Unconventional Secretion of Nigerolysins A from Aspergillus Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Growth Conditions, and Chemicals
2.2. Preparation of Fluorescent Protein Cassettes for Fungal Transformation
2.3. Fungal Transformation, Selection, and Confirmation of Transformants
2.4. Construction of Nigerolysin Gene Deletion Mutants
2.5. Secretion of Nigerolysins A
2.6. Live-Cell Imaging
2.7. Immunolocalization and Cell Imaging
2.8. Comparative Genomics, Prediction, and Phylogenetic Methods
3. Results
3.1. Alignment of Nigerolysins A
3.2. Prediction of Localization for Nigerolysins A
3.3. Secretion of Nigerolysins A
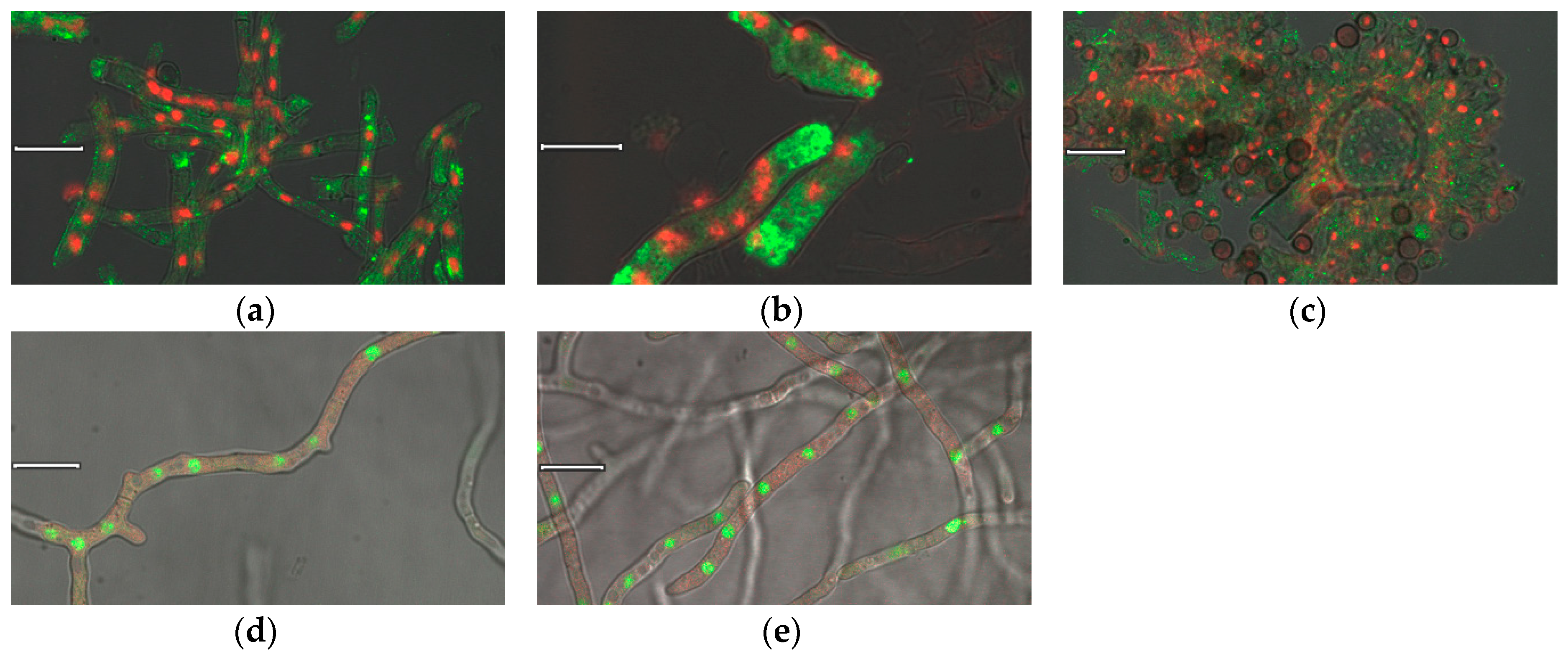
3.4. Immunocytochemistry and Live Cell Imaging of Nigerolysin A Proteins in Hyphae
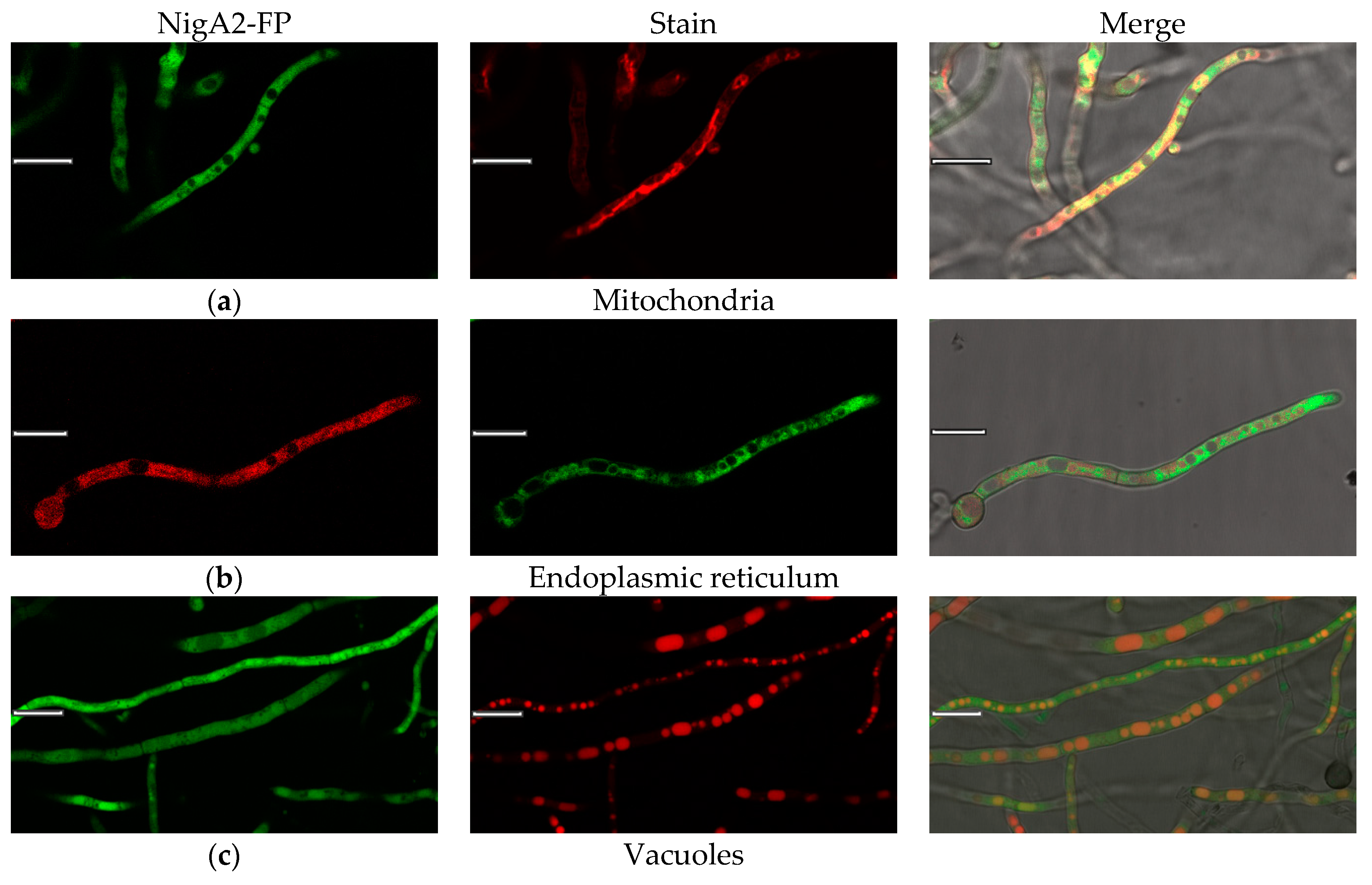
3.5. Live Cell Colocalization of Nigerolysin A Proteins in Hyphae
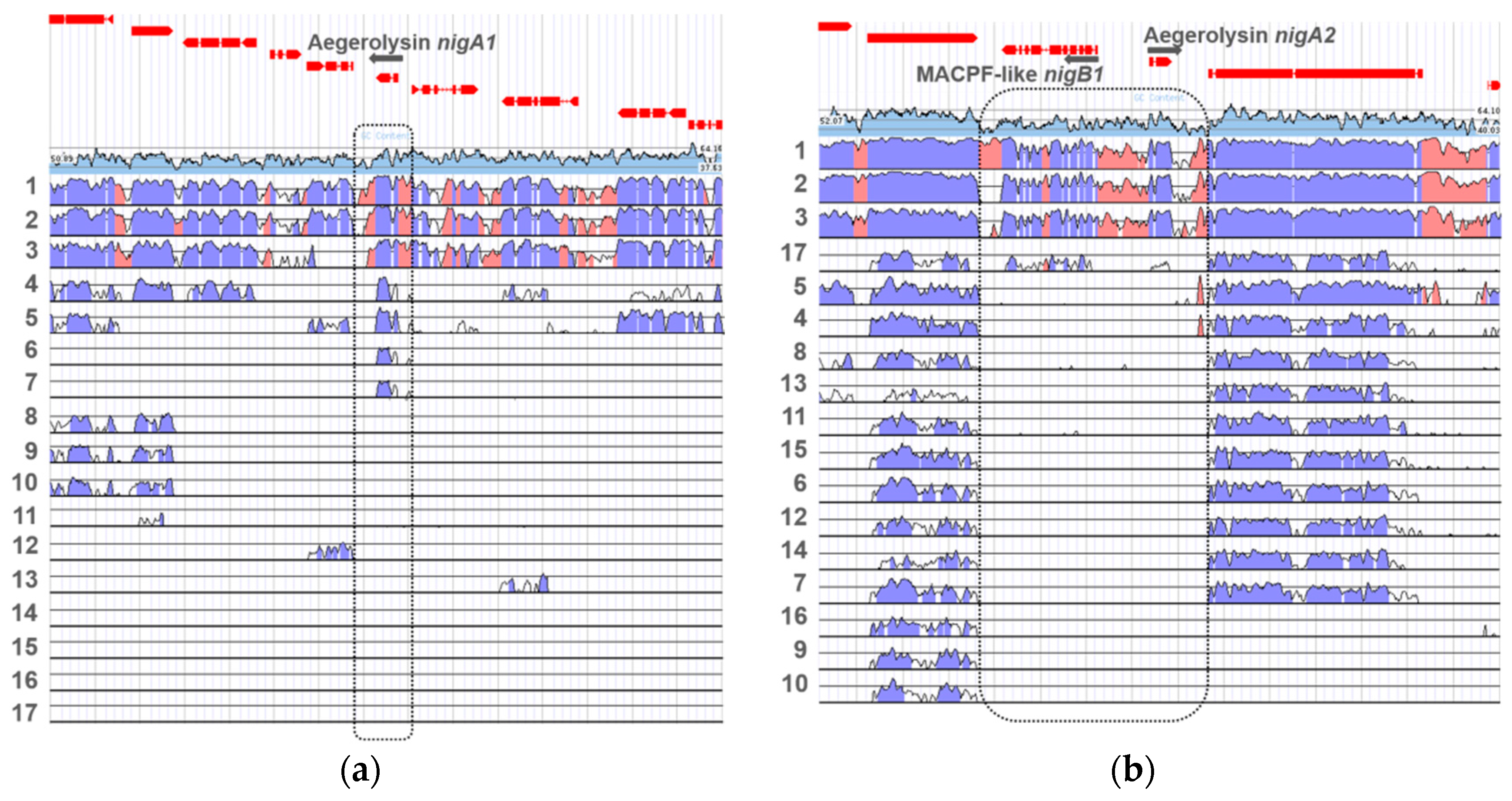
3.6. Chromosomal Loci of Aegerolysins Genes
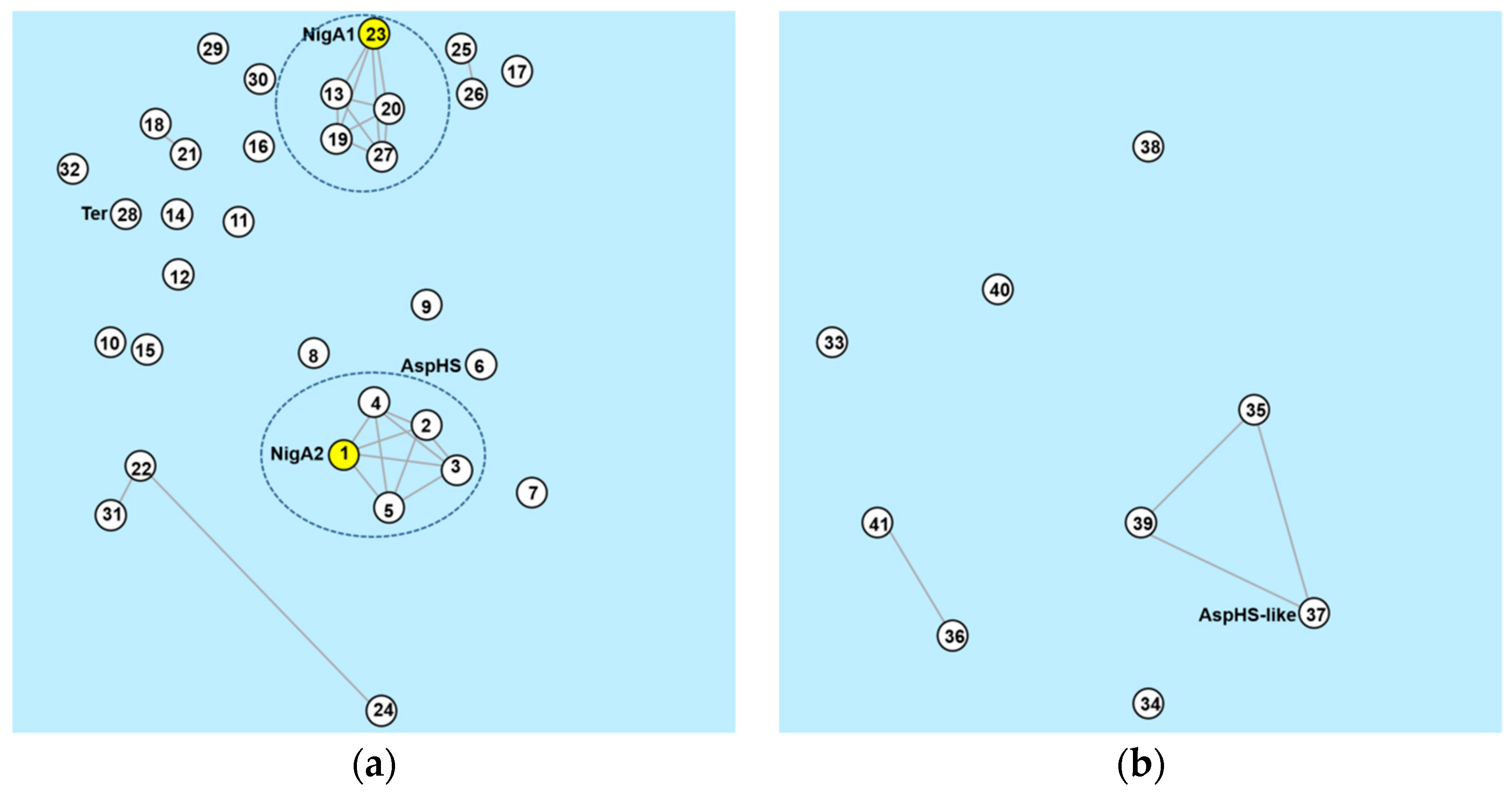
3.7. Aegerolysin Orthologues in Aspergillus Species
3.8. Aegerolysin Synteny among Aspergillus Species
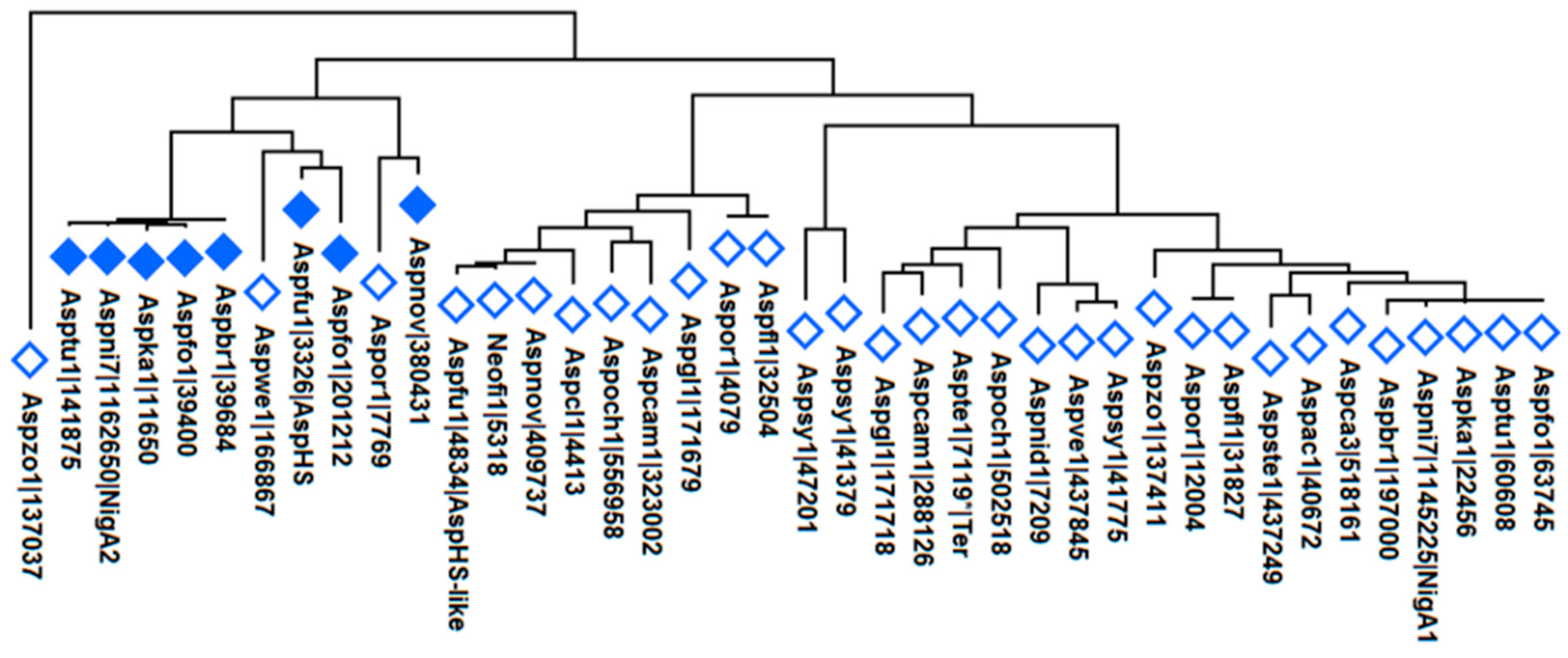
3.9. Aegerolysin Phylogeny of the Aspergillus Species
4. Discussion
4.1. Non-Conventional Protein Secretion (i.e., Not Signal Peptide Triggered Secretion)
4.2. Application of Aegerolysin Secretion Pathway in the Production of Non-Fungal Proteins
4.3. Function of Aergerolysins
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berne, S.; Lah, L.; Sepčić, K. Aegerolysins: Structure, function, and putative biological role. Protein Sci. 2009, 18, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Novak, M.; Kraševec, N.; Skočaj, M.; Maček, P.; Anderluh, G.; Sepčić, K. Fungal aegerolysin-like proteins: Distribution, activities, and applications. Appl. Microbiol. Biotechnol. 2015, 99, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Butala, M.; Novak, M.; Kraševec, N.; Skočaj, M.; Veranič, P.; Maček, P.; Sepčić, K. Aegerolysins: Lipid-binding proteins with versatile functions. Semin. Cell Dev. Biol. 2017, 72, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Sepčić, K.; Berne, S.; Rebolj, K.; Batista, U.; Plemenitaš, A.; Šentjurc, M.; Maček, P. Ostreolysin, a pore-forming protein from the oyster mushroom, interacts specifically with membrane cholesterol-rich lipid domains. FEBS Lett. 2004, 575, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.; Leonardi, A.; Mikelj, M.; Skočaj, M.; Wohlschlager, T.; Künzler, M.; Aebi, M.; Narat, M.; Križaj, I.; Anderluh, G.; et al. Membrane cholesterol