First Report of Filamentous Phages Isolated from Tunisian Orchards to Control Erwinia amylovora
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Bacteriophage Isolation, Purification and Transmission Electron Microscopy
2.3. Host Range Analysis
2.4. Temperature and pH Stability
2.5. One-Step Growth Curve
2.6. Motility Assay
2.7. Pear Slice Assay
2.8. DNA Isolation, Manipulation and Sequencing
3. Results
3.1. Isolation of Six E. amylovora-Specific Filamentous Phages
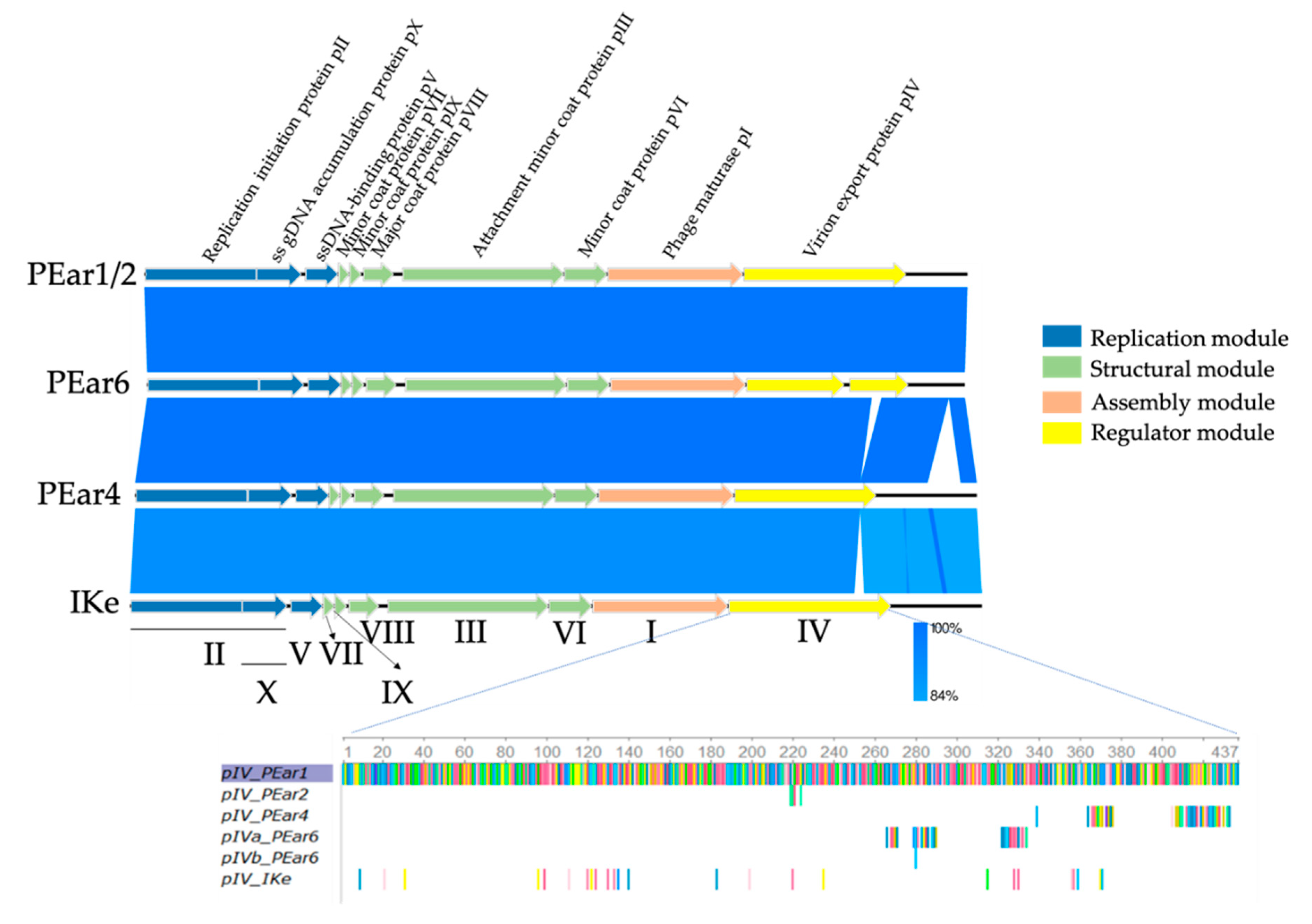
3.2. Sequence Analysis Shows That PEar1-6 Resemble Filamentous Phage IKe
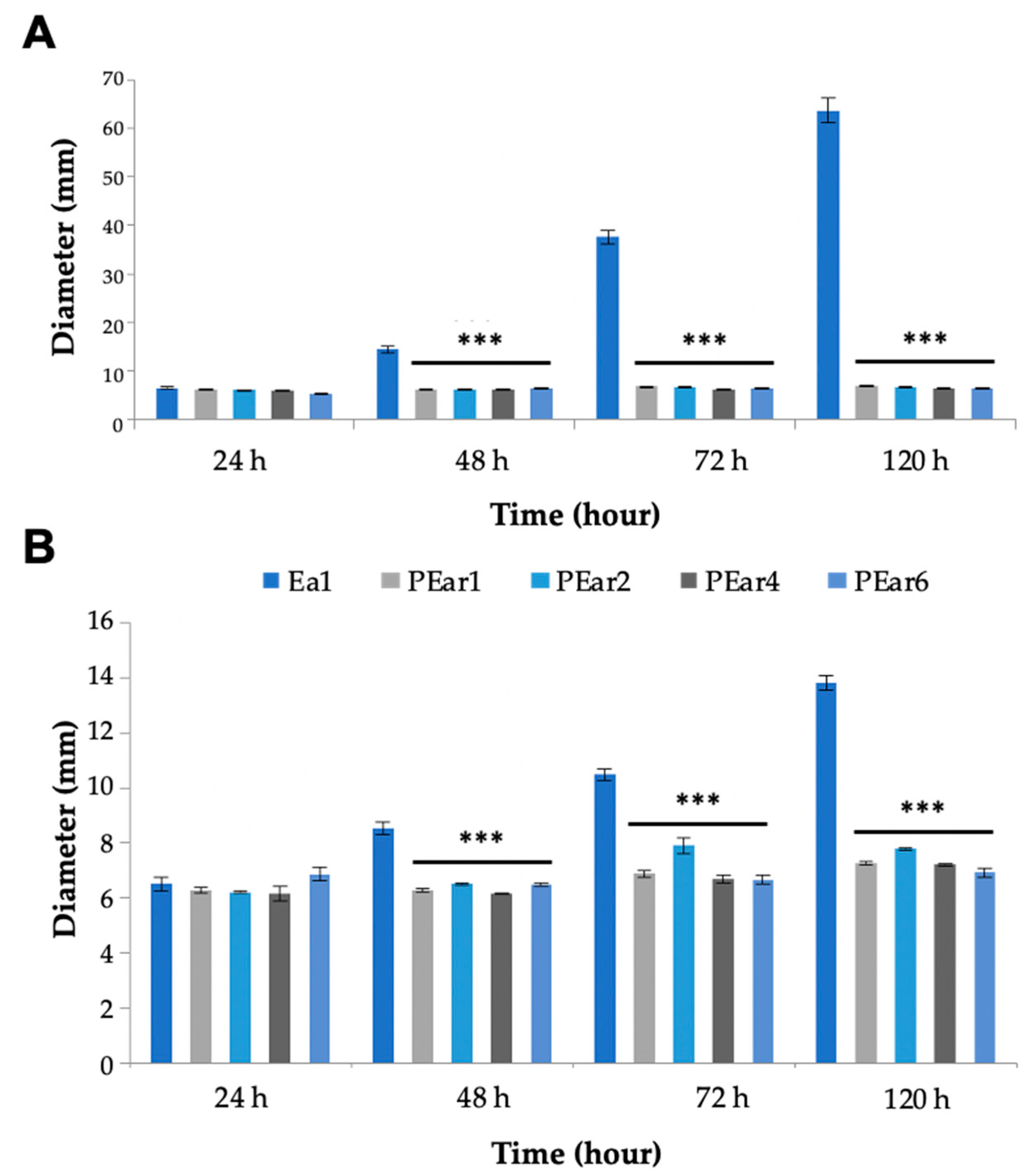
3.3. Temperature and pH Stability Makes the Isolated Phages Suitable as Biocontrol Agents
3.4. PEar Phages Influence the Swimming and Swarming Motility of Their Host Drastically
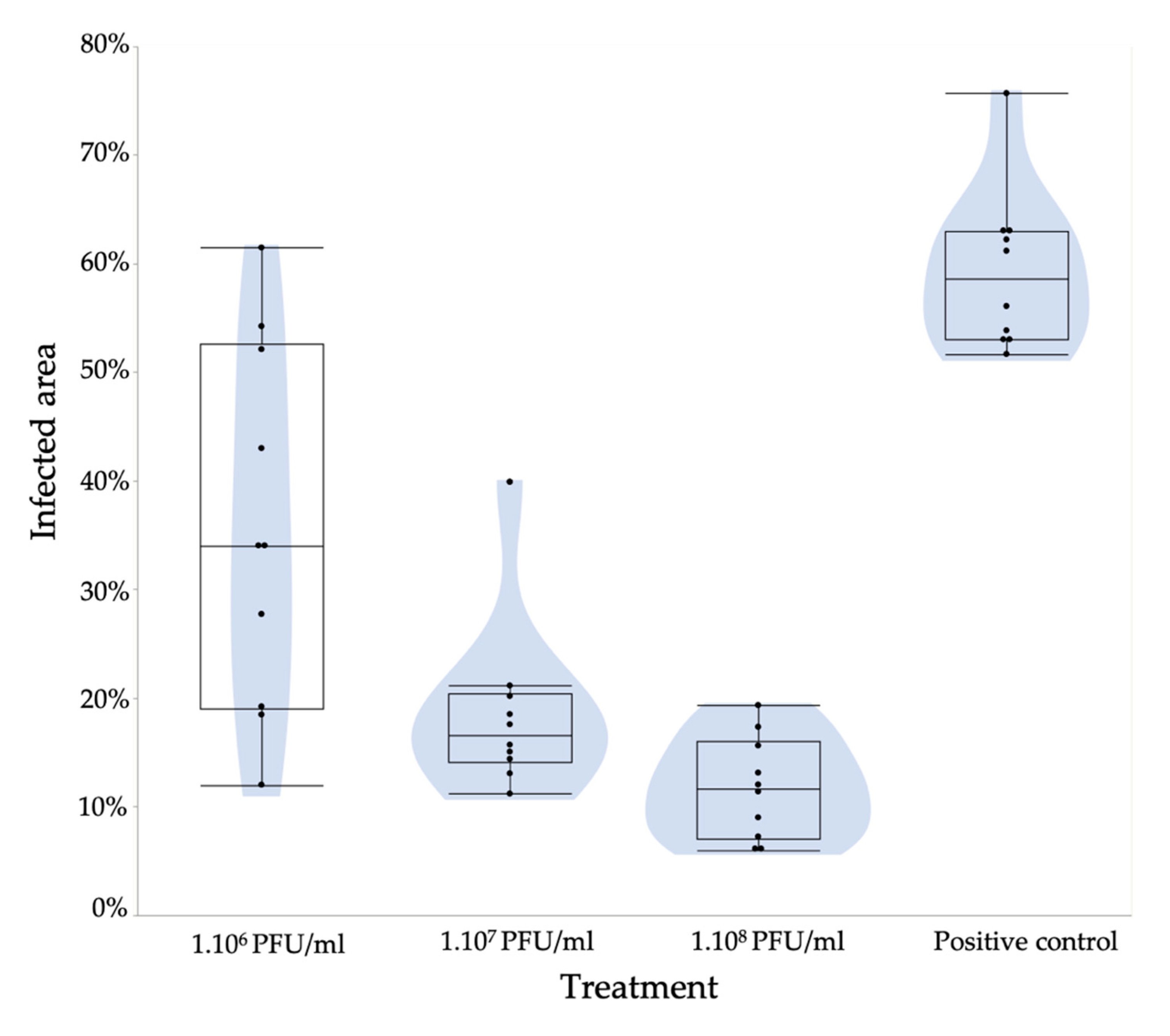
3.5. A Pear Slice Assay Shows the Potential of These Filamentous Phages to Control Fire Blight
4. Discussion
4.1. Understanding the Genome Organization of the PEar Phages
4.2. The Use of Filamentous Phages as Biotechnological Tools
4.3. Assessing the Potential of PEar Phages as Biocontrol Agents
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Van der Zwet, T.; Orolaza-Halbrendt, N.; Zeller, W. Losses due to fire blight and economic importance of the disease. In Fire Blight: History, Biology, and Management; The American Phytopathological Society: St. Paul, MN, USA, 2016; pp. 37–41. [Google Scholar]
- Gaaliche, B.; Chehimi, S.; Dardouri, S.; Hajlaoui, M.R. Health status of the pear tree following the establishment of fire blight in Northern Tunisia. Int. J. Fruit Sci. 2017, 18, 85–98. [Google Scholar] [CrossRef]
- Rhouma, A.; Helali, F.; Chettaoui, M.; Hajjouji, M.; Hajlaoui, M.R. First report of fire blight caused by Erwinia amylovora on pear in Tunisia. Plant Dis. 2014, 98, 158. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, J. Practice of applying streptomycin to control fireblight in Hungary. EPPO Bull. 2004, 34, 381–382. [Google Scholar] [CrossRef]
- McManus, P.S.; Stockwell, V.O.; Sundin, G.W.; Jones, A.L. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 2002, 40, 443–465. [Google Scholar] [CrossRef]
