Influence of Lab Adapted Natural Diet and Microbiota on Life History and Metabolic Phenotype of Drosophila melanogaster
Abstract
:1. Introduction
2. Materials and Methods
2.1. Food Preparation
2.2. Drosophila Stocks and Husbandry
2.3. Drosophila Embryos Sterilization
2.4. Larvae Rearing and Collection
2.5. Measuring Experimental Phenotypes
2.6. DNA Extraction and Sequencing
2.7. Statistical Analysis
3. Results
3.1. There Are Substantial Differences in the Life History and Metabolic Phenotypes for Larvae Raised on a Natural Peach Diet with a Naturally Occurring and/or Maternally Inherited Community of Bacteria Relative to a Standard Lab Diet
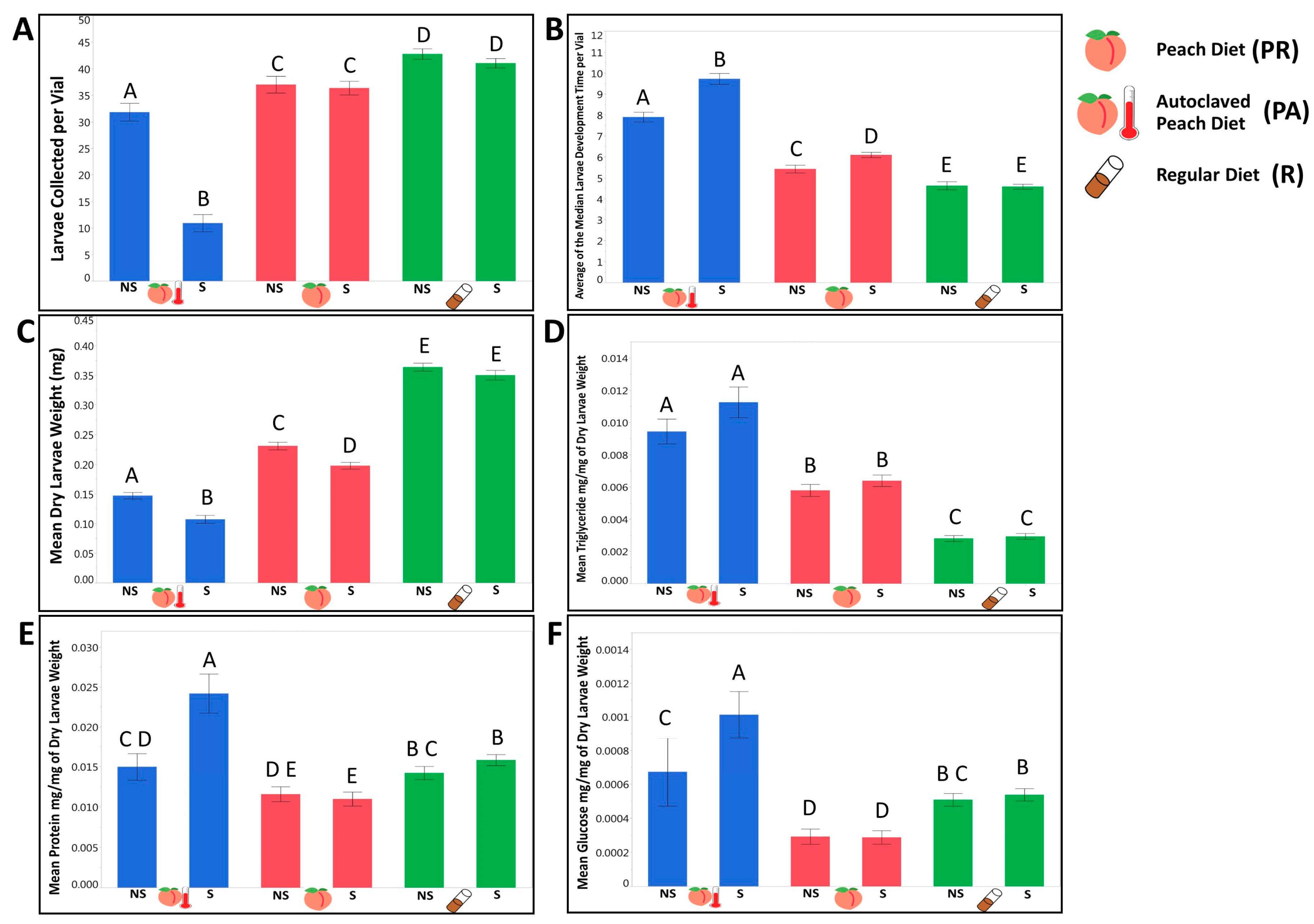
3.1.1. Larvae Raised on a Natural Diet Exhibited Different Life History Traits and Metabolic Phenotypes from Larvae Raised on a Standard Lab Diet
3.1.2. We Observed That the Presence of a Maternally Transmitted Bacteria Significantly Impacted Larvae Phenotypes, and That Impact Varied across Dietary Treatments
3.1.3. Evaluating the Contribution of Tested Independent Variables on Larvae Phenotypes, We Observed a Genetic Variation in Most of the Tested Life History Traits and Phenotypes that Interacted with the Dietary Conditions and the Availability of Maternally Transmitted Microbiota
3.2. The Symbiotic Bacterial Community Composition of the Larvae Raised on the Natural Diet Was Different from the Lab Food Raised Larvae and Was Influenced by Maternally Inherited Bacteria and the Host’s Genotype
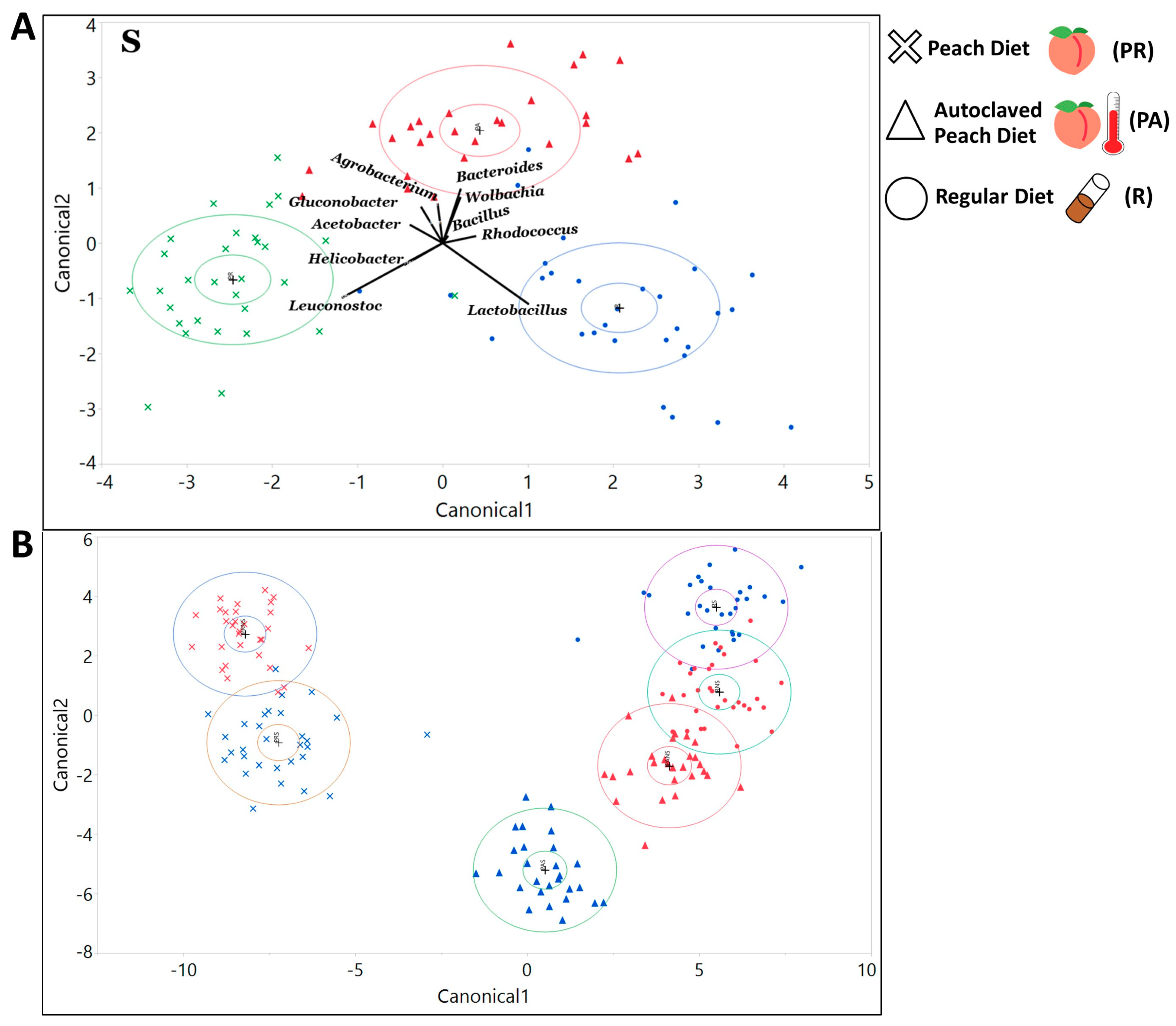
3.2.1. The Gut Bacterial Community Composition and Diversity Varied Substantially across Dietary and Treatment Conditions
3.2.2. The Maternally Transmitted Microbiota Influenced the Composition of the Larvae’s Symbiotic Bacterial Communities
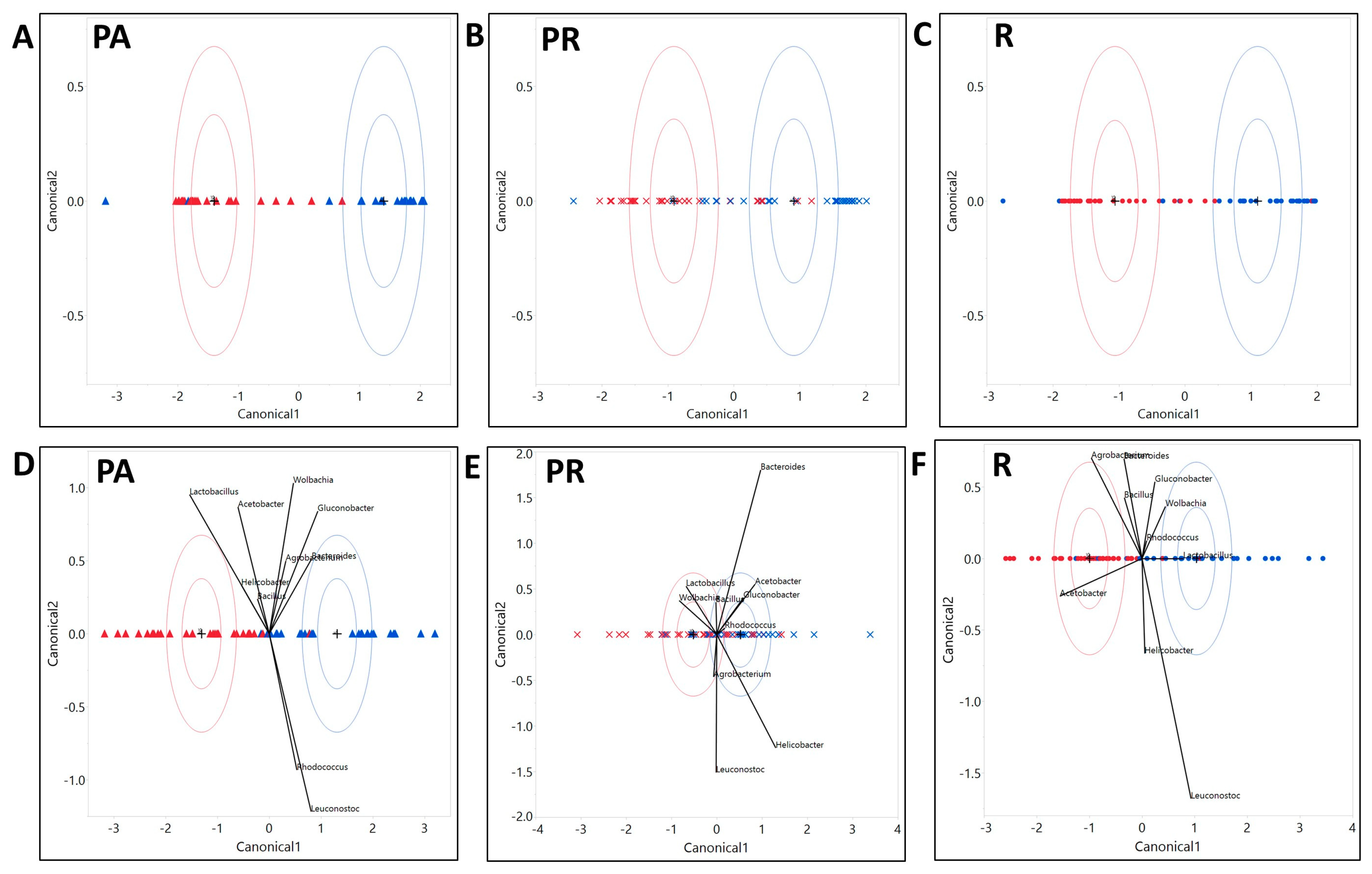
3.2.3. The Composition of the Microbial Community Exhibited Variation with Host Genotype, Which Further Exhibited a Significant Interactive Effect with Diet and Treatment
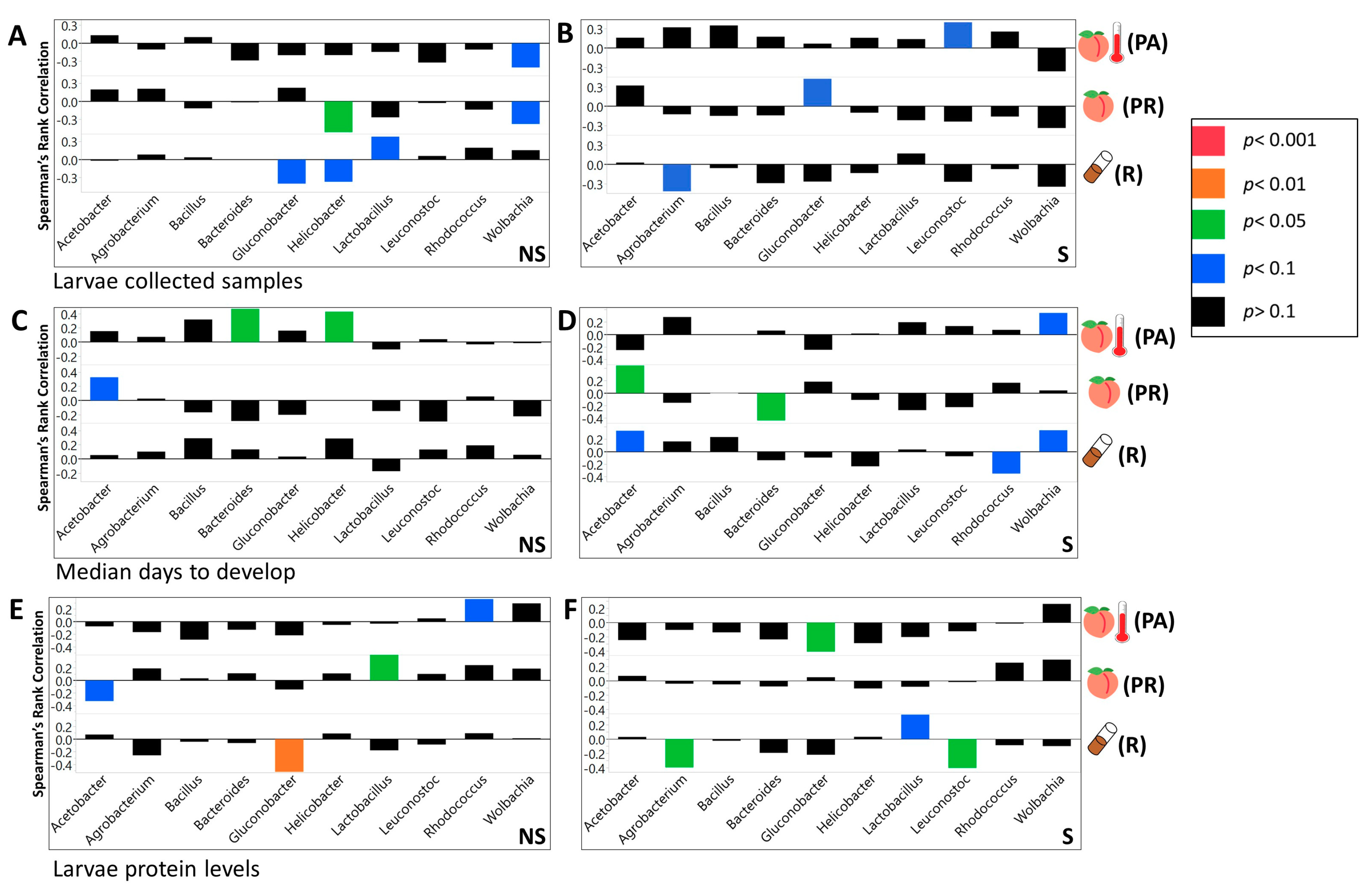
3.3. We Identified Microbial Taxa That Exhibited Correlations with Host Phenotype across Diets and Treatments, with Many that Had a Diet, Treatment, or Genotype Specific Relationship
4. Discussion
4.1. Frozen Peach Food Was Capable of Providing Nutritional Conditions Similar to the Natural Ones and Can Preserve Key Microbial Taxa Necessary for Survival and Development of Drosophila Larvae
4.2. Maternally Deposited Microbes Produced Positive Effects on Larvae that Were Raised on the Peach Diets
4.3. Genotype Was One of the Key Factors that Influenced Larvae Phenotypes
4.4. Bacteria of the Larvae Raised on PR Food Exhibit a Distinct Community Structure
4.5. Community Structure of Symbiotic Bacteria Were Correlated with Diet, Treatment, Host Genotype, and Their Specific Interactive Effects