and sphingomyelin, and ostreolysin A are obligatory for pore-formation by a MACPF/CDC-like pore-forming protein, pleurotolysin B. Biochimie 2013, 95, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Skočaj, M.; Resnik, N.; Grundner, M.; Ota, K.; Rojko, N.; Hodnik, V.; Anderluh, G.; Sobota, A.; Maček, P.; Veranič, P.; et al. Tracking cholesterol/sphingomyelin-rich membrane domains with the ostreolysin A-mCherry protein. PLoS ONE 2014, 9, e92783. [Google Scholar] [CrossRef] [PubMed]
- Bhat, H.B.; Ishitsuka, R.; Inaba, T.; Murate, M.; Abe, M.; Makino, A.; Kohyama-Koganeya, A.; Nagao, K.; Kurahashi, A.; Kishimoto, T.; et al. Evaluation of aegerolysins as novel tools to detect and visualize ceramide phosphoethanolamine, a major sphingolipid in invertebrates. FASEB J. 2015, 29, 3920–3934. [Google Scholar] [CrossRef]
- Bando, H.; Hisada, H.; Ishida, H.; Hata, Y.; Katakura, Y.; Kondo, A. Isolation of a novel promoter for efficient protein expression by Aspergillus oryzae in solid-state culture. Appl. Microbiol. Biotechnol. 2011, 92, 561–569. [Google Scholar] [CrossRef]
- Hisada, H.; Tsutsumi, H.; Ishida, H.; Hata, Y. High production of llama variable heavy-chain antibody fragment (VHH) fused to various reader proteins by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2013, 97, 761–766. [Google Scholar] [CrossRef]
- Hatzmann, M. Constitutive Promoter Application. European Patent EP 2 388 331 A1, 23 November 2011. [Google Scholar]
- Tomita, T.; Noguchi, K.; Mimuro, H.; Ukaji, F.; Ito, K.; Sugawara-Tomita, N.; Hashimoto, Y. Pleurotolysin, a Novel Sphingomyelin-specific Two-component Cytolysin from the Edible Mushroom Pleurotus ostreatus, Assembles into a Transmembrane Pore Complex. J. Biol. Chem. 2004, 279, 26975–26982. [Google Scholar] [CrossRef] [PubMed]
- Bhat, H.B.; Kishimoto, T.; Abe, M.; Makino, A.; Inaba, T.; Murate, M.; Dohmae, N.; Kurahashi, A.; Nishibori, K.; Fujimori, F.; et al. Binding of a pleurotolysin ortholog from Pleurotus eryngii to sphingomyelin and cholesterol-rich membrane domains. J. Lipid Res. 2013, 54, 2933–2943. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Kudou, M.; Hoshi, Y.; Kudo, A.; Nanashima, N.; Miyairi, K. Isolation and characterization of a novel two-component hemolysin, erylysin A and B, from an edible mushroom, Pleurotus eryngii. Toxicon 2010, 56, 1436–1442. [Google Scholar] [CrossRef] [PubMed]
- Lukoyanova, N.; Kondos, S.C.; Farabella, I.; Law, R.H.P.; Reboul, C.F.; Caradoc-Davies, T.T.; Spicer, B.A.; Kleifeld, O.; Traore, D.A.K.; Ekkel, S.M.; et al. Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin. PLoS Biol. 2015, 13, e1002049. [Google Scholar] [CrossRef] [PubMed]