- Tancos, K.A.; Cox, K.D. Exploring diversity and origins of streptomycin-resistant Erwinia amylovora isolates in New York through CRISPR spacer arrays. Plant Dis. 2016, 100, 1307–1313. [Google Scholar] [CrossRef] [Green Version]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and bacterial plant diseases. Front. Microbiol. 2017, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Gill, J.J. Bacteriophages of Erwinia amylovora and their Potential Use in Biological Control; Brock University: St. Catharines, ON, Canada, 2000. [Google Scholar]
- Müller, I.; Lurz, R.; Kube, M.; Quedenau, C.; Jelkmann, W.; Geider, K. Molecular and physiological properties of bacteriophages from North America and Germany affecting the fire blight pathogen Erwinia amylovora. Microb. Biotechnol. 2011, 4, 735–745. [Google Scholar] [CrossRef] [Green Version]
- Boulé, J.; Sholberg, P.L.; Lehman, S.M.; O’gorman, D.T.; Svircev, A.M. Isolation and characterization of eight bacteriophages infecting Erwinia amylovora and their potential as biological control agents in British Columbia, Canada. Can. J. Plant Pathol. 2011, 33, 308–317. [Google Scholar] [CrossRef]
- Parcey, M.; Gayder, S.; Castle, A.J.; Svircev, A.M. Molecular profile of phage infection: A novel approach for the characterization of Erwinia phages through qPCR. Int. J. Mol. Sci. 2020, 21, 553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarczinger, I.; Kiss, E.; Süle, S.; Tóth, M.; Hevesi, M. Control of fire blight by bacteriophages on apple flowers. Proc. XII Int. Work. Fire Blight 2011, 896, 457–462. [Google Scholar] [CrossRef]
- Schwarczinger, I.; Kolozsváriné Nagy, J.; Künstler, A.; Szabó, L.; Geider, K.; Király, L.; Pogány, M.; Nagy, J.K.; Kunstler, A.; Szabo, L.; et al. Characterization of Myoviridae and Podoviridae family bacteriophages of Erwinia amylovora from Hungary—potential of application in biological control of fire blight. Eur. J. Plant Pathol. 2017, 149, 639–652. [Google Scholar] [CrossRef] [Green Version]
- Piekarowicz, A.; Klyz, A.; Majchrzak, M.; Szczesna, E.; Piechucki, M.; Kwiatek, A.; Maugel, T.K.; Stein, D.C. Neisseria gonorrhoeae filamentous phage Ngo6 is capable of infecting a variety of Gram-negative bacteria. J. Virol. 2013, 88, 1002–1010. [Google Scholar] [CrossRef] [Green Version]
- Stassen, A.P.M.; Folmer, R.H.A.; Hilbers, C.W.; Konings, R.N.H. Single-stranded DNA binding protein encoded by the filamentous bacteriophage M13: Structural and functional characteristics. Mol. Biol. Rep. 1995, 20, 109–127. [Google Scholar] [CrossRef]
