Soluble Expression of hFGF19 without Fusion Protein through Synonymous Codon Substitutions and DsbC Co-Expression in E. coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Chemicals
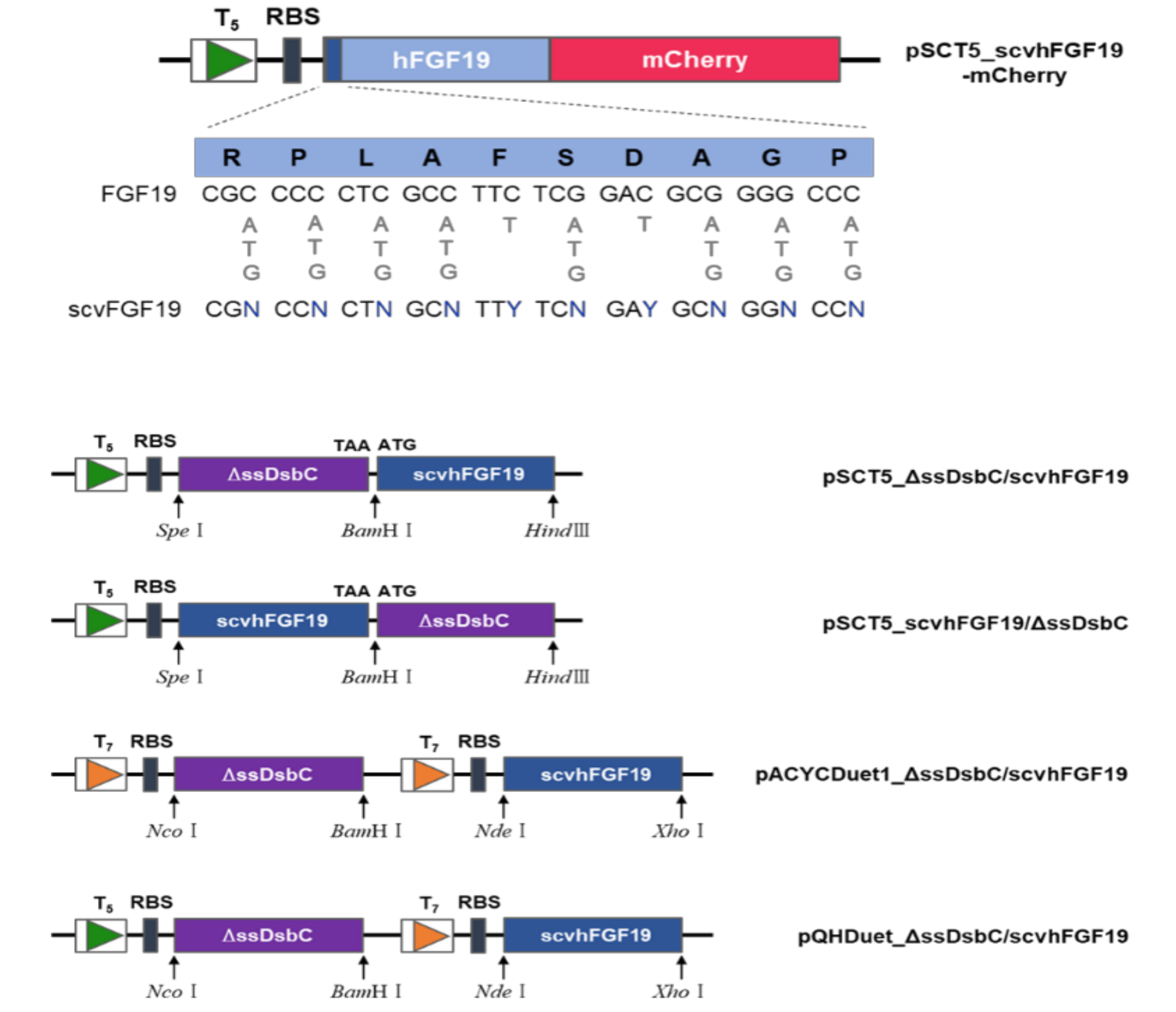
2.2. Library Construction and Screening of Synonymous Codon Variants
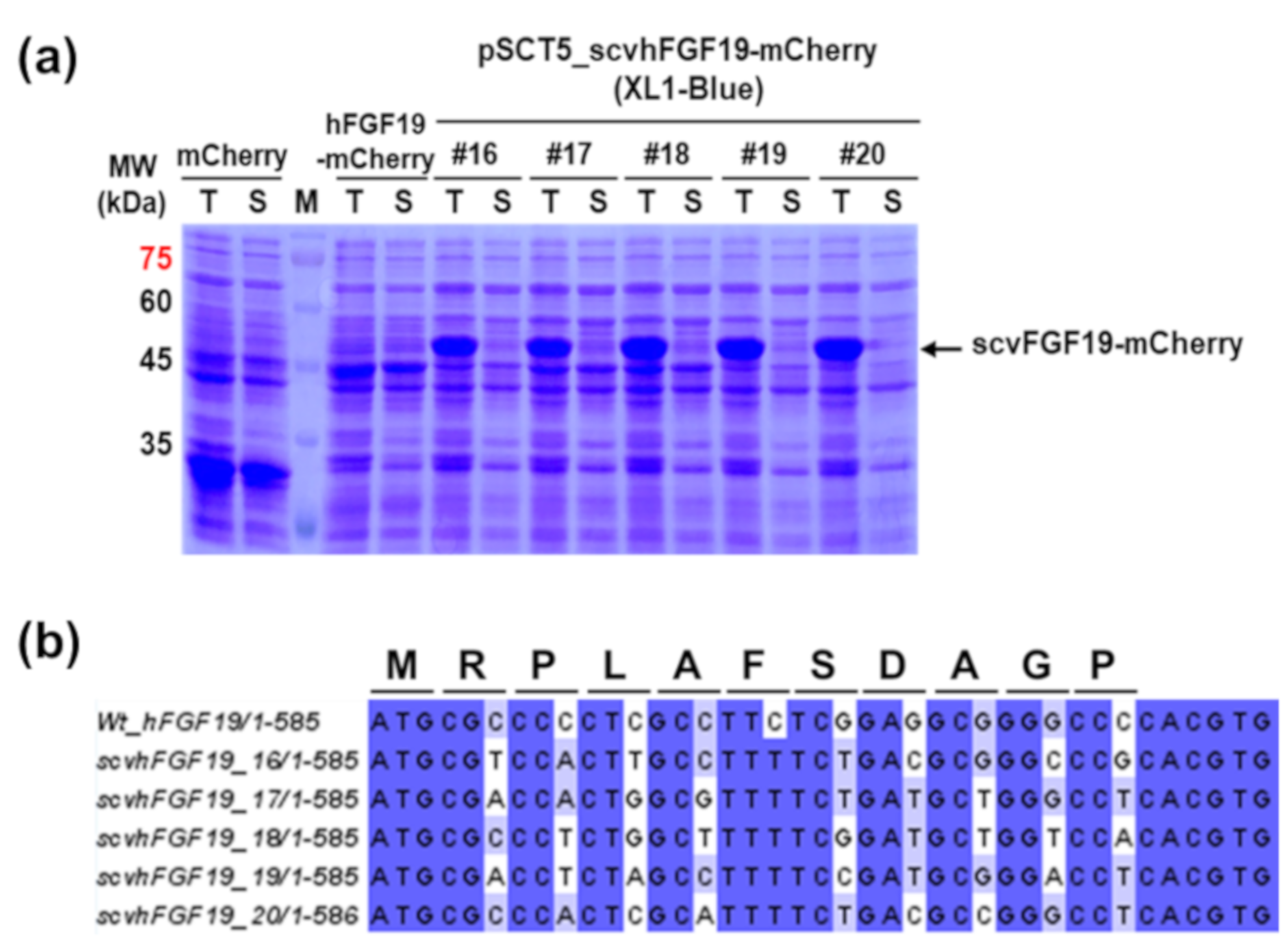
2.3. Analysis of Expression Pattern of mCherry-Fused scvhFGF19 Variants
2.4. Analysis of the Expression Pattern of scvhFGF19 Variants without Fusion Partner
2.5. Construction of Plasmids for Co-Expression of DsbC and scvhFGF19
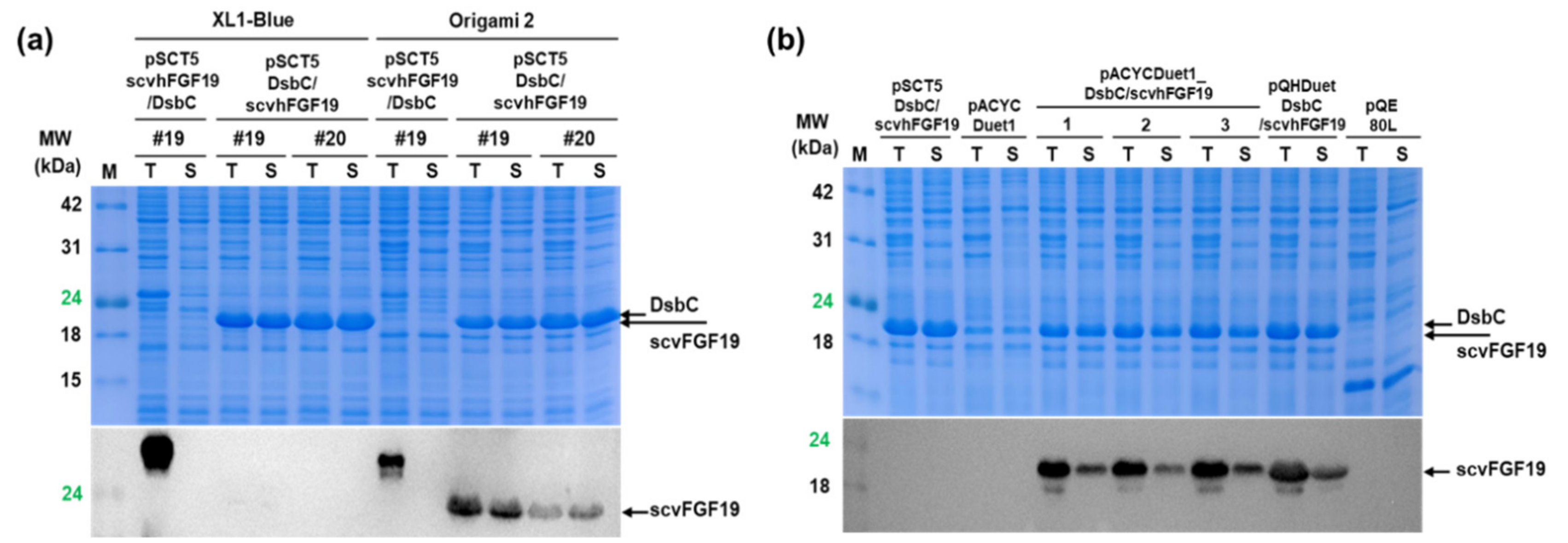
2.6. Analysis of scvhFGF19 Expression Patterns Due to Co-Expression of ΔssDsbC in E. coli Cytoplasm
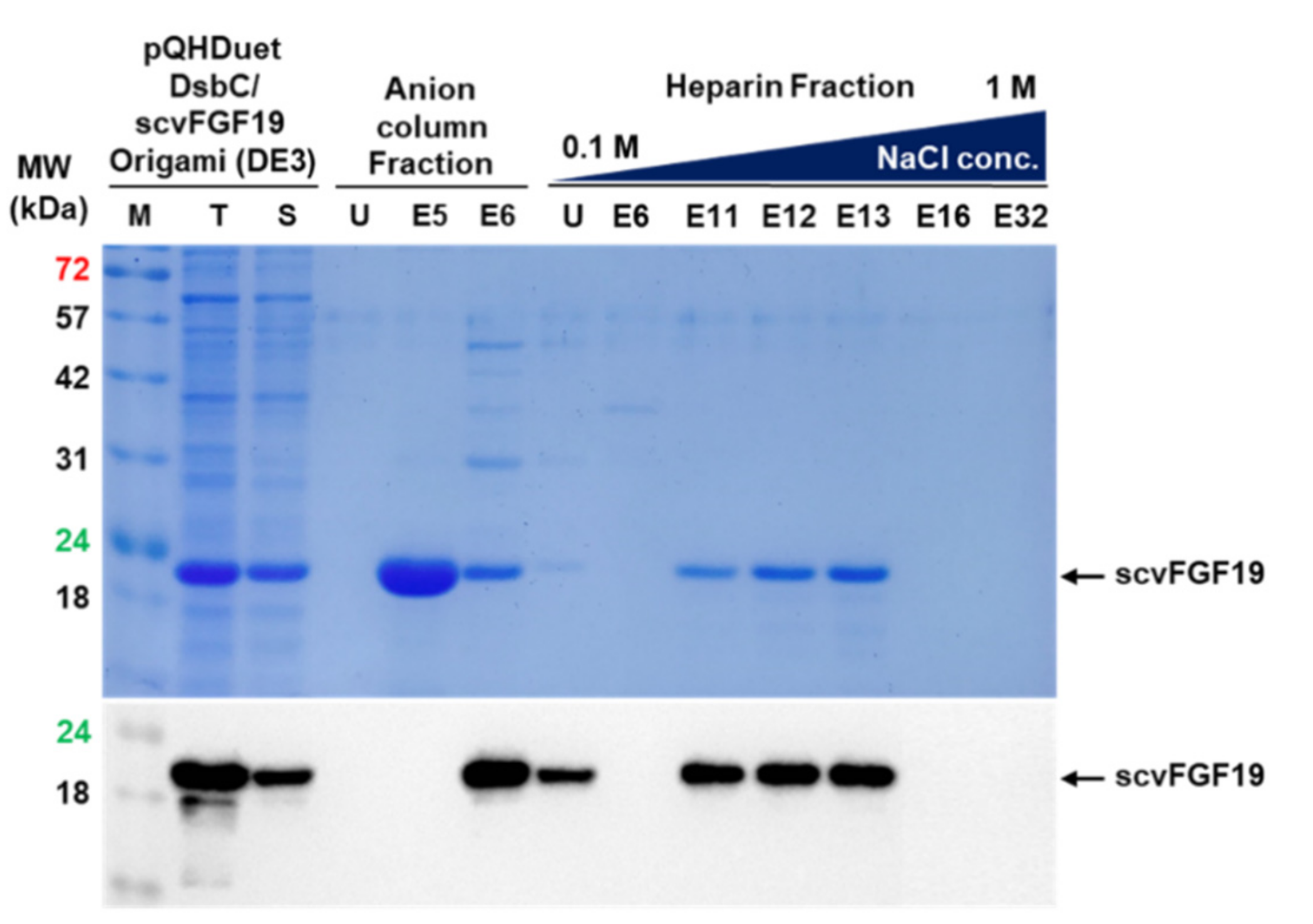
2.7. Purification of scvhFGF19
2.8. Endotoxin Removal
2.9. Activity Assay of scvhFGF19
3. Results
3.1. Expression of scvhFGF19 Screened from Synonymous Codon Variant Library
3.2. Enhanced Soluble Expression of scvhFGF19 Due to Co-Expression of DsbC in E. coli Cytoplasm
3.3. Improved Total Expression of scvhFGF19
3.4. Purification of scvhFGF19
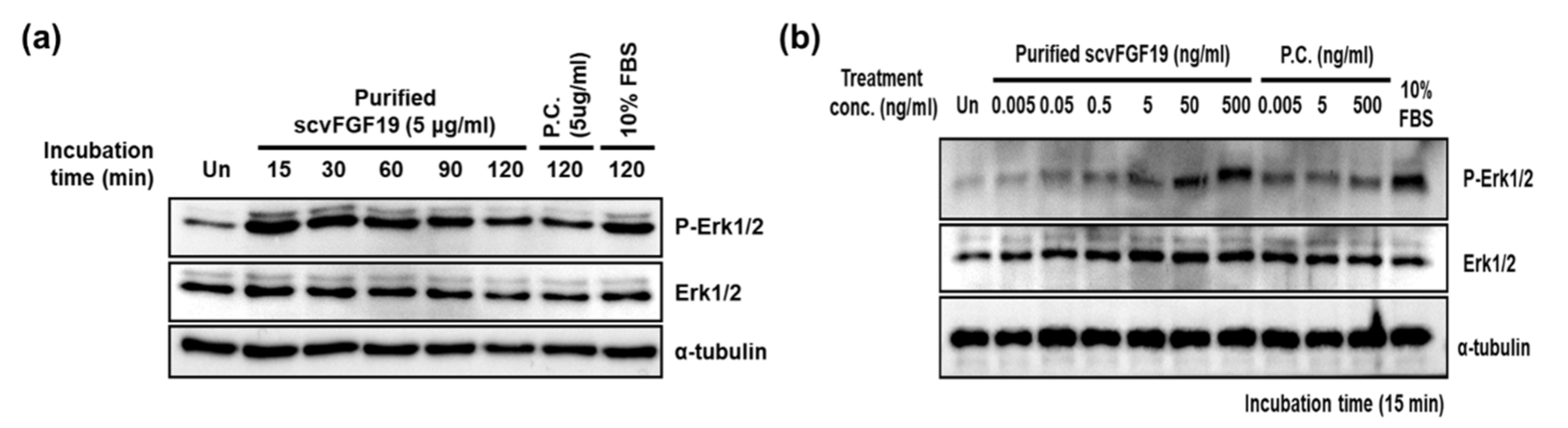
3.5. Biological Activity of Purified scvhFGF19