- Resnik, N.; Repnik, U.; Kreft, M.E.; Sepčić, K.; Maček, P.; Turk, B.; Veranič, P. Highly Selective Anti-Cancer Activity of Cholesterol-Interacting Agents Methyl-β-Cyclodextrin and Ostreolysin A/Pleurotolysin B Protein Complex on Urothelial Cancer Cells. PLoS ONE 2015, 10, e0137878. [Google Scholar] [CrossRef]
- Novak, M.; Krpan, T.; Panevska, A.; Shewell, L.K.; Day, C.J.; Jennings, M.P.; Guella, G.; Sepčić, K. Binding specificity of ostreolysin A6 towards Sf9 insect cell lipids. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183307. [Google Scholar] [CrossRef]
- Panevska, A.; Hodnik, V.; Skočaj, M.; Novak, M.; Modic, Š.; Pavlic, I.; Podržaj, S.; Zarić, M.; Resnik, N.; Maček, P.; et al. Pore-forming protein complexes from Pleurotus mushrooms kill western corn rootworm and Colorado potato beetle through targeting membrane ceramide phosphoethanolamine. Sci. Rep. 2019, 9, 5073. [Google Scholar] [CrossRef]
- Panevska, A.; Skočaj, M.; Križaj, I.; Maček, P.; Sepčić, K. Ceramide phosphoethanolamine, an enigmatic cellular membrane sphingolipid. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1284–1292. [Google Scholar] [CrossRef]
- Meyer, V.; Andersen, M.R.; Brakhage, A.A.; Braus, G.H.; Caddick, M.X.; Cairns, T.C.; de Vries, R.P.; Haarmann, T.; Hansen, K.; Hertz-Fowler, C.; et al. Current challenges of research on filamentous fungi in relation to human welfare and a sustainable bio-economy: A white paper. Fungal Biol. Biotechnol. 2016, 3. [Google Scholar] [CrossRef]
- Meyer, V.; Basenko, E.Y.; Benz, J.P.; Braus, G.H.; Caddick, M.X.; Csukai, M.; de Vries, R.P.; Endy, D.; Frisvad, J.C.; Gunde-Cimerman, N.; et al. Growing a circular economy with fungal biotechnology: A white paper. Fungal Biol. Biotechnol. 2020, 7, 5. [Google Scholar] [CrossRef]
- de Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef] [PubMed]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.-C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef] [PubMed]
- Kjærbølling, I.; Vesth, T.C.; Frisvad, J.C.; Nybo, J.L.; Theobald, S.; Kuo, A.; Bowyer, P.; Matsuda, Y.; Mondo, S.; Lyhne, E.K.; et al. Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proc. Natl. Acad. Sci. USA 2018, 115, E753–E761. [Google Scholar] [CrossRef] [PubMed]
- Vesth, T.C.; Nybo, J.L.; Theobald, S.; Frisvad, J.C.; Larsen, T.O.; Nielsen, K.F.; Hoof, J.B.; Brandl, J.; Salamov, A.; Riley, R.; et al. Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat. Genet. 2018, 50, 1688–1695. [Google Scholar] [CrossRef]
