Burkholderia gladioli CGB10: A Novel Strain Biocontrolling the Sugarcane Smut Disease
Abstract
1. Introduction
2. Methods and Materials
2.1. Isolation of Endophytes from Sugarcane Leaves
2.2. Genome Sequencing
2.3. Genomic Sequence Analysis
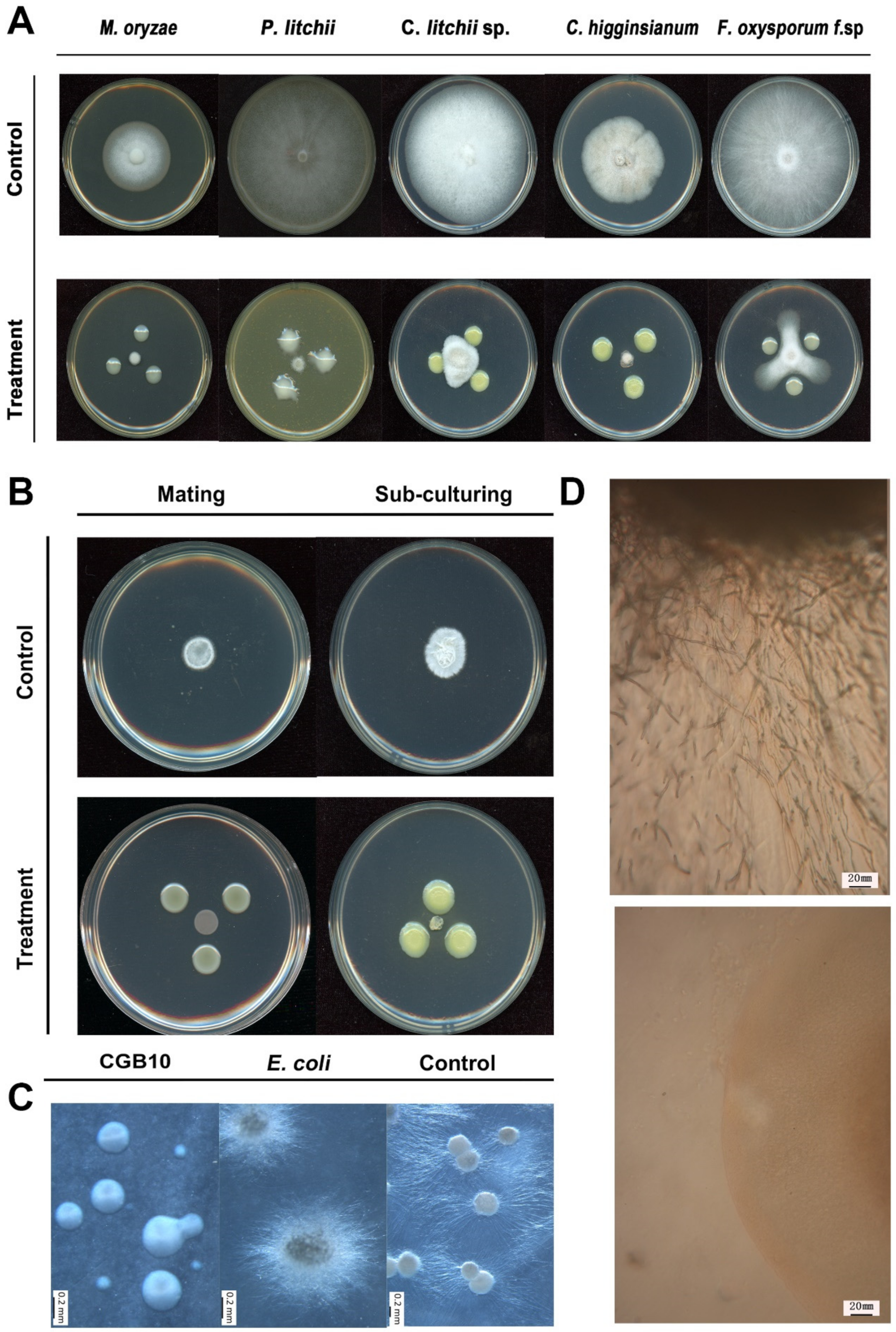
2.4. Fungal Strains and Culture Conditions
2.5. Assays for Antifungal Activity
2.6. S. Scitamineum Teliospore Germination Assay
2.7. Detection of Toxoflavin in B. gladioli CGB10
2.8. Extracellular Hydrolytic Enzymatic Activity Test
2.9. Assessment of Bacterial Pathogenicity to Plants
2.10. Field Experiment for CGB10 Biocontrol of Sugarcane Smut
3. Results
3.1. Genome Sequencing and Phylogenetic Analysis of CGB10
3.2. Antifungal Activity of CGB10 under In Vitro Culture Condition
3.3. Antifungal Activity of CGB10 under Field Conditions
3.4. Toxoflavin is the Major Filamentation-Suppressing Compound Produced by CGB10
3.5. Identification of Quorum-Sensing Genes in CGB10
3.6. Analysis of CGB10 Pathogenicity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zuo, W.; Okmen, B.; Depotter, J.R.L.; Ebert, M.K.; Redkar, A.; Misas Villamil, J.; Doehlemann, G. Molecular Interactions Between Smut Fungi and Their Host Plants. Annu. Rev. Phytopathol. 2019, 57, 411–430. [Google Scholar] [CrossRef] [PubMed]
- Sundar, A.R.; Barnabas, E.L.; Malathi, P.; Viswanathan, R. A Mini-Review on Smut Disease of Sugarcane Caused by Sporisorium scitamineum. Botany 2014, 2014, 226. [Google Scholar]
- Strobel, G.; Daisy, B.; Castillo, U.; Harper, J. Natural products from endophytic microorganisms. J. Nat. Prod. 2004, 67, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Kloeppe, J.W.; Rodríguez-Kábana, R.; Zehnder, A.W.; Murphy, J.F.; Sikora, E.; Fernández, C. Plant root-bacterial interactions in biological control of soilborne diseases and potential extension to systemic and foliar diseases. Australas. Plant Pathol. 1999, 28, 21–26. [Google Scholar] [CrossRef]
- Posada, F.; Vega, F.E. Establishment of the fungal entomopathogen Beauveria bassiana (Ascomycota: Hypocreales) as an endophyte in cocoa seedlings (Theobroma cacao). Mycologia 2005, 97, 1195–1200. [Google Scholar] [CrossRef]
- Lodewyckx, C.; Vangronsveld, J.; Porteous, F.; Moore, E.R.B.; Taghavi, S.; Mezgeay, M.; van der Lelie, D. Endophytic bacteria and their potential applications. Crit. Rev. Plant Sci. 2002, 21, 583–606. [Google Scholar] [CrossRef]
- Mannisto, M.K.; Tiirola, M.A.; Puhakka, J.A. Degradation of 2,3,4,6-tetrachlorophenol at low temperature and low dioxygen concentrations by phylogenetically different groundwater and bioreactor bacteria. Biodegradation 2001, 12, 291–301. [Google Scholar] [CrossRef]
