Levilactobacillus brevis Lb3 | 6.10 | 585 ± 137 | 118 ± 1 | 23.57 ± 0.85 |
| Latilactobacillus curvatus Lcu4 | 6.52 | 190 ± 12 | 101 ± 1 | 27.67 ± 1.57 |
| Loigolactobacillus coryniformis Lco4 | 4.44 | 1189 ± 38 | 101 ± 1 | 8.01 ± 0.78 |
| Lactiplantibacillus plantarum Lpl5 | 4.54 | 1108 ± 7 | 103 ± 1 | 25.39 ± 0.46 |
| Lactiplantibacillus plantarum Lpl7 | 4.12 | >1250 | 102 ± 9 | 20.50 ± 0.47 |
| Lacticaseibacillus paracasei Lpa4 | 4.02 | >1250 | 101 ± 1 | 25.51 ± 2.35 |
| Leuconostoc pseudomesenteroides Leps1 | 4.21 | >1250 | 146 ± 17 | 23.76 ± 3.41 |
| Pediococcus pentosaceus Pp3 | 6.68 | 178 ± 1 | 95 ± 4 | 43.02 ± 1.44 |
| Leuconostoc mesenteroides Lm3 | 6.59 | 274 ± 87 | 128 ± 17 | 32.21 ± 1.79 |
| Fructilactobacillus sanfranciscensis Lsa3 | 6.71 | <35 | 93 ± 1 | 33.21 ± 5.73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fraberger, V.; Ladurner, M.; Nemec, A.; Grunwald-Gruber, C.; Call, L.M.; Hochegger, R.; Domig, K.J.; D’Amico, S. Insights into the Potential of Sourdough-Related Lactic Acid Bacteria to Degrade Proteins in Wheat. Microorganisms 2020, 8, 1689. https://doi.org/10.3390/microorganisms8111689
Fraberger V, Ladurner M, Nemec A, Grunwald-Gruber C, Call LM, Hochegger R, Domig KJ, D’Amico S. Insights into the Potential of Sourdough-Related Lactic Acid Bacteria to Degrade Proteins in Wheat. Microorganisms. 2020; 8(11):1689. https://doi.org/10.3390/microorganisms8111689
Chicago/Turabian StyleFraberger, Vera, Martin Ladurner, Alexandra Nemec, Clemens Grunwald-Gruber, Lisa M. Call, Rupert Hochegger, Konrad J. Domig, and Stefano D’Amico. 2020. "Insights into the Potential of Sourdough-Related Lactic Acid Bacteria to Degrade Proteins in Wheat" Microorganisms 8, no. 11: 1689. https://doi.org/10.3390/microorganisms8111689