4.6. The Correlations between Microbial Taxa as Well as the Correlation between the Whole Microbial Community and the Host May Vary with the Diet and Other Environmental and Genetic Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rosenberg, E.; Sharon, G.; Atad, I.; Zilber-Rosenberg, I. The evolution of animals and plants via symbiosis with microorganisms. Environ. Microbiol. Rep. 2010, 2, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Bordenstein, S.R.; Theis, K.R. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PLoS Biol. 2015, 13, e1002226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Read, M.N.; Holmes, A.J. towards an integrative Understanding of Diet–Host–Gut Microbiome interactions. Front. Immunol. 2017, 8, 538. [Google Scholar] [CrossRef] [PubMed]
- Leitão-Gonçalves, R.; Carvalho-Santos, Z.; Francisco, A.P.; Fioreze, G.T.; Anjos, M.; Baltazar, C.; Elias, A.P.; Itskov, P.M.; Piper, M.D.; Ribeiro, C. Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol. 2017, 15, e2000862. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1131. [Google Scholar] [CrossRef]
- Flint, H.J.; Duncan, S.H.; Louis, P. The impact of nutrition on intestinal bacterial communities. Curr. Opin. Microbiol. 2017, 38, 59–65. [Google Scholar] [CrossRef]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [Green Version]
- Tilg, H.; Moschen, A.R. Gut Microbiome, Obesity, and Metabolic Syndrome. In Metabolic Syndrome: A Comprehensive Textbook; Ahima, R.S., Ed.; Springer: Cham, Switzerland, 2016; pp. 447–459. [Google Scholar]
- Morais, P.B.; Martins, M.B.; Klaczko, L.B.; Mendonça-Hagler, L.C.; Hagler, A.N. Yeast succession in the Amazon fruit Parahancornia amapa as resource partitioning among Drosophila spp. Appl. Environ. Microbiol. 1995, 61, 4251–4257. [Google Scholar] [CrossRef] [Green Version]
- Ryu, J.-H.; Kim, S.-H.; Lee, H.-Y.; Bai, J.Y.; Nam, Y.-D.; Bae, J.-W.; Lee, D.G.; Shin, S.C.; Ha, E.-M.; Lee, W.-J. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 2008, 319, 777–782. [Google Scholar] [CrossRef] [Green Version]
- Ridley, E.V.; Wong, A.C.; Westmiller, S.; Douglas, A.E. Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS ONE 2012, 7, e36765. [Google Scholar] [CrossRef]
- Wong, A.C.-N.; Luo, Y.; Jing, X.; Franzenburg, S.; Bost, A.; Douglas, A.E. The host as the driver of the microbiota in the gut and external environment of Drosophila melanogaster. Appl. Environ. Microbiol. 2015, 81, 6232–6240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newell, P.D.; Douglas, A.E. Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl. Environ. Microbiol. 2014, 80, 788–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobson, A.J.; Chaston, J.M.; Newell, P.D.; Donahue, L.; Hermann, S.L.; Sannino, D.R.; Westmiller, S.; Wong, A.C.-N.; Clark, A.G.; Lazzaro, B.P. Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nat. Commun. 2015, 6, 6312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.-H.; Douglas, A.E. Consumption of dietary sugar by gut bacteria determines Drosophila lipid content. Biol. Lett. 2015, 11, 20150469. [Google Scholar] [CrossRef] [Green Version]