- Shehata, H.R.; Lyons, E.M.; Jordan, K.S.; Raizada, M.N. Bacterial endophytes from wild and ancient maize are able to suppress the fungal pathogen Sclerotinia homoeocarpa. J. Appl. Microbiol. 2016, 120, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.M.; Miller, R.V.; Garton-Kenny, D.; Redgrave, B.; Sears, J.; Condron, M.M.; Teplow, D.B.; Strobel, G.A. Ecomycins, unique antimycotics from Pseudomonas viridiflava. J. Appl. Microbiol. 1998, 84, 937–944. [Google Scholar] [CrossRef]
- Cui, L.; Yang, C.; Wei, L.; Li, T.; Chen, X. Isolation and identification of an endophytic bacteria Bacillus velezensis 8-4 exhibiting biocontrol activity against potato scab. Biol. Control 2020, 141. [Google Scholar] [CrossRef]
- Mendes, R.; Pizzirani-Kleiner, A.A.; Araujo, W.L.; Raaijmakers, J.M. Diversity of cultivated endophytic bacteria from sugarcane: Genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 2007, 73, 7259–7267. [Google Scholar] [CrossRef] [PubMed]
- Lufeng, L.; Haichun, C.; Pengfei, H.; Yining, D.; Yixin, W.; Lilian, H.; Fusheng, L.; Yueqiu, H. Isolation, Identification and Multiple Function Analyses of Sugarcane Endophytes. Chin. J. Trop. Crops 2019, 40, 1144–1152. [Google Scholar]
- Vial, L.; Groleau, M.C.; Dekimpe, V.; Deziel, E. Burkholderia diversity and versatility: An inventory of the extracellular products. J. Microbiol. Biotechnol. 2007, 17, 1407–1429. [Google Scholar] [PubMed]
- Schmidt, S.; Blom, J.F.; Pernthaler, J.; Berg, G.; Baldwin, A.; Mahenthiralingam, E.; Eberl, L. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ. Microbiol. 2009, 11, 1422–1437. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Zhu, G.; Lin, S.; Xian, X.; Chang, C.; Xi, P.; Shen, W.; Huang, W.; Cai, E.; Jiang, Z.; et al. The mating-type locus b of the sugarcane smut Sporisorium scitamineum is essential for mating, filamentous growth and pathogenicity. Fungal Genet. Biol. 2016, 86, 1–8. [Google Scholar] [CrossRef]
- Yan, M.; Cai, E.; Zhou, J.; Chang, C.; Xi, P.; Shen, W.; Li, L.; Jiang, Z.; Deng, Y.Z.; Zhang, L.H. A Dual-Color Imaging System for Sugarcane Smut Fungus Sporisorium scitamineum. Plant Dis. 2016, 100, 2357–2362. [Google Scholar] [CrossRef]
- Deng, S.; Sun, W.; Dong, L.; Cui, G.; Deng, Y.Z. MoGT2 Is Essential for Morphogenesis and Pathogenicity of Magnaporthe oryzae. Msphere 2019, 4, e00309-19. [Google Scholar] [CrossRef]
- Jiang, L.; Situ, J.; Deng, Y.Z.; Wan, L.; Xu, D.; Chen, Y.; Xi, P.; Jiang, Z. PlMAPK10, a Mitogen-Activated Protein Kinase (MAPK) in Peronophythora litchii, Is Required for Mycelial Growth, Sporulation, Laccase Activity, and Plant Infection. Front. Microbiol. 2018, 9, 426. [Google Scholar] [CrossRef]
- Lv, M.; Hu, M.; Li, P.; Jiang, Z.; Zhang, L.H.; Zhou, J. A two-component regulatory system VfmIH modulates multiple virulence traits in Dickeya zeae. Mol. Microbiol. 2019, 111, 1493–1509. [Google Scholar] [CrossRef]