- Kjærbølling, I.; Vesth, T.; Frisvad, J.C.; Nybo, J.L.; Theobald, S.; Kildgaard, S.; Petersen, T.I.; Kuo, A.; Sato, A.; Lyhne, E.K.; et al. A comparative genomics study of 23 Aspergillus species from section Flavi. Nat. Commun. 2020, 11, 1106. [Google Scholar] [CrossRef]
- Sakaguchi, O.; Shimada, H.; Yokota, K. Proceedings: Purification and characteristics of hemolytic toxin from Aspergillus fumigatus. Jpn. J. Med. Sci. Biol. 1975, 28, 328–331. [Google Scholar]
- Ebina, K.; Yokota, K.; Sakaguchi, O. Studies on toxin of Aspergillus fumigatus. XIV. Relationship between Asp-hemolysin and experimental infection for mice. Jpn. J. Med. Mycol. 1982, 23, 246–252. [Google Scholar] [CrossRef]
- Wartenberg, D.; Lapp, K.; Jacobsen, I.D.; Dahse, H.-M.; Kniemeyer, O.; Heinekamp, T.; Brakhage, A.A. Secretome analysis of Aspergillus fumigatus reveals Asp-hemolysin as a major secreted protein. Int. J. Med. Microbiol. 2011, 301, 602–611. [Google Scholar] [CrossRef]
- Rementeria, A.; López-Molina, N.; Ludwig, A.; Vivanco, A.B.; Bikandi, J.; Pontón, J.; Garaizar, J. Genes and molecules involved in Aspergillus fumigatus virulence. Rev. Iberoam. Micol. 2005, 22, 1–23. [Google Scholar] [CrossRef]
- Nayak, A.P.; Blachere, F.M.; Hettick, J.M.; Lukomski, S.; Schmechel, D.; Beezhold, D.H. Characterization of recombinant terrelysin, a hemolysin of Aspergillus terreus. Mycopathologia 2011, 171, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Braaksma, M.; Martens-Uzunova, E.S.; Punt, P.J.; Schaap, P.J. An inventory of the Aspergillus niger secretome by combining in silico predictions with shotgun proteomics data. BMC Genom. 2010, 11, 584. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Sun, J.; Nimtz, M.; Wissing, J.; Zeng, A.-P.; Rinas, U. The intra- and extracellular proteome of Aspergillus niger growing on defined medium with xylose or maltose as carbon substrate. Microb. Cell Fact. 2010, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Novak, M.; Čepin, U.; Hodnik, V.; Narat, M.; Jamnik, M.; Kraševec, N.; Sepčić, K.; Anderluh, G. Functional studies of aegerolysin and MACPF-like proteins in Aspergillus niger. Mol. Microbiol. 2019, 112, 1253–1269. [Google Scholar] [CrossRef]
- Levin, A.M.; de Vries, R.P.; Conesa, A.; de Bekker, C.; Talon, M.; Menke, H.H.; van Peij, N.N.M.E.; Wösten, H.A.B. Spatial Differentiation in the Vegetative Mycelium of Aspergillus niger. Eukaryot. Cell 2007, 6, 2311–2322. [Google Scholar] [CrossRef]