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yun, Y.R.; Won, J.E.; Jeon, E.; Lee, S.; Kang, W.; Jo, H.; Jang, J.H.; Shin, U.S.; Kim, H.W. Fibroblast growth factors: Biology, function, and application for tissue regeneration. J. Tissue Eng. 2010, 2010, 218142. [Google Scholar] [CrossRef] [PubMed]
- Ornitz, D.M.; Itoh, N. Fibroblast growth factors. Genome Biol. 2001, 2, Reviews3005. [Google Scholar] [CrossRef] [PubMed]
- Harmer, N.J.; Pellegrini, L.; Chirgadze, D.; Fernandez-Recio, J.; Blundell, T.L. The crystal structure of fibroblast growth factor (FGF) 19 reveals novel features of the FGF family and offers a structural basis for its unusual receptor affinity. Biochemistry 2004, 43, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Ornitz, D.M. Fibroblast growth factors: From molecular evolution to roles in development, metabolism and disease. J. Biochem. 2011, 149, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Walters, J.R.F.; Appleby, R.N. A variant of FGF19 for treatment of disorders of cholestasis and bile acid metabolism. Ann. Transl. Med. 2015, 3, S7. [Google Scholar] [PubMed]
- Zhou, M.; Learned, R.M.; Rossi, S.J.; DePaoli, A.M.; Tian, H.; Ling, L. Engineered FGF19 eliminates bile acid toxicity and lipotoxicity leading to resolution of steatohepatitis and fibrosis in mice. Hepatol. Commun. 2017, 1, 1024–1042. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, T.; Choi, M.; Moschetta, A.; Peng, L.; Cummins, C.L.; McDonald, J.G.; Luo, G.; Jones, S.A.; Goodwin, B.; Richardson, J.A.; et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005, 2, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.; Guo, G.L. Soluble expression of disulfide bond containing proteins FGF15 and FGF19 in the cytoplasm of Escherichia coli. PLoS ONE 2014, 9, e85890. [Google Scholar] [CrossRef] [PubMed]
- Cheong, D.E.; Ko, K.C.; Han, Y.; Jeon, H.G.; Sung, B.H.; Kim, G.J.; Choi, J.H.; Song, J.J. Enhancing functional expression of heterologous proteins through random substitution of genetic codes in the 5’ coding region. Biotechnol. Bioeng. 2015, 112, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Cheong, D.E.; Lee, J.; Choi, H.J.; Yoo, S.K.; Lee, D.H.; Kim, G.J. Soluble overexpression of a flagellin derivative from Salmonella enterica using synonymous codon substitutions of 5’-coding region in Escherichia coli. Biotechnol. Lett. 2019, 41, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.G.; Cheong, D.E.; Han, Y.; Song, J.J.; Choi, J.H. Itaconic