- Chatterjee, A.; Cui, Y.; Liu, Y.; Dumenyo, C.K.; Chatterjee, A.K. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Appl. Environ. Microbiol. 1995, 61, 1959–1967. [Google Scholar] [CrossRef]
- Scott-Craig, J.S.; Panaccione, D.G.; Cervone, F.; Walton, J.D. Endopolygalacturonase is not required for pathogenicity of Cochliobolus carbonum on maize. Plant Cell 1990, 2, 1191–1200. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chatterjee, A.K.; Thurn, K.K.; Tyrell, D.J. Isolation and characterization of Tn5 insertion mutants of Erwinia chrysanthemi that are deficient in polygalacturonate catabolic enzymes oligogalacturonate lyase and 3-deoxy-D-glycero-2,5-hexodiulosonate dehydrogenase. J. Bacteriol. 1985, 162, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Barras, F.; Thurn, K.K.; Chatterjee, A.K. Resolution of four pectate lyase structural genes of Erwinia chrysanthemi (EC16) and characterization of the enzymes produced in Escherichia coli. Mol. Gen. Genet. 1987, 209, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, J.; Kim, S.; Park, I.; Seo, Y.S. Differential regulation of toxoflavin production and its role in the enhanced virulence of Burkholderia gladioli. Mol. Plant Pathol. 2016, 17, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Kim, J.; Kim, S.; Kang, Y.; Nagamatsu, T.; Hwang, I. Toxoflavin Produced by Burkholderia glumae Causing Rice Grain Rot Is Responsible for Inducing Bacterial Wilt in Many Field Crops. Plant Dis. 2003, 87, 890–895. [Google Scholar] [CrossRef]
- Wan-Kuan, S.; Zhan-Duan, Y.; Fu-Ye, L. Identification and evaluation of some sugarcane varieties or clones for smut resistance. J. Huazhong Agric. Univ. 2014, 33, 40–44. [Google Scholar]
- Eberl, L.; Vandamme, P. Members of the genus Burkholderia: Good and bad guys. F1000Res 2016, 5. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Johnson, S.L.; Bishop-Lilly, K.A.; Ladner, J.T.; Daligault, H.E.; Davenport, K.W.; Jaissle, J.; Frey, K.G.; Koroleva, G.I.; Bruce, D.C.; Coyne, S.R.; et al. Complete genome sequences for 59 burkholderia isolates, both pathogenic and near neighbor. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Jha, G.; Tyagi, I.; Kumar, R.; Ghosh, S. Draft Genome Sequence of Broad-Spectrum Antifungal Bacterium Burkholderia gladioli Strain NGJ1, Isolated from Healthy Rice Seeds. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Shehata, H.R.; Ettinger, C.L.; Eisen, J.A.; Raizada, M.N. Genes Required for the Anti-fungal Activity of a Bacterial Endophyte Isolated from a Corn Landrace Grown Continuously by Subsistence Farmers Since 1000 BC. Front. Microbiol. 2016, 7, 1548. [Google Scholar] [CrossRef]