- Wösten, H.A.B.; Moukha, S.M.; Sietsma, J.H.; Wessels, J.G. Localization of growth and secretion of proteins in Aspergillus niger. J. Gen. Microbiol. 1991, 137, 2017–2023. [Google Scholar] [CrossRef]
- Nitsche, B.M.; Jørgensen, T.R.; Akeroyd, M.; Meyer, V.; Ram, A.F. The carbon starvation response of Aspergillus niger during submerged cultivation: Insights from the transcriptome and secretome. BMC Genom. 2012, 13, 380. [Google Scholar] [CrossRef]
- Nayak, A.P.; Green, B.J.; Janotka, E.; Hettick, J.M.; Friend, S.; Vesper, S.J.; Schmechel, D.; Beezhold, D.H. Monoclonal antibodies to hyphal exoantigens derived from the opportunistic pathogen Aspergillus terreus. Clin. Vaccine Immunol. 2011, 18, 1568–1576. [Google Scholar] [CrossRef]
- Punt, P.J.; van den Hondel, C.A.M.J.J. Transformation of filamentous fungi based on hygromycin b and phleomycin resistance markers. Methods Enzymol. 1992, 216, 447–457. [Google Scholar]
- Bagar, T.; Altenbach, K.; Read, N.D.; Benčina, M. Live-Cell Imaging and Measurement of Intracellular pH in Filamentous Fungi Using a Genetically Encoded Ratiometric Probe. Eukaryot. Cell 2009, 8, 703–712. [Google Scholar] [CrossRef]
- Bos, C.J.; Debets, A.J.; Swart, K.; Huybers, A.; Kobus, G.; Slakhorst, S.M. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 1988, 14, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Mustalahti, E.; Saloheimo, M.; Joensuu, J.J. Intracellular protein production in Trichoderma reesei (Hypocrea jecorina) with hydrophobin fusion technology. N. Biotechnol. 2013, 30, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Podobnik, B.; Stojan, J.; Lah, L.; Krasevec, N.; Seliskar, M.; Rizner, T.L.; Rozman, D.; Komel, R. CYP53A15 of Cochliobolus lunatus, a Target for Natural Antifungal Compounds. J. Med. Chem. 2008, 51, 3480–3486. [Google Scholar] [CrossRef] [PubMed]
- Novak, M.; Sepčić, K.; Kraševec, N.; Križaj, I.; Maček, P.; Anderluh, G.; Guella, G.; Mancini, I. Targeted Lipid Analysis of Haemolytic Mycelial Extracts of Aspergillus niger. Molecules 2014, 19, 9051–9069. [Google Scholar] [CrossRef] [PubMed]
- Hickey, P.C.; Read, N.D. Imaging living cells of Aspergillus in vitro. Med. Mycol. 2009, 47, S110–S119. [Google Scholar] [CrossRef][Green Version]
- Cerqueira, G.C.; Arnaud, M.B.; Inglis, D.O.; Skrzypek, M.S.; Binkley, G.; Simison, M.; Miyasato, S.R.; Binkley, J.; Orvis, J.; Shah, P.; et al. The Aspergillus Genome Database: Multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014, 42, D705–D710. [Google Scholar] [CrossRef]