acid production from glycerol using Escherichia coli harboring a random synonymous codon-substituted 5’-coding region variant of the cadA gene. Biotechnol. Bioeng. 2016, 113, 1504–1510. [Google Scholar] [CrossRef]
- Uemura, E.; Niwa, T.; Minami, S.; Takemoto, K.; Fukuchi, S.; Machida, K.; Imataka, H.; Ueda, T.; Ota, M.; Taguchi, H. Large-scale aggregation analysis of eukaryotic proteins reveals an involvement of intrinsically disordered regions in protein folding. Sci. Rep. 2018, 8, 678. [Google Scholar] [CrossRef]
- Rietsch, A.; Bessette, P.; Georgiou, G.; Beckwith, J. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J. Bacteriol. 1997, 179, 6602–6608. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Song, J.L.; Zhang, S.; Wang, Y.; Cui, D.F.; Wang, C.C. Chaperone activity of DsbC. J. Biol. Chem. 1999, 274, 19601–19605. [Google Scholar] [CrossRef] [PubMed]
- Raja, A.; Park, I.; Haq, F.; Ahn, S.M. FGF19-FGFR4 signaling in hepatocellular carcinoma. Cells. 2019, 8, 536. [Google Scholar] [CrossRef]
- Jahn, D.; Rau, M.; Hermanns, H.M.; Geier, A. Mechanisms of enterohepatic fibroblast growth factor 15/19 signaling in health and disease. Cytokine Growth Factor Rev. 2015, 26, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Owen, B.M.; Mangelsdorf, D.J.; Kliewer, S.A. Tissue-specific actions of the metabolic hormones FGF15/19 and FGF21. Trends Endocrinol. Metab. 2015, 26, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, E.; Fu, L.; John, L.; Hultgren, B.; Huang, X.; Renz, M.; Stephan, J.P.; Tsai, S.P.; Powell-Braxton, L.; French, D.; et al. Transgenic mice expressing human fibroblast growth factor-19 display increased metabolic rate and decreased adiposity. Endocrinology 2002, 143, 1741–1747. [Google Scholar] [CrossRef] [PubMed]
- Raghunathan, G.; Munussami, G.; Moon, H.; Paik, H.J.; An, S.S.A.; Kim, Y.S.; Kang, S.; Lee, S.G. A variant of green fluorescent protein exclusively deposited to active intracellular inclusion bodies. Microb. Cell Fact. 2014, 13, 989. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, S.; Li, Y.; Jiang, C.; Tian, H. Fibroblast growth factor 9 subfamily and the heart. Appl. Microbiol. Biotechnol. 2018, 102, 605–613. [Google Scholar] [CrossRef] [PubMed]

| Primers | Sequence (5′→3′) | Cloning 1 |
|---|---|---|
| pSCT5_FGF19-mC-fw | ATAACTAGTATGCGCCCCCTCGCCTTC | SpeⅠ |