- Webster, G.; Jones, C.; Mullins, A.J.; Mahenthiralingam, E. A rapid screening method for the detection of specialised metabolites from bacteria: Induction and suppression of metabolites from Burkholderia species. J. Microbiol. Methods 2020, 178, 106057. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Jenner, M.; Masschelein, J.; Jones, C.; Bull, M.J.; Harris, S.R.; Hartkoorn, R.C.; Vocat, A.; Romero-Canelon, I.; Coupland, P.; et al. Discovery and Biosynthesis of Gladiolin: A Burkholderia gladioli Antibiotic with Promising Activity against Mycobacterium tuberculosis. J. Am. Chem. Soc. 2017, 139, 7974–7981. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Wang, R.; Wang, Q.; Lu, L. Toxoflavin Produced by Burkholderia gladioli from Lycoris aurea Is a New Broad-Spectrum Fungicide. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef] [PubMed]
- Jenner, M.; Jian, X.; Dashti, Y.; Masschelein, J.; Hobson, C.; Roberts, D.M.; Jones, C.; Harris, S.; Parkhill, J.; Raja, H.A.; et al. An unusual Burkholderia gladioli double chain-initiating nonribosomal peptide synthetase assembles ‘fungal’ icosalide antibiotics. Chem. Sci. 2019, 10, 5489–5494. [Google Scholar] [CrossRef] [PubMed]
- Philmus, B.; Shaffer, B.T.; Kidarsa, T.A.; Yan, Q.; Raaijmakers, J.M.; Begley, T.P.; Loper, J.E. Investigations into the Biosynthesis, Regulation, and Self-Resistance of Toxoflavin in Pseudomonas protegens Pf-5. Chembiochem 2015, 16, 1782–1790. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, J.G.; Kang, Y.; Jang, J.Y.; Jog, G.J.; Lim, J.Y.; Kim, S.; Suga, H.; Nagamatsu, T.; Hwang, I. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol. Microbiol. 2004, 54, 921–934. [Google Scholar] [CrossRef]
- Hussain, A.; Shahbaz, M.; Tariq, M.; Ibrahim, M.; Hong, X.; Naeem, F.; Khalid, Z.; Raza, H.M.Z.; Bo, Z.; Bin, L. Genome re-seqeunce and analysis of Burkholderia glumae strain AU6208 and evidence of toxoflavin: A potential bacterial toxin. Comput. Biol. Chem. 2020, 86, 107245. [Google Scholar] [CrossRef]
- Chen, R.; Barphagha, I.K.; Karki, H.S.; Ham, J.H. Dissection of quorum-sensing genes in Burkholderia glumae reveals non-canonical regulation and the new regulatory gene tofM for toxoflavin production. PLoS ONE 2012, 7, e52150. [Google Scholar] [CrossRef]
- Naughton, L.M.; An, S.Q.; Hwang, I.; Chou, S.H.; He, Y.Q.; Tang, J.L.; Ryan, R.P.; Dow, J.M. Functional and genomic insights into the pathogenesis of Burkholderia species to rice. Environ. Microbiol. 2016, 18, 780–790. [Google Scholar] [CrossRef]
- Zhou, F.; Ning, H.; Chen, F.; Wu, W.; Chen, A.; Zhang, J. Burkholderia gladioli infection isolated from the blood cultures of newborns in the neonatal intensive care unit. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1533–1537. [Google Scholar] [CrossRef] [PubMed]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef]