- Samire, P.; Serrano, B.; Duché, D.; Lemarié, E.; Lloubès, R.; Houot, L. Decoupling filamentous phage uptake and energy of the TolQRA motor in Escherichia coli. J. Bacteriol. 2019, 202. [Google Scholar] [CrossRef]
- Sharma, R.S.; Karmakar, S.; Kumar, P.; Mishra, V. Application of filamentous phages in environment: A tectonic shift in the science and practice of ecorestoration. Ecol. Evol. 2019, 9, 2263–2304. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, T.; Nagata, S.; Fujiwara, A.; Satsuma, H.; Fujie, M.; Usami, S.; Yamada, T. Genomic characterization of the filamentous integrative bacteriophages phiRSS1 and phiRSM1, which infect Ralstonia solanacearum. J. Bacteriol. 2007, 189, 5792–5802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murugaiyan, S.; Bae, J.Y.; Wu, J.; Lee, S.D.; Um, H.Y.; Choi, H.K.; Chung, E.; Lee, J.-H.; Lee, S.-W. Characterization of filamentous bacteriophage PE226 infecting Ralstonia solanacearum strains. J. Appl. Microbiol. 2010, 110, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959. [Google Scholar]
- Addy, H.S.; Ahmad, A.A.; Huang, Q. Molecular and biological characterization of Ralstonia phage RsoM1USA, a new species of P2virus, isolated in the United States. Front. Microbiol. 2019, 10, 267. [Google Scholar] [CrossRef]
- Pajunen, M.I.; Kiljunen, S.J.; Söderholm, M.E.-L.; Skurnik, M. Complete genomic sequence of the lytic bacteriophage phiYeO3-12 of Yersinia enterocolitica serotype O:3. J. Bacteriol. 2001, 183, 1928–1937. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, A.A.; Askora, A.; Kawasaki, T.; Fujie, M.; Yamada, T. The filamentous phage XacF1 causes loss of virulence in Xanthomonas axonopodis pv. citri, the causative agent of citrus canker disease. Front. Microbiol. 2014, 5, 321. [Google Scholar] [CrossRef] [Green Version]
- Tsonos, J.; Oosterik, L.H.; Tuntufye, H.N.; Klumpp, J.; Butaye, P.; De Greve, H.; Hernalsteens, J.P.; Lavigne, R.; Goddeeris, B.M. A cocktail of in vitro efficient phages is not a guarantee for in vivo therapeutic results against avian colibacillosis. Vet. Microbiol. 2014, 171, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2001; ISBN 0879695773. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2016, 45, D535–D542. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Peeters, B.P.H.; Peters, R.M.; Schoenmakers, J.G.G.; Konings, R.N.H. Nucleotide sequence and genetic organization of the genome of the N-specific filamentous bacteriophage IKe. J. Mol. Biol. 1985, 181, 27–39. [Google Scholar] [CrossRef]