| pSCT5_scvFGF19-mC-fw | ATAACTAGTATGCGNCCNCTNGCNTTYTCNGAYGCNGGNCCNCACGTGCACT | SpeⅠ |
| pSCT5_FGF19-mC-rv | ATAGGATCCCTTCTCAAAGCTGGGACTCCTCAC | BamHⅠ |
| pSCT5_scvFGF19-rv | ATAAAGCTTTTACTTCTCAAAGCTGGGACTCCTC | HindⅢ |
| pSCT5_ΔssDsbC-fw | ATAACTAGTATGGATGACGCGGCAATTC | SpeⅠ |
| pSCT5_ΔssDsbC-rv | ATAGGATCCTTATTTACCGCTGGTCATTTTTTGGTG | BamHⅠ |
| pSCT5_FGF19/ΔssDsbC-fw | ATAGGATCCTAAATGGATGACGCGGCAATTCAAC | BamHⅠ |
| pSCT5_FGF19/ΔssDsbC-rv | ATAAAGCTTTTATTTACCGCTGGTCATTTTTTGGTGTTC | HindⅢ |
| pSCT5_scvFGF19-fw | ATACAATTTCACACAGAATTCATTAAAGAGGAGAAAGGATCCATGCG | BamHⅠ |
| pSCT5_scvFGF19-rv | ATAAAGCTTTTACTTCTCAAAGCTGGGACTCCTC | HindⅢ |
| pACYCDuet1_ΔssDsbC-fw | AGGAGATATACCATGGATGACGCGGCAATTCAACAAACG | NcoⅠ |
| pACYCDuet1_ΔssDsbC-rv | ATAGGATCCTTATTTACCGCTGGTCATTTTTTGGTGTTC | BamHⅠ |
| pACYCDuet1_scvFGF19-fw | CAATTTCACACAGAATTCATTAAAGAGGAGAAACATATGCG | NdeⅠ |
| pACYCDuet1_scvFGF19-rv | ATACTCGAGTTACTTCTCAAAGCTGGGACTCCTCACG | XhoⅠ |
| pQE80L V-fw | AGTTAATTTCTCCTCTTTAATGAATTCTGTGTG | Infusion |
| pQE80L V-rv | CCGCCCTCTAGATTACGTGC | Infusion |
| pQHDuet_InfuΔssDsbC-fw | GAGGAGAAATTAACTATGGATGACGCGGCAATTC | Infusion |