- Reed, L.K.; Lee, K.; Zhang, Z.; Rashid, L.; Poe, A.; Hsieh, B.; Deighton, N.; Glassbrook, N.; Bodmer, R.; Gibson, G. Systems genomics of metabolic phenotypes in wild-type Drosophila melanogaster. Genetics 2014, 197, 781–793. [Google Scholar] [CrossRef] [Green Version]
- Birse, R.T.; Choi, J.; Reardon, K.; Rodriguez, J.; Graham, S.; Diop, S.; Ocorr, K.; Bodmer, R.; Oldham, S. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 2010, 12, 533–544. [Google Scholar] [CrossRef] [Green Version]
- Dew-Budd, K.; Jarnigan, J.; Reed, L.K. Genetic and sex-specific transgenerational effects of a high fat diet in Drosophila melanogaster. PLoS ONE 2016, 11, e0160857. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.C.-N.; Dobson, A.J.; Douglas, A.E. Gut microbiota dictates the metabolic response of Drosophila to diet. J. Exp. Biol. 2014, 217, 1894–1901. [Google Scholar] [CrossRef] [Green Version]
- Chaston, J.M.; Newell, P.D.; Douglas, A.E. Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. MBio 2014, 5, e01631-14. [Google Scholar] [CrossRef] [Green Version]
- Chaston, J.M.; Dobson, A.J.; Newell, P.D.; Douglas, A.E. Host genetic control of the microbiota mediates the Drosophila nutritional phenotype. Appl. Environ. Microbiol. 2016, 82, 671–679. [Google Scholar] [CrossRef] [Green Version]
- Early, A.M.; Shanmugarajah, N.; Buchon, N.; Clark, A.G. Drosophila Genotype Influences Commensal Bacterial Levels. PLoS ONE 2017, 12, e0170332. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.C.; Kim, S.-H.; You, H.; Kim, B.; Kim, A.C.; Lee, K.-A.; Yoon, J.-H.; Ryu, J.-H.; Lee, W.-J. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 2011, 334, 670–674. [Google Scholar] [CrossRef] [Green Version]
- Chandler, J.A.; Lang, J.M.; Bhatnagar, S.; Eisen, J.A.; Kopp, A. Bacterial communities of diverse Drosophila species: Ecological context of a host–microbe model system. PLoS Genet. 2011, 7, e1002272. [Google Scholar] [CrossRef] [PubMed]
- Vacchini, V.; Gonella, E.; Crotti, E.; Prosdocimi, E.M.; Mazzetto, F.; Chouaia, B.; Callegari, M.; Mapelli, F.; Mandrioli, M.; Alma, A. Bacterial diversity shift determined by different diets in the gut of the spotted wing fly Drosophila suzukii is primarily reflected on acetic acid bacteria. Environ. Microbiol. Rep. 2017, 9, 91–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tefit, M.; Gillet, B.; Joncour, P.; Hughes, S.; Leulier, F. Stable association of a Drosophila-derived microbiota with its animal partner and the nutritional environment upon transfer between populations and generations. bioRxiv 2017, arXiv:111492. [Google Scholar]
- Mendez, S.; Watanabe, L.; Hill, R.; Owens, M.; Moraczewski, J.; Rowe, G.C.; Riddle, N.C.; Reed, L.K. The TreadWheel: A novel apparatus to measure genetic variation in response to gently induced exercise for Drosophila. PLoS ONE 2016, 11, e0164706. [Google Scholar] [CrossRef]
- Carvalho, G.B.; Kapahi, P.; Benzer, S. Compensatory ingestion upon dietary restriction in Drosophila melanogaster. Nat. Methods 2005, 2, 813. [Google Scholar] [CrossRef]