- Schaller, H. The intergenic region and the origins for filamentous phage DNA replication. Cold Spring Harb. Symp. Quant. Biol. 1979, 43, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; De Jesus, J.G.; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S.; et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019, 20, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Roach, D.R.; Sjaarda, D.R.; Castle, A.J.; Svircev, A.M. Host exopolysaccharide quantity and composition impact Erwinia amylovora bacteriophage pathogenesis. Appl. Environ. Microbiol. 2013, 79, 3249–3256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mai-Prochnow, A.; Hui, J.G.K.; Kjelleberg, S.; Rakonjac, J.; McDougald, D.; Rice, S.A. Big things in small packages: The genetics of filamentous phage and effects on fitness of their host. FEMS Microbiol. Rev. 2015, 39, 465–487. [Google Scholar] [CrossRef] [Green Version]
- Hay, I.D.; Lithgow, T. Filamentous phages: Masters of a microbial sharing economy. EMBO Rep. 2019, 20, e47427. [Google Scholar] [CrossRef]
- Lv, M.; Chen, Y.; Liao, L.; Liang, Z.; Shi, Z.; Tang, Y.; Ye, S.; Zhou, J.; Zhang, L. Fis is a global regulator critical for modulation of virulence factor production and pathogenicity of Dickeya zeae. Sci. Rep. 2018, 8, 341. [Google Scholar] [CrossRef] [Green Version]
- Jakob, R.P.; Geitner, A.-J.; Weininger, U.; Balbach, J.; Dobbek, H.; Schmid, F.X. Structural and energetic basis of infection by the filamentous bacteriophage IKe. Mol. Microbiol. 2012, 84, 1124–1138. [Google Scholar] [CrossRef]
- Stengele, I.; Bross, P.; Garces, X.; Giray, J.; Rasched, I. Dissection of functional domains in phage fd adsorption protein. J. Mol. Biol. 1990, 212, 143–149. [Google Scholar] [CrossRef]
- Horiuchi, K. Initiation mechanisms in replication of filamentous phage DNA. Genes Cells 1997, 2, 425–432. [Google Scholar] [CrossRef] [Green Version]