- Kersey, P.J.; Allen, J.E.; Allot, A.; Barba, M.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Grabmueller, C.; et al. Ensembl Genomes 2018: An integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018, 46, D802–D808. [Google Scholar] [CrossRef]
- Mabey Gilsenan, J.; Cooley, J.; Bowyer, P. CADRE: The Central Aspergillus Data REpository 2012. Nucleic Acids Res. 2012, 40, D660–D666. [Google Scholar] [CrossRef]
- Basenko, E.; Pulman, J.; Shanmugasundram, A.; Harb, O.; Crouch, K.; Starns, D.; Warrenfeltz, S.; Aurrecoechea, C.; Stoeckert, C.; Kissinger, J.; et al. FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes. J. Fungi 2018, 4, 39. [Google Scholar] [CrossRef]
- FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes. Available online: https://fungidb.org/fungidb/app (accessed on 27 August 2020).
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- FungiDB—Fungal & Oomycete Informatics Resouces. Available online: https://beta.orthomcl.org/orthomcl.beta/ (accessed on 13 July 2020).
- Li, L. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, J.; Angiuoli, S.V.; Wortman, J.R.; White, O.R. Sybil: Methods and Software for Multiple Genome Comparison and Visualization. Methods Mol. Biol. 2007, 408, 93–108. [Google Scholar] [CrossRef]
- Arnaud, M.B.; Cerqueira, G.C.; Inglis, D.O.; Skrzypek, M.S.; Binkley, J.; Chibucos, M.C.; Crabtree, J.; Howarth, C.; Orvis, J.; Shah, P.; et al. The Aspergillus Genome Database (AspGD): Recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2012, 40, D653–D659. [Google Scholar] [CrossRef] [PubMed]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Futagami, T.; Mori, K.; Yamashita, A.; Wada, S.; Kajiwara, Y.; Takashita, H.; Omori, T.; Takegawa, K.; Tashiro, K.; Kuhara, S.; et al. Genome announcement: Genome sequence of the white Koji mold Aspergillus kawachii IFO 4308, used for brewing the Japanese distilled spirit shochu. Eukaryot. Cell 2011, 10, 1586–1587. [Google Scholar] [CrossRef]
- Andersen, M.R.; Salazar, M.P.; Schaap, P.J.; Van De Vondervoort, P.J.I.; Culley, D.; Thykaer, J.; Frisvad, J.C.; Nielsen, K.F.; Albang, R.; Albermann, K.; et al. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 2011, 21, 885–897. [Google Scholar] [CrossRef]
- Pel, H.J.; de Winde, J.H.; Archer, D.B.; Dyer, P.S.; Hofmann, G.; Schaap, P.J.; Turner, G.; de Vries, R.P.; Albang, R.; Albermann, K.; et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007, 25, 221–231. [Google Scholar] [CrossRef]
- EffectorP: Predicting Fungal Effector Proteins from Secretomes Using Machine Learning. Available online: http://effectorp.csiro.au (accessed on 13 July 2020).