- Leboffe, M.J.; Pierce, B.E. Microbiology: Laboratory Theory and Application; Morton Publishing Company: Englewood, CO, USA, 2012. [Google Scholar]
- Maturin, L.; Peeler, J.T. BAM Aerobic Plate Count; US Food and Drug Administration: Silver Spring, MD, USA, 2001. [Google Scholar]
- Mackay, T.F.; Richards, S.; Stone, E.A.; Barbadilla, A.; Ayroles, J.F.; Zhu, D.; Casillas, S.; Han, Y.; Magwire, M.M.; Cridland, J.M. The Drosophila melanogaster genetic reference panel. Nature 2012, 482, 173. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Massouras, A.; Inoue, Y.; Peiffer, J.; Ràmia, M.; Tarone, A.M.; Turlapati, L.; Zichner, T.; Zhu, D.; Lyman, R.F. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014, 24, 1193–1208. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M. Drosophila. A laboratory Handbook; Cold Spring Harbor Laboratory Press: Cold Spring, Harbor, NY, USA, 1989. [Google Scholar]
- Clark, A.G.; Keith, L.E. Variation among extracted lines of Drosophila melanogaster in triacylglycerol and carbohydrate storage. Genetics 1988, 119, 595–607. [Google Scholar]
- De Luca, M.; Yi, N.; Allison, D.B.; Leips, J.; Ruden, D.M. Mapping quantitative trait loci affecting variation in Drosophila triacylglycerol storage. Obes. Res. 2005, 13, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.K.; Williams, S.; Springston, M.; Brown, J.; Freeman, K.; DesRoches, C.E.; Sokolowski, M.B.; Gibson, G. Genotype-by-diet interactions drive metabolic phenotype variation in Drosophila melanogaster. Genetics 2010, 185, 1009–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Rulifson, E.J.; Kim, S.K.; Nusse, R.J.S. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science 2002, 296, 1118–1120. [Google Scholar] [CrossRef]
- Kumar, R.; Eipers, P.; Little, R.B.; Crowley, M.; Crossman, D.K.; Lefkowitz, E.J.; Morrow, C.D. Getting started with microbiome analysis: Sample acquisition to bioinformatics. Curr. Protoc. Hum. Genet. 2014, 82, 18.8.1–18.8.29. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B.J.B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.J.B. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R.J.B. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Bruno, D.; Bonelli, M.; De Filippis, F.; Di Lelio, I.; Tettamanti, G.; Casartelli, M.; Ercolini, D.; Caccia, S. The intestinal microbiota of Hermetia illucens larvae is affected by diet and shows a diverse composition in the different midgut regions. Appl. Environ. Microbiol. 2019, 85, e01864-18. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.J.S. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Bruce-Keller, A.J.; Salbaum, J.M.; Luo, M.; Blanchard, E., IV; Taylor, C.M.; Welsh, D.A.; Berthoud, H.-R. Obese-type gut microbiota induce neurobehavioral changes in the absence of obesity. Biol. Psychiatry 2015, 77, 607–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klepsatel, P.; Procházka, E.; Gáliková, M. Crowding of Drosophila larvae affects lifespan and other life-history traits via reduced availability of dietary yeast. Exp. Gerontol. 