- Ettinger, C.L.; Shehata, H.R.; Johnston-Monje, D.; Raizada, M.N.; Eisen, J.A. Draft Genome Sequence of Burkholderia gladioli Strain UCD-UG_CHAPALOTE (Phylum Proteobacteria). Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Tjalsma, H.; Antelmann, H.; Jongbloed, J.D.; Braun, P.G.; Darmon, E.; Dorenbos, R.; Dubois, J.Y.; Westers, H.; Zanen, G.; Quax, W.J.; et al. Proteomics of protein secretion by Bacillus subtilis: Separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 2004, 68, 207–233. [Google Scholar] [CrossRef] [PubMed]
- Mehat, J.W.; Park, S.F.; van Vliet, A.H.M.; La Ragione, R.M. CapC, a Novel Autotransporter and Virulence Factor of Campylobacter jejuni. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Coulthurst, S. The Type VI secretion system: A versatile bacterial weapon. Microbiology 2019, 165, 503–515. [Google Scholar] [CrossRef]
- Cianciotto, N.P.; White, R.C. Expanding Role of Type II Secretion in Bacterial Pathogenesis and Beyond. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef]
- Bai, F.; Li, Z.; Umezawa, A.; Terada, N.; Jin, S. Bacterial type III secretion system as a protein delivery tool for a broad range of biomedical applications. Biotechnol. Adv. 2018, 36, 482–493. [Google Scholar] [CrossRef]
- Feng, L.; Schaefer, A.L.; Hu, M.; Chen, R.; Greenberg, E.P.; Zhou, J. Virulence Factor Identification in the Banana Pathogen Dickeya zeae MS2. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef]
- Jacobs, J.L.; Fasi, A.C.; Ramette, A.; Smith, J.J.; Hammerschmidt, R.; Sundin, G.W. Identification and onion pathogenicity of Burkholderia cepacia complex isolates from the onion rhizosphere and onion field soil. Appl. Environ. Microbiol. 2008, 74, 3121–3129. [Google Scholar] [CrossRef]
- Titball, R.W.; Burtnick, M.N.; Bancroft, G.J.; Brett, P. Burkholderia pseudomallei and Burkholderia mallei vaccines: Are we close to clinical trials? Vaccine 2017, 35, 5981–5989. [Google Scholar] [CrossRef] [PubMed]
- Hemarajata, P.; Baghdadi, J.D.; Hoffman, R.; Humphries, R.M. Burkholderia pseudomallei: Challenges for the Clinical Microbiology Laboratory. J. Clin. Microbiol. 2016, 54, 2866–2873. [Google Scholar] [CrossRef] [PubMed]
- Burkholder, W.H. Sour skin, a bacterial rot of onion bulbs. Phytopathology 1950, 64, 468–475. [Google Scholar]
- Coenye, T.; Henry, D.; Speert, D.P.; Vandamme, P. Burkholderia phenoliruptrix sp. nov., to accommodate the 2,4,5-trichlorophenoxyacetic acid and halophenol-degrading strain AC1100. Syst. Appl. Microbiol. 2004, 27, 623–627. [Google Scholar] [CrossRef]
- Martinez, P.; Agullo, L.; Hernandez, M.; Seeger, M. Chlorobenzoate inhibits growth and induces stress proteins in the PCB-degrading bacterium Burkholderia xenovorans LB400. Arch. Microbiol. 2007, 188, 289–297. [Google Scholar] [CrossRef]
- Kim, S.; Park, J.; Kim, J.H.; Lee, J.; Bang, B.; Hwang, I.; Seo, Y.S. RNAseq-based Transcriptome Analysis of Burkholderia glumae Quorum Sensing. Plant. Pathol. J. 2013, 29, 249–259. [Google Scholar] [CrossRef]