| pQHDuet_InfuFGF19-rv | TAATCTAGAGGGCGGTTTTTAAGGCAGTTATTGGTGCCC | Infusion |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, H.-J.; Cheong, D.-E.; Yoo, S.-K.; Park, J.; Lee, D.-H.; Kim, G.-J. Soluble Expression of hFGF19 without Fusion Protein through Synonymous Codon Substitutions and DsbC Co-Expression in E. coli. Microorganisms 2020, 8, 1942. https://doi.org/10.3390/microorganisms8121942
Choi H-J, Cheong D-E, Yoo S-K, Park J, Lee D-H, Kim G-J. Soluble Expression of hFGF19 without Fusion Protein through Synonymous Codon Substitutions and DsbC Co-Expression in E. coli. Microorganisms. 2020; 8(12):1942. https://doi.org/10.3390/microorganisms8121942
Chicago/Turabian StyleChoi, Hye-Ji, Dae-Eun Cheong, Su-Kyoung Yoo, Jaehong Park, Dong-Hyun Lee, and Geun-Joong Kim. 2020. "Soluble Expression of hFGF19 without Fusion Protein through Synonymous Codon Substitutions and DsbC Co-Expression in E. coli" Microorganisms 8, no. 12: 1942. https://doi.org/10.3390/microorganisms8121942
APA StyleChoi, H.-J., Cheong, D.-E., Yoo, S.-K., Park, J., Lee, D.-H., & Kim, G.-J. (2020). Soluble Expression of hFGF19 without Fusion Protein through Synonymous Codon Substitutions and DsbC Co-Expression in E. coli. Microorganisms, 8(12), 1942. https://doi.org/10.3390/microorganisms8121942