2018, 110, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Bing, X.; Gerlach, J.; Loeb, G.; Buchon, N. Nutrient-dependent impact of microbes on Drosophila suzukii development. MBio 2018, 9, e02199-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skorupa, D.A.; Dervisefendic, A.; Zwiener, J.; Pletcher, S.D. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell 2008, 7, 478–490. [Google Scholar] [CrossRef] [Green Version]
- Sang, J.H. The quantitative nutritional requirements of Drosophila melanogaster. J. Exp. Biol. 1956, 33, 45–72. [Google Scholar]
- Sgro, C.M.; Partridge, L. Evolutionary responses of the life history of wild-caught Drosophila melanogaster to two standard methods of laboratory culture. Am. Nat. 2000, 156, 341–353. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Hallas, R.; Sinclair, C.; Partridge, L. Rapid loss of stress resistance in Drosophila melanogaster under adaptation to laboratory culture. Evolution 2001, 55, 436–438. [Google Scholar] [CrossRef]
- Russell, T.; Kurtz, R. A Comparison of Laboratory-Reared Stock and Captured Fruit Flies (Drosophila melanogaster) using Upward Movement, Phototaxic, and Starvation Assays Reveals Significant Behavioral Differences. Staff Rev. 2012, 6. Available online: https://www.commackschools.org/Downloads/Fruit%20Fly%20Trinity%20Russell.pdf (accessed on 4 October 2020).
- Staubach, F.; Baines, J.F.; Künzel, S.; Bik, E.M.; Petrov, D.A. Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment. PLoS ONE 2013, 8, e70749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pais, I.S.; Valente, R.S.; Sporniak, M.; Teixeira, L. Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol. 2018, 16, e2005710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbaniec, K.; Zalewski, P.; Zhu, X.X. A decomposition approach for retrofit design of energy systems in the sugar industry. Appl. Therm. Eng. 2000, 20, 1431–1442. [Google Scholar] [CrossRef]
- Hillier, R.M.; Lyster, R.L.J. Whey protein denaturation in heated milk and cheese whey. J. Dairy Res. 1979, 46, 95–102. [Google Scholar] [CrossRef]
- Adair, K.L.; Wilson, M.; Bost, A.; Douglas, A.E. Microbial community assembly in wild populations of the fruit fly Drosophila melanogaster. ISME J. 2018, 1, 959–972. [Google Scholar] [CrossRef]
- Douglas, A.E. The Drosophila model for microbiome research. Lab Anim. 2018, 47, 157–164. [Google Scholar] [CrossRef]
- Wong, A.C.; Chaston, J.M.; Douglas, A.E. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013, 7, 1922. [Google Scholar] [CrossRef] [Green Version]
- Corby-Harris, V.; Pontaroli, A.C.; Shimkets, L.J.; Bennetzen, J.L.; Habel, K.E.; Promislow, D.E. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl. Environ. Microbiol. 2007, 73, 3470–3479. [Google Scholar] [CrossRef] [Green Version]