- Kokoska, R.J.; Steege, D.A. Appropriate expression of filamentous phage f1 DNA replication genes II and X requires RNase E-dependent processing and separate mRNAs. J. Bacteriol. 1998, 180, 3245–3249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fulford, W.; Model, P. Regulation of bacteriophage f1 DNA replication. J. Mol. Biol. 1988, 203, 49–62. [Google Scholar] [CrossRef]
- Russel, M. Filamentous phage assembly. Mol. Microbiol. 1991, 5, 1607–1613. [Google Scholar] [CrossRef]
- Kazmierczak, B.I.; Mielke, D.L.; Russel, M.; Model, P. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J. Mol. Biol. 1994, 238, 187–198. [Google Scholar] [CrossRef]
- Feng, J.; Model, P.; Russel, M. A trans-envelope protein complex needed for filamentous phage assembly and export. Mol. Microbiol. 1999, 34, 745–755. [Google Scholar] [CrossRef]
- Russel, M.; Model, P. Genetic analysis of the filamentous bacteriophage packaging signal and of the proteins that interact with it. J. Virol. 1989, 63, 3284–3295. [Google Scholar] [CrossRef] [Green Version]
- Michel, B.; Zinder, N.D. Translational repression in bacteriophage f1: Characterization of the gene V protein target on the gene II mRNA. Proc. Natl. Acad. Sci. USA 1989, 86, 4002–4006. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Dayan, N.; Goldbourt, A.; Xiang, Y. Cryo-electron microscopy structure of the filamentous bacteriophage IKe. Proc. Natl. Acad. Sci. USA 2019, 116, 5493–5498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivey-Hoyle, M.; Steege, D.A. Mutational analysis of an inherently defective translation initiation site. J. Mol. Biol. 1992, 224, 1039–1054. [Google Scholar] [CrossRef]
- Born, Y.; Fieseler, L.; Marazzi, J.; Lurz, R.; Duffy, B.; Loessner, M.J. Novel virulent and broad-host-range Erwinia amylovora bacteriophages reveal a high degree of mosaicism and a relationship to Enterobacteriaceae phages. Appl. Environ. Microbiol. 2011, 77, 5945–5954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puławska, J.; Kałużna, M.; Warabieda, W.; Mikiciński, A. Comparative transcriptome analysis of a lowly virulent strain of Erwinia amylovora in shoots of two apple cultivars susceptible and resistant to fire blight. BMC Genom. 2017, 18, 868. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; Chaudhry, W.N.; Akhtar, M.N.; Andleeb, S.; Qadri, I. Bacteriophages and their implications on future biotechnology: A review. Virol. J. 2012, 9, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrenko, V.A. Landscape phage as a molecular recognition interface for detection devices. Microelectron. J. 2008, 39, 202–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holtappels, M.; Vrancken, K.; Schoofs, H.; Deckers, T.; Remans, T.; Noben, J.P.; Valcke, R. A comparative proteome analysis reveals flagellin, chemotaxis regulated proteins and amylovoran to be involved in virulence differences between Erwinia amylovora strains. J. Proteom. 2015, 123, 54–69. [Google Scholar] [CrossRef]
- Addy, H.S.; Askora, A.; Kawasaki, T.; Fujie, M.; Yamada, T. The filamentous phage phiRSS1 enhances virulence of phytopathogenic Ralstonia solanacearum on tomato. Phytopathology 2012, 102, 244–251. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Q.; Sundin, G.W. Genome-wide identification of Hfq-regulated small RNAs in the fire blight pathogen Erwinia amylovora discovered small RNAs with virulence regulatory function. BMC Genom. 2014, 15, 414. [Google Scholar] [CrossRef] [Green Version]
- Schachterle, J.K.; Zeng, Q.; Sundin, G.W. Three Hfq-dependent small RNAs regulate flagellar motility in the fire blight pathogen Erwinia amylovora. Mol. Microbiol. 2019, 111, 1476–1492. [Google Scholar] [CrossRef]
- Jian, H.; Xiong, L.; Xu, G.; Xiao, X.; Wang, F. Long 5’ untranslated regions regulate the RNA stability of the deep-sea filamentous phage SW1. Sci. Rep. 2016, 6, 21908. [Google Scholar] [CrossRef] [Green Version]
- Balogh, B.; Nga, N.T.T.; Jones, J.B. Relative level of bacteriophage multiplication in vitro or in phyllosphere may not predict in planta efficacy for controlling bacterial leaf spot on tomato caused by Xanthomonas perforans. Front. Microbiol. 2018, 9, 2176. [Google Scholar] [CrossRef] [Green Version]
- Svircev, A.; Roach, D.; Castle, A. Framing the future with bacteriophages in agriculture. Viruses 2018, 10, 218. [Google Scholar] [CrossRef] [Green Version]
- Pusey, P.L.; Stockwell, V.O.; Reardon, C.L.; Smits, T.H.M.; Duffy, B. Antibiosis activity of Pantoea agglomerans biocontrol strain E325 against Erwinia amylovora on apple flower stigmas. Phytopathology 2011, 101, 1234–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Species | Strain | PEar1 | PEar2 | PEar3 | PEar4 | PEar5 | PEar6 |