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Singh, K.B.; Taylor, J.M. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant. Pathol. 2018, 19, 2094–2110. [Google Scholar] [CrossRef]
- OCTOPUS: Prediction of Membrane Protein Topology. Available online: http://octopus.cbr.su.se/ (accessed on 13 July 2020).
- Viklund, H.; Elofsson, A. OCTOPUS: Improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 2008, 24, 1662–1668. [Google Scholar] [CrossRef]
- Phobius a Combined Transmembrane Topology and Signal Peptide Predictor. Available online: http://phobius.binf.ku.dk/index.html (accessed on 13 July 2020).
- Kall, L.; Krogh, A.; Sonnhammer, E.L.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef] [PubMed]
- PredGPI GPI-Anchor Predictor. Available online: http://gpcr2.biocomp.unibo.it/gpipe/pred.htm (accessed on 13 July 2020).
- Pierleoni, A.; Martelli, P.; Casadio, R. PredGPI: A GPI-anchor predictor. BMC Bioinform. 2008, 9, 392. [Google Scholar] [CrossRef] [PubMed]
- PrediSi PREDIction of SIgnal Peptides. Available online: http://www.predisi.de/ (accessed on 13 July 2020).
- Hiller, K.; Grote, A.; Scheer, M.; Munch, R.; Jahn, D. PrediSi: Prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004, 32, W375–W379. [Google Scholar] [CrossRef] [PubMed]
- PredαTM and PredβTM: Transmembrane Region Predictors. Available online: https://www.ki.si/odseki/d01-teoreticni-odsek/laboratorij-za-kemijsko-informatiko/transmembrane-region-predictors/ (accessed on 13 July 2020).
- Perdih, A.; Roy Choudhury, A.; Župerl, Š.; Sikorska, E.; Zhukov, I.; Solmajer, T.; Novič, M. Structural Analysis of a Peptide Fragment of Transmembrane Transporter Protein Bilitranslocase. PLoS ONE 2012, 7, e38967. [Google Scholar] [CrossRef] [PubMed]
- Roy Choudhury, A.; Novič, M. PredβTM: A Novel β-Transmembrane Region Prediction Algorithm. PLoS ONE 2015, 10, e0145564. [Google Scholar] [CrossRef] [PubMed]
- SecretomeP 2.0 Server. Prediction of Non-Classical Protein Secretion. Available online: http://www.cbs.dtu.dk/services/SecretomeP (accessed on 13 July 2020).
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; von Heijne, G.; Brunak, S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004, 17, 349–356. [Google Scholar] [CrossRef] [PubMed]
- SignalP-5.0 Server. Available online: http://www.cbs.dtu.dk/services/SignalP/ (accessed on 13 July 2020).
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- TargetP-2.0 Server. Available online: http://www.cbs.dtu.dk/services/TargetP/ (accessed on 13 July 2020).
- Almagro Armenteros, J.J.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef]
- TMHMM Server v. 2.0. Prediction of Transmembrane Helices in Proteins. Available online: http://www.cbs.dtu.dk/services/TMHMM-2.0/ (accessed on 13 July 2020).
- WOLF PSORT Advanced Computational Tool for Protein Subcellular Localization Prediction. Available online: http://www.genscript.com/wolf-psort.html (accessed on 13 July 2020).
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Endapally, S.; Frias, D.; Grzemska, M.; Gay, A.; Tomchick, D.R.; Radhakrishnan, A. Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes. Cell 2019, 176, 1040–1053. [Google Scholar] [CrossRef]
- Anderluh, G.; Kisovec, M.; Kraševec, N.; Gilbert, R.J.C. Distribution of MACPF/CDC Proteins. In MACPF/CDC Proteins—Agents of Defence, Attack and Invasion; Subcellular Biochemistry; Springer: Dordrecht, The Netherlands, 2014; Volume 80, pp. 7–30. ISBN 9789401788816. [Google Scholar]
- McDonagh, A.; Fedorova, N.D.; Crabtree, J.; Yu, Y.; Kim, S.; Chen, D.; Loss, O.; Cairns, T.; Goldman, G.; Armstrong-James, D.; et al. Sub-Telomere Directed Gene Expression during Initiation of Invasive Aspergillosis. PLoS Pathog. 2008, 4, e1000154. [Google Scholar] [CrossRef] [PubMed]
- Shoji, J.; Kikuma, T.; Kitamoto, K. Vesicle trafficking, organelle functions, and unconventional secretion in fungal physiology and pathogenicity. Curr. Opin. Microbiol. 2014, 20, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Miura, N.; Ueda, M. Evaluation of Unconventional Protein Secretion by Saccharomyces cerevisiae and other Fungi. Cells 2018, 7, 128. [Google Scholar] [CrossRef] [PubMed]
- Reindl, M.; Hänsch, S.; Weidtkamp-Peters, S.; Schipper, K. A Potential Lock-Type Mechanism for Unconventional Secretion in Fungi. Int. J. Mol. Sci. 2019, 20, 460. [Google Scholar] [CrossRef] [PubMed]
- Kraševec, N.; Benčina, M. Gene Expression in Filamentous Fungi: Advantages and Disadvantages Compared to Other Systems. In Gene Expression Systems in Fungi: Advancements and Applications, Fungal Biology; Schmoll, M., Dattenböck, C., Eds.; Springer: Cham, Switzerland, 2016; pp. 201–226. [Google Scholar] [CrossRef]