- Jehrke, L.; Stewart, F.A.; Droste, A.; Beller, M. The impact of genome variation and diet on the metabolic phenotype and microbiome composition of Drosophila melanogaster. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.N.A.; Ng, P.; Douglas, A.E. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 2011, 13, 1889–1900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moghadam, N.N.; Thorshauge, P.M.; Kristensen, T.N.; de Jonge, N.; Bahrndorff, S.; Kjeldal, H.; Nielsen, J.L. Strong responses of Drosophila melanogaster microbiota to developmental temperature. Fly 2018, 12, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Porchas, M.; Villalpando-Canchola, E.; Suarez, L.E.O.; Vargas-Albores, F. How conserved are the conserved 16S-rRNA regions? PeerJ 2017, 5, e3036. [Google Scholar] [CrossRef] [PubMed]
- Behar, A.; Jurkevitch, E.; Yuval, B. Bringing back the fruit into fruit fly–bacteria interactions. Mol. Ecol. 2008, 17, 1375–1386. [Google Scholar] [CrossRef]
- Ferguson, L.V.; Dhakal, P.; Lebenzon, J.E.; Heinrichs, D.E.; Bucking, C.; Sinclair, B.J.J.F.E. Seasonal shifts in the insect gut microbiome are concurrent with changes in cold tolerance and immunity. Funct. Ecol. 2018, 32, 2357–2368. [Google Scholar] [CrossRef]
- Tong, Q.; Zhang, J. Effects of captivity and season on the gut microbiota of the brown frog (Rana dybowskii). Front. Microbiol. 2019, 10, 1912. [Google Scholar] [CrossRef] [Green Version]
- Maurice, C.F.; Knowles, S.C.; Ladau, J.; Pollard, K.S.; Fenton, A.; Pedersen, A.B.; Turnbaugh, P.J. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015, 9, 2423–2434. [Google Scholar] [CrossRef] [Green Version]
- Parks, B.W.; Nam, E.; Org, E.; Kostem, E.; Norheim, F.; Hui, S.T.; Pan, C.; Civelek, M.; Rau, C.D.; Bennett, B.J. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013, 17, 141–152. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Zhang, M.; Wang, S.; Han, R.; Cao, Y.; Hua, W.; Mao, Y.; Zhang, X.; Pang, X.; Wei, C. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010, 4, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Kreznar, J.H.; Keller, M.P.; Traeger, L.L.; Rabaglia, M.E.; Schueler, K.L.; Stapleton, D.S.; Zhao, W.; Vivas, E.I.; Yandell, B.S.; Broman, A.T. Host genotype and gut microbiome modulate insulin secretion and diet-induced metabolic phenotypes. Cell Rep. 2017, 18, 1739–1750. [Google Scholar] [CrossRef]
- Rosenthal, N.; Brown, S. The mouse ascending: Perspectives for human-disease models. Nat. Cell Biol. 2007, 9, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Berger, Z.; Ttofi, E.K.; Michel, C.H.; Pasco, M.Y.; Tenant, S.; Rubinsztein, D.C.; O’Kane, C.J. Lithium rescues toxicity of aggregate-prone proteins in Drosophila by perturbing Wnt pathway. Hum. Mol. Genet. 2005, 14, 3003–3011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, T.L.; Balser, S.R.; Young, G.S.; Lewis, S.D. Cost and effectiveness of commercially available nesting substrates for Deer Mice (Peromyscus maniculatus). J. Am. Assoc. Lab. Anim. Sci. 2016, 55, 412–418. [Google Scholar] [PubMed]