- Elshafie, H.S.; Devescovi, G.; Venturi, V.; Camele, I.; Bufo, S.A. Study of the Regulatory Role of N-Acyl Homoserine Lactones Mediated Quorum Sensing in the Biological Activity of Burkholderia gladioli pv. agaricicola Causing Soft Rot of Agaricus spp. Front. Microbiol. 2019, 10, 2695. [Google Scholar] [CrossRef]
- Seo, Y.S.; Lim, J.; Choi, B.S.; Kim, H.; Goo, E.; Lee, B.; Lim, J.S.; Choi, I.Y.; Moon, J.S.; Kim, J.; et al. Complete genome sequence of Burkholderia gladioli BSR3. J. Bacteriol. 2011, 193, 3149. [Google Scholar] [CrossRef]
- Imataki, O.; Kita, N.; Nakayama-Imaohji, H.; Kida, J.I.; Kuwahara, T.; Uemura, M. Bronchiolitis and bacteraemia caused by Burkholderia gladioli in a non-lung transplantation patient. New Microbes New Infect. 2014, 2, 175–176. [Google Scholar] [CrossRef][Green Version]




| Strain * | Biosample | Isolated | tofI-M-R_Chrosome(BSR3) | tofI-M-R_plasmid (BSR3) | Toxoflavin Gene Cluster (BSR3) | toxJ (BSR3) | Producing Toxoflavin | Ref. |
|---|---|---|---|---|---|---|---|---|
| B. gladioli | ||||||||
| KACC11889 | SAMN07253176 | Gladiolus | 0 | 0 | CP022005 (98.359%) | CP022006 (98.758%) | no | [24] |
| BSR3 | SAMN02603164 | Rice | CP002600.1 (100.000%) | CP002601.1 (100.000%) | CP002599.1 (100%) | CP002600.1 (100.000%) | yes | [24] |
| CGB10 | SAMN15158960 | Healthy sugarcane | tig00002 (98.999%) | 0 | tig00001 (99.065%) | tig00002 (98.646%) | yes | this study |
| BCC 0238 | SAMEA6503627 | Sputum of a child with cystic fibrosis | CADEVO010000001.1 (98.904%) | 0 | CADEVO010000027.1 (99.065%) | CADEVO010000022.1 (98.871%) | yes | [32] |
| ATCC 10248 | SAMN03010439 | Gladiolus sp. bulb | 0 | 0 | CP009323.1 (98.359%) | CP009322.1 (98.758%) | no | [29] |
| NGJ1 | SAMN03764558 | Healthy rice seed | LEKY01000068.1 (98.570%) | 0 | LEKY01000021.1 (99.000%) | LEKY01000068.1 (99.438%) | no report | [30] |
| UCD-UG_CHAPALOTE | SAMN03019910 | Seeds of an ancient Mexican landrace of corn | JRGO01000057.1 (98.713%) | 0 | JRGO01000016.1 (99.355%) | JRGO01000062.1 (99.101%) | no report | [31] |
| B. glumae | ||||||||
| BGR1 | SAMN02603166 | Diseased rice panicle | CP001504.2 (88.353%) | 0 | CP001504.2 (96.703%) | CP001504.2 (82.163%) | yes | [24] |
| Sequence_ID | Cluster_ID | Cluster_type | Start | End | Length | Most Similar Known Cluster | Similarity |
|---|---|---|---|---|---|---|---|
| tig00001 | 1 | Terpene | 145,180 | 166,019 | 20,840 | Lasalocid (t1pks) | 7% |
| tig00001 | 2 | NRPS | 565,719 | 705,139 | 139,421 | Xenoamicins | 25% |
| tig00001 | 3 | NRPS, Bacteriocin | 1,172,186 | 1,235,801 | 63,616 | - | - |
| tig00001 | 4 | NRPS | 1,263,608 | 1,318,740 | 55,133 | Sulfazecin | 100% |
| tig00001 | 5 | T1PKS | 2,056,963 | 2,102,119 | 45,157 | Capsular polysaccharide (Saccharide) | 25% |
| Gene Name | Strain | Gene ID | Location |