- Maraun, M.; Martens, H.; Migge, S.; Theenhaus, A.; Scheu, S. Adding to ‘the enigma of soil animal diversity’: Fungal feeders and saprophagous soil invertebrates prefer similar food substrates. Eur. J. Soil Biol. 2003, 39, 85–95. [Google Scholar] [CrossRef]
- Dubey, M.; Jensen, D.F.; Karlsson, M. Functional characterization of the AGL1 aegerolysin in the mycoparasitic fungus Trichoderma atroviride reveals a role in conidiation and antagonism. Mol. Genet. Genom. 2020. [Google Scholar] [CrossRef] [PubMed]






| Primers | Sequence (5′→3′) |
|---|---|
| nigA1NotIF | agtacaggcggccgcatgcctgcagccggcc |
| nigA1EcoRIR | gctcgaattctttgctattcttcttcttctgctgc |
| nigA2NotIF | agtacaggcggccgcatggccgagcgagcagaag |
| nigA2EcoRIR | gctcgaattcaagacttgaccgaatcgcacg |
| Dye/Fluorescent Protein | Selectivity | Working Concentration | Incubation Time | Absorption/Emission (nm) |
|---|---|---|---|---|
| Alexa Fluor 488 Dye | NigA2 antibody | n. a. | n. a. | 495/519 |
| Cell Tracker™ CMAC Blue (Molecular probes C2110) | Vacuole | 0.5–5 µM | 30 min | 353/466 |
| DAPI (Termo Fisher 62248) | Nucleus, nucleic acids | 0.3 µM | 5 min | 360/460 |
| eYFP | NigA2-FP protein | n. a. | n. a. | 514/520–570 |
| ER-Tracker™ Blue-White DPX (Invitrogen E12353) | Endoplasmic reticulum | 0.1 µM–1 µM | 20 min | 374/430–640 |
| Filipin (Sigma F9765) | Sterol rich domains | 0.05 mg/mL | 15 min | 340–380/385–470 |
| mCherry | NigA2-FP protein | n. a. | n. a. | 514/587–610 |
| Mito Tracker® Deep Red (Molecular probes M22426) | Mitochondria | 0.1 µM–0.5 µM | 15–45 min | 644/665 |
| Nile Red (Invitrogen N1142) | Lipids, lipid droplets | 1–10 µg/mL | 15–30 min | 552/636 |
| Synapto Red™ (Biotium 70027) | Cell membranes | 0.02 µM–0.2 µM | 15–45 min | 510/750 |
| Sytox Green (Invitrogen S7020) | Nucleus, nucleic acids | 0.01 µM–1 µM | 10 min | 504/523 |
| Tool | NigA1 | Probability/Specificity | NigA2 | Probability/Specificity | OlyA6 | Probability/Specificity |
|---|---|---|---|---|---|---|
| SignalP-5.0 (Eukarya) [76,77] | Sec/SPI Other SP | 0.0006 0.9994 | Sec/SPI Other SP | 0.0008 0.9992 | Sec/SPI Other SP | 0.0129 0.9871 |
| PrediSi (Eukaryotic) [69,70] | Cleavage position: 37; not predicted for secretion | 0 | Cleavage position: 33; not predicted for secretion | 0.043 | Cleavage position: 16; not predicted for secretion | 0.1810 |
| SecretomeP 2.0 (Mammalia) [74,75] | Non-classically secreted 0.688 | 2.126 | Non-classically secreted 0.875 | 4.910 | Non-classically secreted 0.329 | 0.616 |
| Octopus [63,64] | No TM-regions; in a globular region, further than 23Å from the membrane | / | No TM-regions; in a globular region, further than 23Å from the membrane | / | No TM-regions; in a globular region, further than 23Å from the membrane | / |
| TMHMM-2.0 [80] | No membrane helices | / | No membrane helices | / | No membrane helices | / |
| PredαTM [71,72] | Not probable | Not probable | Not probable | |||
| PredβTM [71,73] | Probable | 20–32 | Probable | 14–20; 27–36; 59–68; 86–97; 128–137 | Not probable | - |
| Phobius [65,66] | Non-cytoplasmic | / | Non-cytoplasmic | / | Non-cytoplasmic | / |
| PredGPI [67,68] | Not GPI-anchored; ω-position: 114 | 24.4% | Not GPI-anchored; ω-position: 123 | 84.6% | Not GPI-anchored; ω-position: 114 | 16.0% |
| TargetP-2.0 (Non-plant) [78,79] | Other SP Mitochondrial transfer peptide | 0.9999 0 0.0001 | Other SP Mitochondrial transfer peptide | 0.9947 0.0022 0.0032 | Other SP Mitochondrial transfer peptide | 0.7305 0.2678 0.0017 |
| Wolfpsort (Fungi) [81,82] | Nucleus Cytosol/nucleus Cytosol Peroxisome Mitochondria Extracellular Vacuole | 12.5 11 8.5 3 1 1 1 | Extracellular Cytosol Cytosol/nucleus Cytosol/mitochondria Nucleus Mitochondria | 14 8.5 6.833 5.666 3 1.5 | Extracellular Cytosol Cytosol/nucleus Mitochondria Vacuole Nucleus | 12 9.5 6 2 2 1.5 |
| EffectorP 2.0 [61,62] | Non-effector | 0.701 | Effector | 0.731 | Effector | 0.577 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kraševec, N.; Novak, M.; Barat, S.; Skočaj, M.; Sepčić, K.; Anderluh, G. Unconventional Secretion of Nigerolysins A from Aspergillus Species. Microorganisms 2020, 8, 1973. https://doi.org/10.3390/microorganisms8121973
Kraševec N, Novak M, Barat S, Skočaj M, Sepčić K, Anderluh G. Unconventional Secretion of Nigerolysins A from Aspergillus Species. Microorganisms. 2020; 8(12):1973. https://doi.org/10.3390/microorganisms8121973
Chicago/Turabian StyleKraševec, Nada, Maruša Novak, Simona Barat, Matej Skočaj, Kristina Sepčić, and Gregor Anderluh. 2020. "Unconventional Secretion of Nigerolysins A from Aspergillus Species" Microorganisms 8, no. 12: 1973. https://doi.org/10.3390/microorganisms8121973
APA StyleKraševec, N., Novak, M., Barat, S., Skočaj, M., Sepčić, K., & Anderluh, G. (2020). Unconventional Secretion of Nigerolysins A from Aspergillus Species. Microorganisms, 8(12), 1973. https://doi.org/10.3390/microorganisms8121973