- King, E.G.; Macdonald, S.J.; Long, A.D. Properties and power of the Drosophila Synthetic Population Resource for the routine dissection of complex traits. Genetics 2012, 191, 935–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, S.; Dew-Budd, K.; Davis, K.; Anderson, J.; Bishop, R.; Freeman, K.; Davis, D.; Bray, K.; Perkins, L.; Hubickey, J. Metabolomic and gene expression profiles exhibit modular genetic and dietary structure linking metabolic syndrome phenotypes in Drosophila. G3 Genes Genomes Genet. 2015, 5, 2817–2829. [Google Scholar] [CrossRef] [Green Version]
- Martyn, J.J.; Kaneki, M.; Yasuhara, S.; Warner, D.S.; Warner, M.A. Obesity-induced insulin resistance and hyperglycemia: Etiological factors and molecular mechanisms. J. Am. Soc. Anesthesiol. 2008, 109, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Akter, R.; Nessa, A.; Husain, M.; Wahed, F.; Khatun, N.; Yesmin, M.; Nasreen, S.; Tajkia, T. Effect of Obesity on Fasting Blood Sugar. Mymensingh Med. J. 2017, 26, 7–11. [Google Scholar]
- Million, M.; Maraninchi, M.; Henry, M.; Armougom, F.; Richet, H.; Carrieri, P.; Valero, R.; Raccah, D.; Vialettes, B.; Raoult, D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int. J. Obes. 2012, 36, 817–825. [Google Scholar] [CrossRef] [Green Version]
- Ignacio, A.; Fernandes, M.R.; Rodrigues, V.A.; Groppo, F.C.; Cardoso, A.L.; Avila-Campos, M.; Nakano, V. Correlation between body mass index and faecal microbiota from children. Clin. Microbiol. Infect. 2016, 22, 258.e1–258.e8. [Google Scholar] [CrossRef] [Green Version]
- Murugesan, S.; Ulloa-Martínez, M.; Martínez-Rojano, H.; Galván-Rodríguez, F.; Miranda-Brito, C.; Romano, M.; Piña-Escobedo, A.; Pizano-Zárate, M.; Hoyo-Vadillo, C.; García-Mena, J. Study of the diversity and short-chain fatty acids production by the bacterial community in overweight and obese Mexican children. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1337–1346. [Google Scholar] [CrossRef]
- Armougom, F.; Henry, M.; Vialettes, B.; Raccah, D.; Raoult, D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PLoS ONE 2009, 4, e7125. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Furet, J.-P.; Kong, L.-C.; Tap, J.; Poitou, C.; Basdevant, A.; Bouillot, J.-L.; Mariat, D.; Corthier, G.; Doré, J.; Henegar, C. Differential adaptation of human gut microbiota to bariatric surgery–induced weight loss. Diabetes 2010, 59, 3049–3057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storelli, G.; Defaye, A.; Erkosar, B.; Hols, P.; Royet, J.; Leulier, F.J.C.m. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011, 14, 403–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawitan, Y.; Michiels, S.; Koscielny, S.; Gusnanto, A.; Ploner, A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics 2005, 21, 3017–3024. [Google Scholar] [CrossRef] [PubMed]



| Treatment | Survival | Development | Weight | Triglyceride | Protein | Glucose |