|---|---|---|---|
| ToxA | BSR3 | AEA59150.1 | tig2 |
| CGB10 | NPGAP_10495 | tig00001 | |
| BGR1 | AAV52806.1 | chromesome1 | |
| ToxB | BSR3 | AEA59151.1 | tig2 |
| CGB10 | NPGAP_10490 | tig00001 | |
| BGR1 | AAV52807.1 | chromesome1 | |
| ToxC | BSR3 | AEA59152.1 | tig2 |
| CGB10 | NPGAP_10485 | tig00001 | |
| BGR1 | AAV52808.1 | chromesome1 | |
| ToxD | BSR3 | AEA59153.1 | tig2 |
| CGB10 | NPGAP_10480 | tig00001 | |
| BGR1 | AAV52809.1 | chromesome1 | |
| ToxE | BSR3 | AEA59154.1 | tig2 |
| CGB10 | NPGAP_10470&NPGAP_10475 | tig00001 | |
| BGR1 | AAV52810.1 | chromesome1 | |
| ToxF | BSR3 | AEA59148.1 | tig2 |
| CGB10 | NPGAP_10505 | tig00001 | |
| BGR1 | AAV52811.1 | chromesome1 | |
| ToxG | BSR3 | AEA59147.1 | tig2 |
| CGB10 | NPGAP_10510 | tig00001 | |
| BGR1 | AAV52812.1 | chromesome1 | |
| ToxH | BSR3 | AEA59146.1 | tig2 |
| CGB10 | NPGAP_10515 | tig00001 | |
| BGR1 | AAV52813.1 | chromesome1 | |
| ToxI | BSR3 | AEA59145.1 | tig2 |
| CGB10 | NPGAP_10520 | tig00001 | |
| BGR1 | AAV52814.1 | chromesome1 | |
| ToxJ | BSR3 | AEA63365.1 | chromesome2 |
| CGB10 | NPGAP_26430&NPGAP_34680 | tig00002 | |
| BGR1 | AAV52815.1 | chromesome1 | |
| ToxR | BSR3 | AEA59149.1 | tig2 |
| CGB10 | NPGAP_10500 | tig00001 | |
| BGR1 | AAV52816.1 | chromesome1 | |
| Tof I | BSR3 | bgla_2g11050 | Chromesome2 |
| CGB10 | NPGAP_35850 | tig2 | |
| BGR1 | ACR31808.1 | Chromesome2 | |
| Tof M | BSR3 | bgla_2g11060 | Chromesome2 |
| CGB10 | NPGAP_35855 | tig2 | |
| BGR1 | ACR31807.1 | Chromesome2 | |
| Tof R | BSR3 | bgla_2g11070 | Chromesome2 |
| CGB10 | NPGAP_35860 | tig2 | |
| BGR1 | ACR31806.1 | Chromesome2 | |
| Tof I-M-R (set 2) | BSR3 | bgla_1p1740 | plasmid1 |
| bgla_1p1750 | |||
| bgla_1p1760 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, G.; Yin, K.; Lin, N.; Liang, M.; Huang, C.; Chang, C.; Xi, P.; Deng, Y.Z. Burkholderia gladioli CGB10: A Novel Strain Biocontrolling the Sugarcane Smut Disease. Microorganisms 2020, 8, 1943. https://doi.org/10.3390/microorganisms8121943
Cui G, Yin K, Lin N, Liang M, Huang C, Chang C, Xi P, Deng YZ. Burkholderia gladioli CGB10: A Novel Strain Biocontrolling the Sugarcane Smut Disease. Microorganisms. 2020; 8(12):1943. https://doi.org/10.3390/microorganisms8121943
Chicago/Turabian StyleCui, Guobing, Kai Yin, Nuoqiao Lin, Meiling Liang, Chengwei Huang, Changqing Chang, Pinggen Xi, and Yi Zhen Deng. 2020. "Burkholderia gladioli CGB10: A Novel Strain Biocontrolling the Sugarcane Smut Disease" Microorganisms 8, no. 12: 1943. https://doi.org/10.3390/microorganisms8121943
APA StyleCui, G., Yin, K., Lin, N., Liang, M., Huang, C., Chang, C., Xi, P., & Deng, Y. Z. (2020). Burkholderia gladioli CGB10: A Novel Strain Biocontrolling the Sugarcane Smut Disease. Microorganisms, 8(12), 1943. https://doi.org/10.3390/microorganisms8121943