|---|---|---|---|---|---|---|---|
| Erwinia amylovora | Ea1 | + | + | + | + | + | + |
| Er4 | - | - | - | + | - | + | |
| Er7 | - | - | - | - | - | + | |
| Eaw | - | + | + | + | + | + | |
| EaJd | + | - | - | - | - | + | |
| Erw | - | - | - | - | - | + | |
| LMG 2024 | - | - | - | + | - | + | |
| GBBC 403 | + | + | + | - | + | + | |
| Erwinia mallotivora | LMG 1271 | - | - | - | - | - | - |
| Pantoea agglomerans | LMG 2660 | - | - | - | - | - | - |
| LMG 2570 | - | - | - | - | - | - | |
| Erwinia carotovora | sp. | - | - | - | - | - | - |
| Dickeya dianthicola | LMG 2485 | - | - | - | - | - | - |
| Dickeya dadantii | CFBP 3855 | - | - | - | - | - | - |
| Mutation (Amino Acid) | Gene Product | Frequency of Mutation (%) | e-Value |
|---|---|---|---|
| L219 → F219 | pIV | 59.6 | 1.81865e-217 |
| T220 → F220 | pIV | 59.8 | 1.00557e-239 |
| N221 → G221 | pIV | 66.2 | 0 |
| D224 → Y224 | pIV | 85 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akremi, I.; Holtappels, D.; Brabra, W.; Jlidi, M.; Hadj Ibrahim, A.; Ben Ali, M.; Fortuna, K.; Ahmed, M.; Meerbeek, B.V.; Rhouma, A.; et al. First Report of Filamentous Phages Isolated from Tunisian Orchards to Control Erwinia amylovora. Microorganisms 2020, 8, 1762. https://doi.org/10.3390/microorganisms8111762
Akremi I, Holtappels D, Brabra W, Jlidi M, Hadj Ibrahim A, Ben Ali M, Fortuna K, Ahmed M, Meerbeek BV, Rhouma A, et al. First Report of Filamentous Phages Isolated from Tunisian Orchards to Control Erwinia amylovora. Microorganisms. 2020; 8(11):1762. https://doi.org/10.3390/microorganisms8111762
Chicago/Turabian StyleAkremi, Ismahen, Dominique Holtappels, Wided Brabra, Mouna Jlidi, Adel Hadj Ibrahim, Manel Ben Ali, Kiandro Fortuna, Mohammed Ahmed, Bart Van Meerbeek, Ali Rhouma, and et al. 2020. "First Report of Filamentous Phages Isolated from Tunisian Orchards to Control Erwinia amylovora" Microorganisms 8, no. 11: 1762. https://doi.org/10.3390/microorganisms8111762
APA StyleAkremi, I., Holtappels, D., Brabra, W., Jlidi, M., Hadj Ibrahim, A., Ben Ali, M., Fortuna, K., Ahmed, M., Meerbeek, B. V., Rhouma, A., Lavigne, R., Ben Ali, M., & Wagemans, J. (2020). First Report of Filamentous Phages Isolated from Tunisian Orchards to Control Erwinia amylovora. Microorganisms, 8(11), 1762. https://doi.org/10.3390/microorganisms8111762