|---|---|---|---|---|---|---|
| NS | R > PR ** | R < PR *** | R > PR *** | R < PR *** | R > PR ** | R > PR *** |
| S | R > PR ** | R < PR *** | R > PR *** | R < PR *** | R > PR *** | R > PR *** |
| NS | PR > PA *** | PR < PA *** | PR > PA *** | PR < PA *** | PR < PA | PR < PA ** |
| S | PR > PA *** | PR < PA *** | PR > PA *** | PR < PA *** | PR < PA *** | PR < PA *** |
| Genetic Line | Diet | Survival | Development | Weight | Triglyceride | Protein | Glucose |
|---|---|---|---|---|---|---|---|
| All | PA | S < NS *** | S > NS *** | S < NS *** | S > NS | S > NS ** | S > NS ** |
| All | PR | S < NS | S > NS *** | S < NS *** | S > NS | S < NS | S > NS |
| All | R | S < NS | S > NS | S < NS | S > NS | S > NS | S > NS |
| Independent Variable | Survival | Development | Weight | Triglyceride | Protein | Glucose |
|---|---|---|---|---|---|---|
| Diet | VE = 28.4% *** | VE = 47.5% *** | VE = 31.4% *** | VE = 41.4% *** | VE = 4.24% *** | VE = 17.8% *** |
| Genetic line | VE = 5.13% *** | VE = 4.98% *** | VE = 7.44% *** | VE = 2.36% ** | VE = 5.71% *** | VE = 3.15% |
| Treatment | VE = 4.69% *** | VE = 2.41% *** | VE = 0.74% *** | VE = 0.36% | VE = 0.40% * | VE = 2.61% *** |
| Diet*Genetic line | VE = 3.13% *** | VE = 0.96% | VE = 2.88% *** | VE = 5.03% *** | VE = 5.09% *** | VE = 5.52% |
| Diet*Treatment | VE = 8.02% *** | VE = 1.41% *** | VE = 0.10% | VE = 0.07% | VE = 0.45% | VE = 3.55% *** |
| Genetic line*Treatment | VE = 0.98% * | VE = 2.82% *** | VE = 1.06% *** | VE = 1.81% * | VE = 3.53% *** | VE = 0.72% |
| Diet*Treatment*Genetic line | VE = 2.37% *** | VE = 0.82% | VE = 2.75% *** | VE = 4.81% *** | VE = 4.92% *** | VE = 2.37% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bombin, A.; Cunneely, O.; Eickman, K.; Bombin, S.; Ruesy, A.; Su, M.; Myers, A.; Cowan, R.; Reed, L. Influence of Lab Adapted Natural Diet and Microbiota on Life History and Metabolic Phenotype of Drosophila melanogaster. Microorganisms 2020, 8, 1972. https://doi.org/10.3390/microorganisms8121972
Bombin A, Cunneely O, Eickman K, Bombin S, Ruesy A, Su M, Myers A, Cowan R, Reed L. Influence of Lab Adapted Natural Diet and Microbiota on Life History and Metabolic Phenotype of Drosophila melanogaster. Microorganisms. 2020; 8(12):1972. https://doi.org/10.3390/microorganisms8121972
Chicago/Turabian StyleBombin, Andrei, Owen Cunneely, Kira Eickman, Sergei Bombin, Abigail Ruesy, Mengting Su, Abigail Myers, Rachael Cowan, and Laura Reed. 2020. "Influence of Lab Adapted Natural Diet and Microbiota on Life History and Metabolic Phenotype of Drosophila melanogaster" Microorganisms 8, no. 12: 1972. https://doi.org/10.3390/microorganisms8121972
APA StyleBombin, A., Cunneely, O., Eickman, K., Bombin, S., Ruesy, A., Su, M., Myers, A., Cowan, R., & Reed, L. (2020). Influence of Lab Adapted Natural Diet and Microbiota on Life History and Metabolic Phenotype of Drosophila melanogaster. Microorganisms, 8(12), 1972. https://doi.org/10.3390/microorganisms8121972









